Abstract
The NLRP3 inflammasome is activated by a variety of external or host-derived stimuli and its activation initiates an inflammatory response through caspase-1 activation, resulting in inflammatory cytokine IL-1β maturation and secretion. The NLRP3 inflammasome activation is a kind of innate immune response, most likely mediated by myeloid cells acting as a host defense mechanism. However, if this activation is not properly regulated, excessive inflammation induced by overactivated NLRP3 inflammasome can be detrimental to the host, causing tissue damage and organ dysfunction, eventually causing several diseases. Previous studies have suggested that mitochondrial damage may be a cause of NLRP3 inflammasome activation and autophagy, which is a conserved self-degradation process that negatively regulates NLRP3 inflammasome activation. Recently, mitochondria-selective autophagy, termed mitophagy, has emerged as a central player for maintaining mitochondrial homeostasis through the elimination of damaged mitochondria, leading to the prevention of hyperinflammation triggered by NLRP3 inflammasome activation. In this review, we will first focus on the molecular mechanisms of NLRP3 inflammasome activation and NLRP3 inflammasome-related diseases. We will then discuss autophagy, especially mitophagy, as a negative regulator of NLPP3 inflammasome activation by examining recent advances in research. [BMB Reports 2016; 49(10): 529-535]
Keywords: Autophagy, Inflammation, Mitophagy, NLRP3 inflammasome
INTRODUCTION
The immune system is a host protection mechanism against invading pathogens, such as viruses and bacteria. Macrophages, the front line defenders who recognize invading pathogens, not only kill pathogens by phagocytosis, but also initiate inflammation producing inflammatory cytokines upon infections. The maturation and secretion of IL-1β, one of the strongest pro-inflammatory cytokines, is enhanced by the inflammasome in macrophages, leading to inflammation and cell death (1, 2).
Inflammasomes are molecular platforms that trigger activation of caspase-1, leading to pro-inflammatory cytokine maturation. One of the best characterized inflammasome is the NLRP3 inflammasome. Even though NLRP3 inflammasome activation is important for protecting cells from pathogenic microbes, excessive NLRP3 inflammasome may cause hyperinflammation resulting in tissue damage and organ failure. NLRP3 inflammasome is also activated by danger signals released from stressed cells and dysregulation of NLRP3 inflammasome activation causes many diseases, such as neurodegenerative diseases, metabolic diseases and sepsis. Therefore, mechanisms to control NLRP3 inflammasome activation are necessary for health.
Upon bacterial infection, innate immune cells, such as macrophages and neutrophils, take up bacteria and generate reactive oxygen species (ROS) (3-5). The 2 main sources of ROS are NADPH oxidase-mediated ROS and mitochondria-derived ROS in innate immune cells (6-8). Some studies have proposed that mitochondrial dysfunction is closely associated with NLRP3 inflammasome activation. Infection causes mitochondrial damage through an unknown mechanism and the damaged mitochondria release mitochondrial DNA (mtDNA) and mitochondrial reactive ROS (mtROS) and are thought to work as danger signals (9, 10). Cells have a regulatory mechanism, ‘mitophagy’, a mitochondria-selective autophagic process to eliminate damaged or unwanted mitochondria, to maintain mitochondrial homeostasis against stress. This review will discuss how the balance between pro-inflammatory response and anti-inflammatory response is maintained in cells focusing on NLRP3 inflammasome activation and mitophagy.
