Abstract
Background
The aims of this study were to analyze the association of LMO4 with non-small-cell lung cancer (NSCLC) survival rate, and to determine its functional role and signaling pathway in lung cancer.
Methods
Immunohistochemistry (IHC) was used to detect the expression of LMO4 in NSCLC cell lines and tumor tissues. Migration and invasion ability was detected respectively by wound healing test and transwell test. Immunofluorescence and western blot were detected of AKT/PI3K pathway related genes MAPK, PI3K, AKT.
Results
LMO4 has high expression level of NSCLC cell lines and tumor tissues, and correlated with a lower survival rate. LMO4 can regulate the migration and invasion of NSCLC cells through the AKT/PI3K pathway.
Conclusions
LMO4 could serve as a promising biomarker and therapeutic target for NSCLC.
Keywords: LMO4, migration, invasion, non-small-cell lung cancer (NSCLC)
Introduction
LIM-only protein 4 (LMO4) is a novel member of the LIM-only (LMO) subfamily of LIM domain-containing transcription factors. The LMO protein mainly localizes to the nucleus (1), where it mediates interactions within multi-protein complexes and DNA. This controls the expression of genes involved in regulating many important biological functions, such as cell proliferation and mammalian development. Therefore, the dysregulation of LMO4 has been implicated in the carcinogenesis and metastasis of a variety of human cancers. Studies have indicated that LMO4 overexpression may disrupt the normal tumor suppressor activities of Tip, promoting breast cancer progression (2), and affecting lung cancer patient survival (3). A number of studies have found LMO4 highly expressed at sites of active epithelial-mesenchymal interactions, and LMO4 can interact with signaling pathways involved in epithelial-mesenchymal signaling (4), leading to an increase in stromal cell invasion and migration. Studies of LMO4 in non-small-cell lung cancer (NSCLC), however, are rare. We hypothesized that lung cancer is a tumor derived from epithelial tissue, and thatLMO4 has a similar functional role in lung cancer.
In the present study, gain and loss of function approaches were used to investigate the role of LMO4 in lung cancer cells. We examined the relationship of LMO4 expression in cancer and adjacent tissues. Our results showed that LMO4 affected lung cancer cell invasion and migration through the AKT/PI3K pathway.
Methods
Cell cultures and patient specimens
The NSCLC cell lines (A549, H1299, H460, SPC-A1, H522, H1250, H1975, and A2) were maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). BEAS-2B, a normal lung epithelial cell line, was cultured in keratinocyte serum-free medium (keratinocyte-SFM; Gibco BRL Life Technologies, Gaithersburg, MD, USA). The 293T cells were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS. All cell lines were maintained at 37 °C in a humidified atmosphere with 5% CO2. All NSCLC specimens were obtained from the Cancer Center of Shandong Cancer Hospital and had confirmed pathological diagnoses. Written informed consents from the patients and approval from the Institutional Research Ethics Committee of the Cancer Center of Guangdong Provincial People’s Hospital were obtained.
Quantitative RT-PCR analyses
Total RNA was extracted from cells with TRIzol® reagent (Invitrogen Life Sciences, Carlsbad, CA, USA). For the detection of LMO4 mRNA, cDNA was reverse transcribed from 1µg of total RNA according to the instructions of the reverse transcription reaction kit (Promega, Fitchburg, WI, USA). The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) amplification was done in parallel as the internal control. The primer sequences used for LMO4 were: forward, 5'-GGA CCG CTT TCT GCT CTA TG-3'; reverse, 5'-AAG CAC CGC TAT TCC CAA AT-3'. The primer sequences used for GAPDH amplification were: 5'-AAC GTG TCA GTG GTG GAC CTG-3' (forward) and 5'-AGT GGG TGT CGC TGT TGA AGT-3' (reverse). The qRT-PCR reactions were performed using a qSYBR-green-containing PCR kit (Invitrogen Life Sciences), and U6small nuclear RNA (snRNA) was used as an endogenous control. The expression of each gene was measured by the cycle threshold (CT) value quantification and normalization, using the 2−∆∆Ct method and compared with the U6 RNA or GAPDH.