THE INFLAMMASOME
Inflammation is one of the first innate immune responses to combat foreign organisms entering the body that are able to cause disease (11). Most foreign organisms express various pathogen-associated molecular patterns (PAMPs) in their cell wall or on their cell surface (12). Therefore, it is possible that a host can distinguish between self and non-self by recognizing PAMPs. Moreover, under stressed conditions, cells release danger signals called danger-associated molecular patterns (DAMPs) to notify their urgent situation to the immune system. PAMPs and DAMPs are recognized by innate immune receptors pattern-recognition receptors (PRRs), such as Toll-like receptors (TLRs) and nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) (13). Activation of PRRs lead to Nf-κB activation and inflammatory cytokine production. Many studies have shown that the inflammasome is a key regulator of inflammation (1, 14-17). Most inflammasomes contain NLR proteins and the NLR family can be classified into 3 subtypes based on their domain structures: the NODs (NOD1-5 and CIITA), the NLRPs (NLRP1-14) and the IPAF (IPAF also known as NLRC4, and NAIP) subtypes (18). The NLR family commonly contains a central nucleotide-binding and oligomerization (NACHT) domain. Most NLRs also contain C-terminal leucine-rich repeats (LRRs) and N-terminal caspase recruitment domains (CARD) or pyrin domains (PYD). NACHT plays an essential role in the activation and formation of signaling complexes and LRR mediates ligand sensing, while CARD and PYD function in homotypic protein-protein interactions for downstream events (18, 19). A non-NLR inflammasome member, absent in melanoma-2 (AIM2), was later identified that can form an inflammasome composed of AIM2, ASC and caspase-1. The function and mechanism of many NLR family members remain poorly understood, but some are well-characterized. For example, NOD1 and NOD2 are receptors for bacterial peptidoglycan fragments. NOD1 and NOD2 recognize D-glutamyl-meso-diaminopimelic acid (DAP) and muramyl dipeptide (MDP), respectively. Upon sensing their ligand, NOD1 and NOD2 oligomerize and recruit RIP2 via CARD-CARD interactions, triggering an inflammatory response (20, 21). CIITA uniquely acts as a transcription factor playing a key role in the regulation of class II MHC genes (22). The IPAF inflammasome is activated in response to gram-negative bacteria containing type III or IV secretion systems, such as Psudomonas aeruginosa, Salmonella typhimurium and Shigella flexneri (23-25). The AIM2 inflammasome is activated by sensing cytosolic double-stranded DNA (dsDNA) through the HIN-200 domain of AIM2 (26, 27). As is demonstrated herein, each inflammasome is activated by different stimuli, but more studies concerning other NLR family members are required. In this review, the NLRP3 inflammasome, which is the most characterized of the inflammasome members, will be discussed in detail.
NLRP3 INFLAMMASOME
The NLRP3 inflammasome is activated in response to abundant stimuli and 2 distinct steps are required for this activation. First, Nuclear factor-κB (NF-κB) activation by TLR ligands increases NLRP3 and pro-1L-1β; this step is called ‘priming’. Second, activating stimuli derived from microbes or the host induce the assembly of NLRP3 inflammasome components (17). The NLRP3 inflammasome is composed of NLRP3, apoptosis-associated speck-like protein containing a CARD (ASC) and pro-caspase-1. ASC is an inflammasome adaptor containing N-terminal PYD and C-terminal CARD. NLRP3 inflammasome is assembled by the following steps: PYD of NLRP3 is oligomerized with ASC through PYD-PYD interactions. Subsequently, pro-caspase-1 is recruited and interacts with ASC through CARD-CARD interactions (28). After the formation of the NLRP3 inflammasome, pro-caspase-1 is autocleaved to active caspase-1 and the active caspase-1 matures cytokines, such as IL-1β, to their bioactive and secreted forms (17). Currently, various sterile DAMP signals and pathogens, including mitochondrial ROS, mitochondrial DNA, potassium efflux, MDP, monosodium urate (MSU), cholesterol crystals, cathepsins, influenza virus, Salmonella typhimurium, Mycobacterium tuberculosis and others, are known to activate the NLRP3 inflammasome (29). Compared to NLRP3 inflammasome activators, negative regulators of the NLRP3 inflammasome are relatively less known. Recent studies suggest that nitric oxide (NO), Ca2+ and cyclic AMP negatively regulate the NLRP3 inflammasome but the detailed mechanisms are unclear (30, 31). If the upregulated NLRP3 inflammasome activation is not downregulated, inflammation will be persistently induced by NLRP3 inflammasome and contribute to a diverse array of diseases.