Plasmid construction and transfection
For the construction of LMO4 protein expression plasmids, we used PCR primers designed for hLMO4 amplification, and joined at the BamH1 and xho1 restriction sites. The LMO4 cDNA sequences were researched from the NCBI database and were amplified using PCR with NSCLC cell cDNA as the templates. The resulting cDNA was spliced into the pcDNA plasmid (Invitrogen Life Sciences) using transient transfection procedures. The A549 and H1299 cells were seeded into six-well plates at 5×105 cells/well. The cells were then transfected with LMO4 plasmid for 48 h. The recombinant clone was screened using kanamycin, and the resulting clone was designated as pcDNA-LMO4.Then, cells were collected for western blotting, immunofluorescence analyses, Transwell® assays, and wound healing assays. The empty pcDNA3 vector was used as a mock transfection control.
Western blots
NSCLC tissues were homogenized in liquid nitrogen and lysed in RIPA buffer containing protease inhibitors, on ice. Cells were directly lysed in RIPA buffer. The protein lysates were resolved on 10% SDS polyacrylamide gels. Proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Invitrogen Life Sciences), and blocked with 5% nonfat milk in Tris-HCl buffer solution, pH 7.5, at room temperature for 1 h. Membranes were immunoblotted at room temperature for 1.5 h with anti-LMO4 antibody (1:1,000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-c-MET antibody (1:1,000 dilution; Santa Cruz Biotechnology), anti-AKT and anti-pAKT antibodies (1:1,000 dilution; Santa Cruz Biotechnology), anti-PI3K and anti-pPI3K antibodies (1:1,000 dilution; Santa Cruz Biotechnology), anti-MAPK and anti-pMAPK antibodies (1:1,000 dilution; Santa Cruz Biotechnology), and anti-GAPDH antibody (Santa Cruz Biotechnology), followed by their respective horseradish peroxidase (HRP)-conjugated secondary antibodies. Signals were detected using enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA).
Transwell® assays
A cell invasion assay was performed using a 24-well Transwell® chamber with a pore size of 8 µm (Costar, Boston, MA, USA). Serum free medium and Matrigel® mixed at 1:5, (BD Bioscience, San Jose, CA, USA) were used. Forty-eight hours after transfection, the cells were digested with 0.25% trypsin, washed three times with serum free medium, and the resulting cell suspension was transferred to the upper chamber in 100 µL of the cell suspension containing 3×105 cells. The lower chamber contained medium with 10% FBS and 500 µL of the chemokines. After conventional culturing for 12 h, the cells were removed from the chamber, washed two times in phosphate-buffered saline (PBS), and then fixed with 5% glutaraldehyde. Cells were stained with Giemsa stain, and then a cotton swab was used to remove the Matrigel® and cells on the upper chamber. Random images were photographed in five fields of view, using a microscope for visualization. The cell counts were averaged, and all analyses were independent of the experiment, and were repeated three times.
Wound healing assays
Cells were seeded into six-well plates, and when the cells covered the bottom plate, a wound was created with a pipette tip, to ensure that all scratch widths were consistent. The cell culture solution was then removed and the cells were washed three times with PBS. Serum-free medium was then added and microscopic images were obtained at 0 and 24 h of incubation.
Immunofluorescence analyses
The cells were transferred to culture dishes with coverslips and cultured until semi-confluent. The coverslips were rinsed with PBS, the cells were fixed with 4% paraformaldehyde for 15 min, and treated for 10 min with 0.25% Triton X-100 to increase the cell membrane permeability. Nonspecific staining was diminished by incubating with 10% heat inactivated (HI) serum from the species in which the second antibody was produced. Slides were incubated with LMO4 antibody (1:200 dilution; Abcam, Cambridge, UK), pAKT antibody (1:100 dilution; Abcam), and pPI3K antibody (1:100 dilution; Abcam), at 4 °C overnight. After washing, the cells were then incubated with DyLight™Alexa Fluor™488-conjugated Affinipure goat anti-mouse secondary antibodies (1:400 dilution; Abcam), at room temperature for 1 h, followed by staining with DAPI nuclear stain (Invitrogen Life Sciences).