NLRP3 INFLAMMASOME IN DISEASE; NEURODEGENERATIVE DISEASE
Dysregulation of NLRP3 inflammasome activation is observed in neurodegenerative disease. Alzheimer’s disease (AD), a representative neurodegenerative disease, is characterized by amyloid-β plaque accumulation. Halle et al., using in vitro and in vivo mouse models, identified that amyloid-β causes lysosomal damage releasing cathepsin B and the cathepsin B activated NLRP3 inflammasome (32). In addition to the mouse study, recently performed human studies showed that increased active caspase-1 expression is observed in the brains of AD patients (33). It has been reported that not only AD but also Parkinson’s disease (PD) are associated with the NLRP3 inflammasome. Accumulation of Lewy bodies (LB) formed by α-synuclein (αSyn) aggregation is a main pathogenesis of PD. A recent study revealed that αSyn induces synthesis of pro-IL-1β by an interaction with TLR2 and activates NLRP3 inflammasome resulting in caspase-1 activation and IL-1β maturation in human primary monocytes (34). In contrast, it is reported that mitochondrial dysfunction may lead to neurodegenerative disease (35). These studies indicate that both the NLRP3 inflammasome and mitochondrial dysfunction are involved in neurodegenerative disease though more studies are required to clarify the relationship among NLRP3 inflammasome, mitochondria and neurodegenerative diseases.
NLRP3 INFLAMMASOME IN DISEASE; METABOLIC DISORDER
It has been known that a High fat diet (HFD) can induce Type 2 diabetes (T2D) and obesity and chronic inflammation is thought to contribute to T2D and obesity (36). As such, there is a possibility that the NLRP3 inflammasome, which can lead to chronic inflammation, is related to these metabolic diseases. Recently Lee et al. observed the activated NLRP3 inflammasome in T2D patients and found that mitochondrial ROS is involved in this phenomenon (37). Other groups showed that saturated fatty acid-induced inflammation causes mitochondrial impairment in adipocytes, while saturated fatty acids can upregulate NLRP3 inflammasome activation (38, 39). Also, a higher level of NLRP3 inflammasome components were detected in obese patients, and active caspase-1 were increased with obesity development in adipose tissues (40). Interestingly, caspase-1 deficient mice gained less weight and they had less adipose tissue formation in the HFD-induced obesity mouse model (41). These data strongly suggest that the NLRP3 inflammasome and mitochondrial dysfunction are closely linked to metabolic disorders.
NLRP3 INFLAMMASOME IN DISEASE NLRP3; SEPSIS
Sepsis is a life-threatening systemic inflammatory condition caused by a host immune response to microbial infection. Normally, increased inflammatory response to infection should be resolved in a timely manner, but when it gets out of control, exaggerated activation of the inflammasome produces excessive inflammatory cytokines leading to sepsis. A few studies provided evidence that mitochondrial dysfunction is associated with sepsis. Damaged mitochondria of the cells treated with NLRP3 inflammasome activators release danger signals, such as mtROS and mtDNA, and these signals can activate the NLRP3 inflammasome (9, 10). Supporting these, increased mtDNA and inflammatory cytokines were detected in septic plasma samples (42). This evidence shows that both the NLRP3 inflammasome and mitochondria contribute to sepsis.
MITOCHONDRIA AND NLRP3 INFLAMMASOME
Mitochondria are double membrane organelles that play pivotal roles in cells, including energy production and regulation of cell death. In addition, recent studies have revealed that mitochondria are also involved in the innate immune response (43, 44). Mitochondria are the main sites of reactive oxygen species (ROS) production. Mitochondrial ROS are not just byproducts of aerobic respiration, but also participate in signaling pathways. However, overproduced ROS can cause cell damage and contribute to pathologies. Some studies have shown that ATP and monosodium urate (MSU) crystals increase ROS production and these activate the inflammasome (45, 46). Silica and asbestos also increase ROS production and activate the NLRP3 inflammasome (47). In another study, increased mtROS by the inhibition of mitochondrial complex I or complex III, triggered NLRP3 inflammasome activation, indicating mtROS can be a main cause of NLRP3 inflammasome activation (48, 49). mtDNA also activates NLRP3 inflammasome. Under normal conditions, mtDNA is located in mitochondria; however, when mitochondria are damaged, mtDNA is released into the cytoplasm. Shimada et al. found that oxidized mtDNA released from damaged mitochondria directly induced the NLRP3 inflammasome (10). Mitochondrial outer membrane proteins are also thought to activate the NLRP3 inflammasome. Mitofusins, mitochondrial outer membrane proteins that are required for mitochondrial fusion, activate the NLRP3 inflammasome in influenza and encephalomyocarditis virus (EMCV) infection, while another mitochondrial outer membrane protein MAVS recruits NLRP3 to mitochondria and activates inflammasome activation in response to ATP, nigericin and poly I:C (50). All of these data indicate that mitochondria play a crucial role in NLRP3 inflammasome activation.