Immunohistochemistry (IHC) analyses
Paraffin sections (3 µm) from the samples were incubated in a Thermotank set at 60 °C, for 20 min. Tissue sections were then immersed in 100% xylene for 10 min, and then in a graded alcohol solution at 100%, 95%, 90%, 80%, and 70% alcohol solution, each for 3–5 min. The sections were finally immersed in distilled water for 3 min. The high-pressure antigen retrieval was in sodium citrate buffer, pH 7.4, for 2 min at 100 °C. Serum was added to block the activity of the endogenous peroxidase. The samples were then incubated in a 37 °C warm box for 30 min. Rabbit anti-human LMO4 antibody (1:200 dilution; Abcam) was added and the samples were incubated overnight at 4 °C. After washing, biotinylated goat anti-rabbit antibody was incubated for 10 min at room temperature. Subsequently, the samples were incubated with HRP-conjugated streptavidin (Maixin Inc., Fuzhou, China), the PBS was removed, and sections were treated with freshly prepared 3,3'-diaminobenzidine (DAB) liquid. Samples were observed under a microscope after 5 min, using a bright field microscope. The immunostained slides were examined and scored in a blinded fashion by two pathologists. The results were based on the percentage of immunoreactivity and intensity of staining in cells. When differences occurred, the respective slides were reinvestigated jointly by both investigators. The proportion of positive tumor cells was evaluated as follows: 0, no positive tumor cells; 1, <10% positive tumor cells; 2, 10–50% positive tumor cells; and 3, >50% positive tumor cells. The scoring criteria for staining intensity were graded as: 0, no staining; 1, light yellow staining; 2, yellow staining; and 3, brown staining. The staining index was calculated by multiplication of the proportion and intensity. Using this assessment method, we measured the expression of LMO4 in NSCLC specimens by determining the staining scores (0, 1, 2, 3, 4, 6, or 9).
Statistical analyses
For statistical analyses, we used SPSS software, version 13 for Windows (SPSS, Chicago, IL, USA). The correlation between LMO4 expression and clinicopathological factors was examined by χ2 tests. The P values were based on a two-sided statistical analysis, and P<0.05 was considered to indicate statistical significance.
Results
The expression of LMO4 in human NSCLC cell lines and tissues indicate spoor prognoses
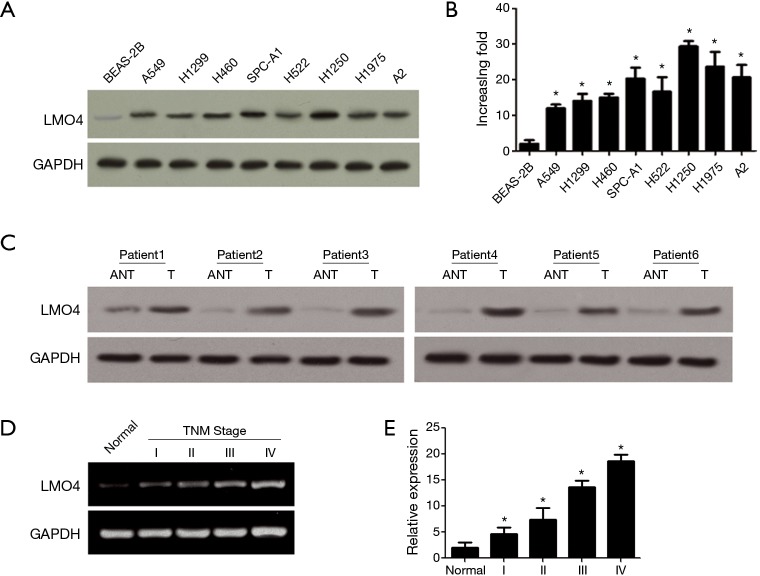
LMO4 is highly expressed in breast cancer, therefore its expression was examined in lung cancer cells. Real-time PCR and western blots were used to detect LMO4 expression in eight NSCLC cell lines and one normal lung epithelial cell line (BEAS-2B). LMO4 expression was higher in the NSCLC cell lines compared to that of the normal lung epithelial cell line (Figure 1A). In addition, LMO4 protein expression in each cell line precisely coincided with mRNA levels (Figure 1B). Furthermore, LMO4 expression was higher in tumor tissues compared to that of the adjacent non-cancerous tissues (ANT). Notably, LMO4 expression was higher in the later stages compared to that of the early stages of NSCLC (Figure 1C,D), then we statistical quantification comparisons of the average LMO4 staining values (Figure 1E). These results indicated that LMO4 may be involved in tumor progression in NSCLC.