AUTOPHAGY AND NLRP3 INFLAMMASOME
As described above, there is a lot of evidence to suggest that damaged mitochondria contribute to disease, including NLRP3 inflammasome-related diseases. Therefore, the importance of mitophagy, which is a process to remove damaged mitochondria, can be considered as a key process to regulate NLRP3 inflammasome activation. Despite the importance of mitophagy, it has mostly only been studied in mitochondria-related gene overexpressed mammalian cell lines in recent years. To investigate its mechanism and function accurately, more in vitro and in vivo studies are required. Knowledge of macroautophagy (hereafter autophagy) is a prerequisite to understanding mitophagy. Autophagy is a self-degradation process to recycle cellular components under stressed conditions, such as lack of nutrients. Autophagy has been regarded as a nonselective process in the past; however, accumulating evidence shows that it can be selective to remove specific proteins and damaged organelles, such as mitochondria under certain conditions (51-53). Autophagy consists of 4 sequential steps: initiation of autophagosome formation, elongation and closure of autophagic membrane, fusion between autophagosome and lysome, and degradation. In many cellular cases, autophagic induction is regulated by the mammalian target of rapamycin (mTOR) and the AMP-activated protein kinase (AMPK). mTOR inactivates unc-51-like kinase 1/2 (ULK1/2) by phosphorylation under basal conditions, but mTOR is inhibited under stress conditions, allowing ULK1/2 to be modified to their active forms by AMPK phosphorylation initiating autophagy. Next, VPS34 lipid kinase complex and phosphatidylinositol 3-phosphate (PI3P) are recruited to complete autophagosome formation. In the step of elongation and closure of the autophagic membrane, the ubiquitin-like proteins, ATG12 and ATG5, are conjugated and then the conjugated forms an E3-like ligase complex with ATG16L1. After that, ATG8 (LC3 and GABARAP subfamilies) is conjugated to the lipid phosphatidylethanolamine by the ATG16L1 ligase complex and in the case of LC3, LC3-I form changes to LC3-II form, which is often used as an autophagosome marker (44, 54). VPS34–Beclin 1 complex containing ultraviolet radiation resistance associated gene protein (UVRAG) is involved in the step of fusion between the autophagosome and lysosome. Once the autophagosome and lysosome are fused to the autolysosome, the cargo in the autolysosome is degraded through lysosomal hydrolase activity (55). When there are certain proteins or organelles to be eliminated, they are ubiquitinated by E3 ubiquitin ligases and the ubiquitin chains serve as ‘eat-me’ signals. Then, adaptor proteins, such as p62 and NBR1, which contain a ubiquitin-associated UBA domain and LC3-interacting region (LIR), capture the ubiquitinated cargos and recruit LC3-II to proceed to selective autophagy (54). Accumulating evidence has shown that autophagy is involved in the innate immune response and NLRP3 inflammasome activation. Inhibition of autophagy by 3MA, which is a PI3K inhibitor increased mtROS production resulting in NLRP3-dependent IL-1β secretion in the absence of inflammasome stimuli (56, 57). Similarly, treatment of 3MA blocking autophagosome formation in Mycobacterium tuberculosis-infected cells led to increased IL-1β secretion (58). Consistent with these data, bone marrow-derived macrophages from LC3 knockout mice and Beclin 1 knockout mice had more damaged mitochondria producing mtROS, and had more caspase-1 activation leading to increased IL-1β secretion upon NLRP3 inflammasome activation (9). Also, Atg16L1 deficient mice were more susceptible to dextran sulfate sodium-induced colitis as a result of enhanced inflammasome activation (58). These data show that autophagy negatively regulates inflammasome activation. Moreover, as damaged mitochondria are detected at higher levels under autophagy inhibition (9, 56, 57), it is plausible to assume that the elimination of damaged mitochondria by autophagy is important to prevent excessive NLRP3 inflammasome activation.