Figure 1.
LMO4 expression in non-small-cell lung cancer (NSCLC) cell lines and tissues. (A) LMO4 protein expression in normal lung epithelial cells and eight NSCLC cell lines analyzed by western blots; (B) LMO4 gene expression in normal lung epithelial cells and eight NSCLC cell lines analyzed by real-time PCR. Error bars represent mean ± SD from three independent experiments. *P<0.05; (C) Western blot analyses of LMO4 protein expression in primary liver cancer tissues (T) and adjacent non-cancerous tissues (ANT) taken from the same patient; (D) LMO4 gene expressions in normal lung tissue (normal) and NSCLC specimens of different tumor-node-metastasis (TNM) stages as analyzed by real-time PCR. Error bars represent the mean ± SD from three independent experiments. *P<0.05; (E) statistical quantification comparisons of the average LMO4 staining values between normal lung tissues (normal) and NSCLC specimens in different TNM stages. Error bars represent the mean ± SD from three independent experiments. *, P<0.05.
The expression pattern and clinical significance of LMO4 in NSCLC specimens
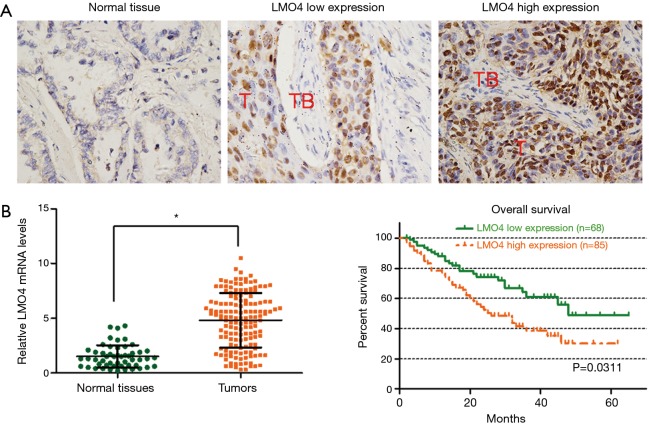
IHC analyses were performed on 153 NSCLC specimens. The characteristics of the patients in the two groups are listed in Table 1. The results from the LMO4 high expression and LMO4 low expression groups are shown (Figure 2A). We further examined the expression levels of LMO4 in specimens obtained from 153 cases of NSCLC and normal tissues. The average expression level of LMO4 was significantly higher in the NSCLC tissues compared to that of the normal tissues (Figure 2B).
Table 1. Correlation between the clinicopathologic characteristics and LMO4 expression in 153 NSCLC patients.
| Factor | No. | LMO4 protein, n (%) | P | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.445 | |||
| <50 | 90 | 38 (42.2) | 52 (57.8) | |
| ≥50 | 63 | 31 (48.4) | 33 (51.6) | |
| Gender | 0.101 | |||
| Male | 82 | 46 (56.1) | 36 (43.9) | |
| Female | 71 | 49 (69.0) | 22 (31.0) | |
| Tumor size (cm) | 0.012 | |||
| <3 | 55 | 25 (45.5) | 30 (54.5) | |
| ≥3 | 98 | 25 (25.5) | 73 (74.5) | |
| Smoking status | 0.092 | |||
| Smoker | 82 | 47 (57.3) | 35 (42.7) | |
| Non-smoker | 71 | 31 (43.7) | 40 (56.3) | |
| Pathology type | 0.021 | |||
| SQC | 81 | 31 (38.3) | 50 (61.7) | |
| ADC | 40 | 11 (27.5) | 29 (72.5) | |
| Other | 32 | 19 (59.4) | 13 (40.6) | |
| TNM stage | 0.000 | |||
| I–II | 41 | 29 (70.7) | 12 (29.3) | |
| III–IV | 112 | 32 (28.6) | 80 (71.4) | |
NSCLC, non-small-cell lung cancer; SQC, squamous carcinoma; ADC, adenocarcinoma.