MITOPHAGY AND NLRP3 INFLAMMASOME
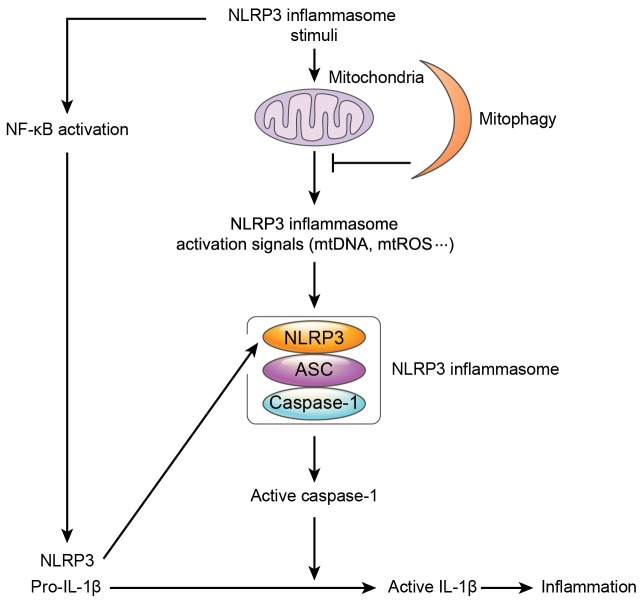
As mentioned above, mitochondrial damage causes NLRP3 inflammasome, and mitophagy can remove damaged mitochondria selectively (Fig. 1). Since mitochondria are evolved from ancient bacteria, a comparison of differences between mitophagy and intracellular bacteria-selective autophagy referred as to xenophagy can provide valuable information to better understand mitophagy. Distinguishing between self and non-self is achieved by different ‘eat-me’ signals and cargo receptors (52, 59). To date, at least 4 cargo receptors have been studied in detail: NDP52, p62, NBR1 and Optineurin (59). Invading bacteria can damage their host vacuoles in which the bacteria replicate exposing glycans from the host vacuoles, and the glycans are recognized by a danger receptor, Galectin-8. Galectin-8 functions as an ‘eat-me’ signal and interacts with NDP52 inducing xenophagy by recruiting LC3 attached to phagophores (60). NDP52 can also directly bind to ubiquitinated bacteria and recruit the autophagic machinery. Similarly, p62, NBR1 and Optineurin serves to connect ubiquitinated bacteria with LC3, facilitating autophagy. How the engulfed bacteria are ubiquitinated is still largely unknown but LRSAM1 has been identified as an E3 ubiquitin ligase that ubiquitinates Salmonella typhimurium recruiting cargo receptors (61). Another E3 ubiquitin ligase, PARKIN, has also been reported to be involved in xenophagy. PARKIN deficient mice and flies were susceptible to intracellular bacterial infections inducing less autophagy, and these indicate that PARKIN plays a crucial role in xenophagy (62). Interestingly, PARKIN is the best known E3 ubiquitin ligase in mitophagy. The PINK-PARKIN pathway is important in the mitophagic process (63, 64). PINK1 is a serine/threonine kinase containing a mitochondrial targeting sequence in the N-terminus. Normally, PINK1 is imported into the mitochondria and anchored at the inner mitochondrial membrane (IMM) and then degraded by mitochondrial proteases. However, when mitochondria are damaged, PINK1 can’t be imported to IMM, but instead accumulates on the outer mitochondria membrane (OMM) (65). The accumulated PINK1 recruits a cytosolic E3 ubiquitin ligase PARKIN and activates PARKIN by phosphorylation. Activated PARKIN ubiquitinates certain mitochondrial proteins or PARKIN itself. Then, the mitochondrial ubiquitination acts as an ‘eat-me’ signal and can be recognized by the adaptor p62 through its ubiquitin binding domain. Since p62 also has an LC3 interacting motif, p62 binds with LC3 on the autophagosome and facilitates the degradation of damaged mitochondria (44). However, not all mitophagy is processed by the PINK-PARKIN pathway. Since mitochondrial proteins BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3), NIP3-like protein X (NIX/BNIP3L) and FUN14 domain containing 1 (FUNDC1) can directly interact with LC3-II, it is thought that other mechanisms exist to enhance mitophagy (66-69). So far we have discussed how mitochondria play a crucial role in the NLRP3 inflammasome activation, however not many studies have been performed concerning the detailed molecular mechanisms of mitophagy. Recently, some studies have shown how mitophagy is related to the control of NLRP3 inflammasome activation. Zhong et al. found that the cargo receptor p62 is increased by NF-κB signaling, and that the increased p62 is translocated to damaged mitochondria, which are ubiquitinated by Parkin and induce mitophagy (70). Consistent with this finding, Kim et al. also observed that p62 is increased and translocated to damaged mitochondria in NLRP3 inflammasome activated cells. Not only p62 but also autophagic inducer Sestrin 2 (SESN2) is increased by NO and is translocated to damaged mitochondria and protects cells from hyperinflammation inducing mitophagy. Additionally, SESN2 increases ULK1 stability leading to the initiation of autophagy (71). These studies indicate that mitophagy is one of the self-limiting systems to protect cells from excessive inflammation.