Figure 2.
LMO4 levels are significantly associated with patient overall survival (OS) in non-small-cell lung cancer (NSCLC). (A) Immunohistochemical staining of LMO4 in NSCLC tissues and normal tissues (magnification, ×400 for all images); (B) the expression levels of LMO4 in NSCLC tissues and normal tissues as detected by q-RT-PCR. *P<0.05; (C) Kaplan-Meier curves with univariate analyses for patients with low LMO4expression versus high LMO4 expression. T, tumor; TB, tumor beside.
We next evaluated the levels of LMO4 expression in predicting the survival of NSCLC patients. Kaplan-Meier and log-rank survival tests showed that NSCLC patients with low LMO4 expression had a significantly increased overall survival (OS) compared to those patients with high LMO4 expression (P<0.05). The median OS in the LMO4 high expression patients and low expression patients were 24.3 and 42.3 months, respectively (Figure 2C).
LMO4 levels significantly associate with patient OS by tumor-node-metastasis (TNM) stage and tumor size
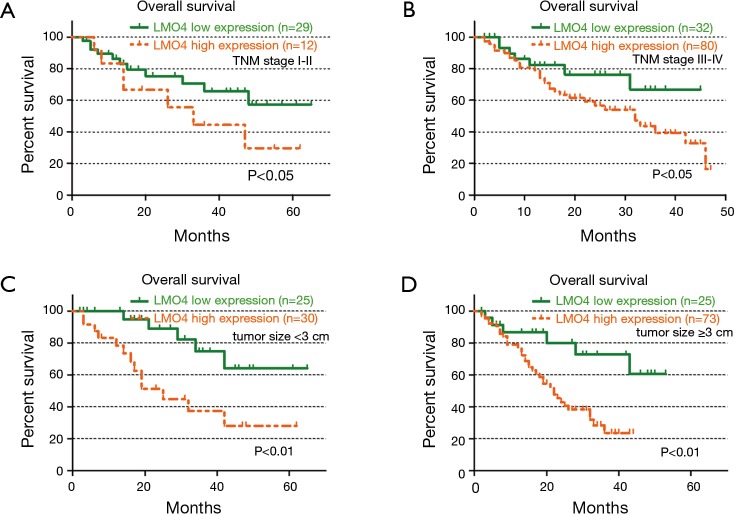
We subgrouped 153 NSCLC patients into different TNM stages (TNM stages I–II and III–IV) and evaluated the prognostic significance of LMO4 in the patient subgroups. In the 41 NSCLC patients in TNM stage I–II, the median OS in the low LMO4 expression group vs. the high LMO expression group was 53.4 and 31.8 months, respectively. Patients with low LMO4 expression also had a longer OS (median OS =36.4 months) compared to those with high LMO4 expression (median OS =24.5 months) in TNM stages III–IV (Figure 3A,B).
Figure 3.
LMO4 levels are significantly associated with patient overall survival (OS) in different tumor stages and sizes. (A,B) OS of patients with high or low-LMO4 expression when further divided into tumor-node-metastasis (TNM) I–II and TNM III–IV subgroups; (C,D) OS rates of patients with tumor size ﹤3 cm and tumor size ≥3 cm, respectively.
We next evaluated the prognostic significance of LMO4 in the patient subgroups by tumor size. Independent of tumor size, the OS was longer in the LMO4 low expression group than in the high expression group (Figure 3C,D). Taken together, our data suggested the promising prognostic value of LMO4 for NSCLC patients in different clinical subgroups.
LMO4 overexpression increases migration and invasion by regulation of AKT signaling
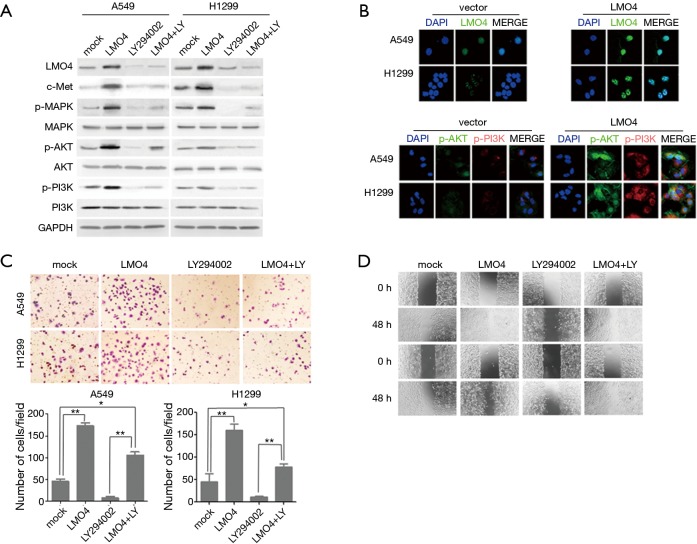
The c-Met and k-Ras genes have been shown to work as driver genes to regulate lung cancer development. AKT has also been shown to regulate LMO4 through inducing mesenchymal-to-epithelial transitions in oral squamous cell carcinoma cells (5,6). To further examine whether LMO4 can drive NSCLC cell functions by regulating the c-met, p-MPAK, or AKT pathway, we overexpressed LMO4 in two NSCLC cell lines (A549 and H1299 cells). The expressions of total MPAK, AKI and PI3K were not influenced by LMO4 overexpression. However, the expression of LMO4 could induce c-met and the phosphorylation of MPAK, AKI and PI3K (Figure 4A). To determine whether a blockade of the AKT pathway with the AKT inhibitor LY294002 could reverse the regulation of AKT by LMO4, LY294002+LMO4-treated NSCLC cell lines were treated with LY294002 (10 µM) for 1 h in advance to block AKT, Western blotting and immunofluorescence assays were used to assess whether AKT inhibition could reverse the expression of p-AKT, pPI3K, and p-MPAK. (Figure 4A,B).
Figure 4.
LMO4 knockdown inhibits non-small-cell lung cancer (NSCLC) cell migration. (A) Protein expression of LMO4, c-met, MPAK, AKT, and PI3K were analyzed by western blots in A549 and H1299 cells; (B) LMO4, p-AKT, p-PI3K protein levels were up regulated in A549 and H1299 cells transfected with LMO4, as detected by immunofluorescence (magnification, ×600); (C) the Boyden assay showed that LMO4 increased NSCLC cell invasion, an effect abrogated by LY294002 (magnification, ×20). Error bars represent mean ± SD from three independent experiments. *, P<0.05; **, P<0.05; (D) wound healing assays of A549 and H1299 cells transfected with control vectors, LMO4, and LMO4 combined with LY294002 at 0 and 48 h after wounding (magnification, ×20).
To determine whether the regulation of the AKT pathway by LMO4 could influence NSCLC cell migration and invasion, we measured the migration and invasion of A549 and H1299 cells using the monolayer scratch and Transwell® assays after the overexpression of LMO4, or in combinations of LMO4 with LY294002 (Figure 4C). The overexpression of LMO4 increased cell migration and invasion, but the combination of LMO4 with LY294002 did not alter cell migration and invasion (Figure 4C,D). In a study found that K-Ras driven tumor growth is mainly mediated through activation of ERK but not AKT, the loss of LMO4 did not affect tumor progression in K-RasG12D-driven mouse model of lung cancer (7). However, our results indicated LMO4 regulated the migration and invasion of NSCLC cells through the AKT pathway, this would explain why no effect was observed when LMO4 was lost in the KRAS mice.
Discussion
In the present study, we investigated the expression patterns of LMO4 and its biological roles in NSCLC. Consistent results from six different assays strongly suggested a correlation between LMO4 levels and the clinical outcome of lung cancer.
LIM domain proteins, with two tandem zinc finger domains, have different but important roles. LMO2 regulates angiogenesis and hematopoiesis (8,9), and LMO1 and LMO3 are expressed during embryogenesis (10). LMO proteins have important roles in cell growth, apoptosis, and differentiation (11-13). A previous study showed that LMO4 is overexpressed during the differentiation of mammary epithelial cells (14), implicating it as an oncogene. Significantly, high LMO4 levels in the cell nucleus may serve as an independent predictor of mortality from breast cancer, pancreatic cancer, and tongue cancer (15-17). Our study showed that LMO4 overexpression was related to NSCLC progression and may therefore predict the prognosis of lung cancer patients, especially those with high-grade/less-differentiated NSCLC (characteristically highly proliferative).