Fig. 1. Mitophagy downregulates aberrant NLRP3 inflammasome activation. NLRP3 inflammasome stimuli activates NF-κB producing NLRP3 and pro-IL-1β. Mitochondria are damaged by NLRP3 inflammasome stimuli and release NLRP3 inflammasome activating signals, such as mtDNA and mtROS, triggering an inflammatory response. Mitophagy downregulates NLRP3 inflammasome activation by eliminating damaged mitochondria, blocking NLRP3 inflammasome activating signals.
CONCLUSION
In the past decade, there have been great advances in understanding of the NLRP3 inflammasome and autophagy. However, knowledge of the link between the NLRP3 inflammasome and autophagy, especially mitophagy, is poorly understood. Nevertheless we have discussed the role of mitophagy as a negative regulator of aberrant NLRP3 inflammasome activation, while detailed mechanisms remain largely unknown. There are questions that remain to be answered. Mitochondrial damage can activate the NLRP3 inflammasome, but inflammatory cytokines can also trigger mitochondrial damage, so it is ambiguous as to which one comes first. Since NLRP3 inflammasome activation has a beneficial effect so how to regulate activation appropriately it should be considered for clinical application. Also, how damaged mitochondria serve as ‘eat-me’ signals in the case where PINK1 is not mediated should be investigated. Further studies are required for a better understanding of the molecular mechanisms of mitophagy, but mitochondria remain possible therapeutic targets to protect cells from aberrant inflammation.
Acknowledgments
This work was supported by NRF grant funded by the Ministry of Science, ICT & Future Planning (2012M3A9C5048709), (2013M3A9D5072550).
References
- 1.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. (2012);481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 2.Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol. (2012);28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 3.Rosen H, Klebanoff SJ. Bactericidal activity of a superoxide anion-generating system. A model for the polymorphonuclear leukocyte. J Exp Med. (1979);149:27–39. doi: 10.1084/jem.149.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Lee KA, Ren X, et al. Synthesis of a highly HOCl-selective fluorescent probe and its use for imaging HOCl in cells and organisms. Nat Protoc. (2016);11:1219–1228. doi: 10.1038/nprot.2016.062. [DOI] [PubMed] [Google Scholar]
- 5.Yin J, Kwon Y, Kim D, et al. Preparation of a cyanine-based fluorescent probe for highly selective detection of glutathione and its use in living cells and tissues of mice. Nat Protoc. (2015);10:1742–1754. doi: 10.1038/nprot.2015.109. [DOI] [PubMed] [Google Scholar]
- 6.Joo JH, Ryu JH, Kim CH, et al. Dual oxidase 2 is essential for the toll-like receptor 5-mediated inflammatory response in airway mucosa. Antioxid Redox Signal. (2012);16:57–70. doi: 10.1089/ars.2011.3898. [DOI] [PubMed] [Google Scholar]
- 7.Kim MJ, Ryu JC, Kwon Y, et al. Dual oxidase 2 in lung epithelia is essential for hyperoxia-induced acute lung injury in mice. Antioxid Redox Signal. (2014);21:1803–1818. doi: 10.1089/ars.2013.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelletier M, Lepow TS, Billingham LK, Murphy MP, Siegel RM. New tricks from an old dog: mitochondrial redox signaling in cellular inflammation. Semin Immunol. (2012);24:384–392. doi: 10.1016/j.smim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakahira K, Haspel JA, Rathinam VA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. (2011);12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimada K, Crother TR, Karlin J, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. (2012);36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. (2007);449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 12.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006);124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. (2010);140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. (2013);39:432–441. doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. (2014);157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Vanaja SK, Rathinam VA, Fitzgerald KA. Mechanisms of inflammasome activation: recent advances and novel insights. Trends Cell Biol. (2015);25:308–315. doi: 10.1016/j.tcb.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. (2015);21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder K, Tschopp J. The inflammasomes. Cell. (2010);140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 19.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. (2013);13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kufer TA, Banks DJ, Philpott DJ. Innate immune sensing of microbes by Nod proteins. Ann N Y Acad Sci. (2006);1072:19–27. doi: 10.1196/annals.1326.020. [DOI] [PubMed] [Google Scholar]