One of the most important tumor suppressor genes, p53, is mutated in over 50%of all human cancers. LMO4 can interact with and direct the transcriptional targeting of p53 (18). Loss of LMO4 subsequently results in reduced cyclin D1 and cyclin E levels. Therefore, LMO4 silencing reduces tumor cell proliferation and induces aG2/M arrest associated with decreased cullin-3, an E3-ubiquitin ligase component important for mitosis (16). LMO4 can inhibit the expression of the fusion proteinengrailed-LMO4 while suppressing cell growth, and can induce the apoptosis of breast cancer cells, indicating that LMO4 contributes to oncogenesis by similar mechanisms, thus enhancing cell survival and proliferation (19).
AKT/PI3K activation is involved in the carcinogenesis and tumor progression of various solid tumors, including NSCLC (20,21). Lu et al. reported that the transcriptional coactivator LMO4 modulated the cytostatic effects of TGF-beta in epithelial cells, and associated with and regulated a prototype Smad target promoter (14). Similar to the other LMO family members, LMO4 is a transcriptional cofactor that functions as a scaffold for the generation of multiprotein complexes. LMO4, with two tandem LIM domains, can independently interact with ERt and metastasis tumor antigen 1 (MTA1). Therefore, if a cell is ERalpha negative, LMO4 may be a component of the MTA1 corepressor complex (22). To detect the relationship between LMO4 and the biological behavior of lung cancer cells, we overexpressed LMO4 in A549 and H1299 cell lines, and found that LMO4 increased cell migration and invasion.
In summary, our study showed that the over expression of LMO4 was related to lung cancer progression. LMO4 levels may predict the prognosis of lung cancer patients, especially those with high grade/less differentiated NSCLC, which are characteristically highly proliferative. We showed that LMO4 promoted cell invasion and migration in vitro, at least in part by activating the PI3K/Akt pathway. This study increases our understanding of the precise role played by LMO4 in lung cancer progression, and could provide a novel therapeutic molecular target for clinical lung cancer patients.
Acknowledgements
Funding: This work was supported by grants from the National Natural Science Foundation of China (grant No. 81301939); and the China Postdoctoral Science Foundation (grant No. 2015M570697).
Ethical Statement: The study was approved by the Institutional Research Ethics Committee of the Cancer Center of Guangdong Provincial People’s Hospital (No GDREC. 2014101A).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Wang N, Lin KK, Lu Z, et al. The LIM-only factor LMO4 regulates expression of the BMP7 gene through an HDAC2-dependent mechanism, and controls cell proliferation and apoptosis of mammary epithelial cells. Oncogene 2007;26:6431-41. 10.1038/sj.onc.1210465 [DOI] [PubMed] [Google Scholar]
- 2.Stokes PH, Liew CW, Kwan AH, et al. Structural basis of the interaction of the breast cancer oncogene LMO4 with the tumour suppressor CtIP/RBBP8. J Mol Biol 2013;425:1101-10. 10.1016/j.jmb.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 3.Taniwaki M, Daigo Y, Ishikawa N, et al. Gene expression profiles of small-cell lung cancers: molecular signatures of lung cancer. Int J Oncol 2006;29:567-75. [PubMed] [Google Scholar]
- 4.Wang N, Kudryavtseva E, Ch'en IL, et al. Expression of an engrailed-LMO4 fusion protein in mammary epithelial cells inhibits mammary gland development in mice. Oncogene 2004;23:1507-13. 10.1038/sj.onc.1207288 [DOI] [PubMed] [Google Scholar]
- 5.Hong KO, Kim JH, Hong JS, et al. Inhibition of Akt activity induces the mesenchymal-to-epithelial reverting transition with restoring E-cadherin expression in KB and KOSCC-25B oral squamous cell carcinoma cells. J Exp Clin Cancer Res 2009;28:28. 10.1186/1756-9966-28-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart DJ. Tumor and host factors that may limit efficacy of chemotherapy in non-small cell and small cell lung cancer. Crit Rev Oncol Hematol 2010;75:173-234. 10.1016/j.critrevonc.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holik AZ, Filby CE, Pasquet J, et al. The LIM-domain only protein 4 contributes to lung epithelial cell proliferation but is not essential for tumor progression. Respir Res 2015;16:67. 10.1186/s12931-015-0228-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada Y, Warren AJ, Dobson C, et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A 1998;95:3890-5. 10.1073/pnas.95.7.3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada Y, Pannell R, Forster A, et al. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc Natl Acad Sci U S A 2000;97:320-4. 10.1073/pnas.97.1.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tse E, Smith AJ, Hunt S, et al. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol 2004;24:2063-73. 10.1128/MCB.24.5.2063-2073.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoyama M, Ozaki T, Inuzuka H, et al. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res 2005;65:4587-97. 10.1158/0008-5472.CAN-04-4630 [DOI] [PubMed] [Google Scholar]
- 12.Wittlin S, Sum EY, Jonas NK, et al. Two promoters within the human LMO4 gene contribute to its overexpression in breast cancer cells. Genomics 2003;82:280-7. 10.1016/S0888-7543(03)00147-2 [DOI] [PubMed] [Google Scholar]
- 13.Mizunuma H, Miyazawa J, Sanada K, et al. The LIM-only protein, LMO4, and the LIM domain-binding protein, LDB1, expression in squamous cell carcinomas of the oral cavity. Br J Cancer 2003;88:1543-8. 10.1038/sj.bjc.6600952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Z, Lam KS, Wang N, et al. LMO4 can interact with Smad proteins and modulate transforming growth factor-beta signaling in epithelial cells. Oncogene 2006;25:2920-30. 10.1038/sj.onc.1209318 [DOI] [PubMed] [Google Scholar]
- 15.Kwong RA, Scarlett CJ, Kalish LH, et al. LMO4 expression in squamous cell carcinoma of the anterior tongue. Histopathology 2011;58:477-80. 10.1111/j.1365-2559.2011.03765.x [DOI] [PubMed] [Google Scholar]
- 16.Yue L, Li L, Liu F, et al. The oncoprotein HBXIP activates transcriptional coregulatory protein LMO4 via Sp1 to promote proliferation of breast cancer cells. Carcinogenesis 2013;34:927-35. 10.1093/carcin/bgs399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Ohuchida K, Nakata K, et al. LIM only 4 is overexpressed in late stage pancreas cancer. Mol Cancer 2008;7:93. 10.1186/1476-4598-7-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Sang M, Liu W, et al. LMO4 inhibits p53-mediated proliferative inhibition of breast cancer cells through interacting p53. Life Sci 2012;91:358-63. 10.1016/j.lfs.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 19.Tian Y, Wang N, Lu Z. Repression of Lim only protein 4-activated transcription inhibits proliferation and induces apoptosis of normal mammary epithelial cells and breast cancer cells. Clin Exp Metastasis 2010;27:455-63. 10.1007/s10585-010-9332-1 [DOI] [PubMed] [Google Scholar]
- 20.Fei F, Li X, Xu L, et al. CD147-CD98hc complex contributes to poor prognosis of non-small cell lung cancer patients through promoting cell proliferation via the PI3K/Akt signaling pathway. Ann Surg Oncol 2014;21:4359-68. 10.1245/s10434-014-3816-1 [DOI] [PubMed] [Google Scholar]
- 21.Zhang E, Feng X, Liu F, et al. Roles of PI3K/Akt and c-Jun signaling pathways in human papillomavirus type 16 oncoprotein-induced HIF-1α, VEGF, and IL-8 expression and in vitro angiogenesis in non-small cell lung cancer cells. PLoS One 2014;9:e103440. 10.1371/journal.pone.0103440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh RR, Barnes CJ, Talukder AH, et al. Negative regulation of estrogen receptor alpha transactivation functions by LIM domain only 4 protein. Cancer Res 2005;65:10594-601. 10.1158/0008-5472.CAN-05-2268 [DOI] [PubMed] [Google Scholar]