- 21.Moreira LO, Zamboni DS. NOD1 and NOD2 Signaling in Infection and Inflammation. Front Immunol. (2012);3:328. doi: 10.3389/fimmu.2012.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. (2000);14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 23.Franchi L, Stoolman J, Kanneganti TD, Verma A, Ramphal R, Nunez G. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. (2007);37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 24.Mariathasan S, Newton K, Monack DM, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. (2004);430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki T, Franchi L, Toma C, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. (2007);3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornung V, Ablasser A, Charrel-Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. (2009);458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts TL, Idris A, Dunn JA, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. (2009);323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 28.Lu A, Magupalli VG, Ruan J, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. (2014);156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. (2012);13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez-Cuellar E, Tsuchiya K, Hara H, et al. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J Immunol. (2012);189:5113–5117. doi: 10.4049/jimmunol.1202479. [DOI] [PubMed] [Google Scholar]
- 31.Lee GS, Subramanian N, Kim AI, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. (2012);492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halle A, Hornung V, Petzold GC, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. (2008);9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heneka MT, Golenbock DT, Latz E. Innate immunity in Alzheimer's disease. Nat Immunol. (2015);16:229–236. doi: 10.1038/ni.3102. [DOI] [PubMed] [Google Scholar]
- 34.Codolo G, Plotegher N, Pozzobon T, et al. Triggering of inflammasome by aggregated alpha-synuclein, an inflammatory response in synucleinopathies. PLoS One. (2013);8:e55375. doi: 10.1371/journal.pone.0055375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voloboueva LA, Giffard RG. Inflammation, mitochondria, and the inhibition of adult neurogenesis. J Neurosci Res. (2011);89:1989–1996. doi: 10.1002/jnr.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotamisligil GS. Inflammation and metabolic disorders. Nature. (2006);444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 37.Lee HM, Kim JJ, Kim HJ, Shong M, Ku BJ, Jo EK. Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes. (2013);62:194–204. doi: 10.2337/db12-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen H, Gris D, Lei Y, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. (2011);12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandanmagsar B, Youm YH, Ravussin A, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. (2011);17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Z, Deng T, Peterson LE, et al. Transcriptome analysis of human adipocytes implicates the NOD-like receptor pathway in obesity-induced adipose inflammation. Mol Cell Endocrinol. (2014);394:80–87. doi: 10.1016/j.mce.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stienstra R, Joosten LA, Koenen T, et al. The inflammasome-mediated caspase-1 activation controls adipocyte differentiation and insulin sensitivity. Cell Metab. (2010);12:593–605. doi: 10.1016/j.cmet.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakahira K, Kyung SY, Rogers AJ, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. (2013);10:e1001577. doi: 10.1371/journal.pmed.1001577. discussion e1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurung P, Lukens JR, Kanneganti TD. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol Med. (2015);21:193–201. doi: 10.1016/j.molmed.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazarou M. Keeping the immune system in check: a role for mitophagy. Immunol Cell Biol. (2015);93:3–10. doi: 10.1038/icb.2014.75. [DOI] [PubMed] [Google Scholar]
- 45.Cruz CM, Rinna A, Forman HJ, Ventura AL, Persechini PM, Ojcius DM. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. (2007);282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. (2007);14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 47.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. (2008);320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang LS, Cobessi D, Tung EY, Berry EA. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol. (2005);351:573–597. doi: 10.1016/j.jmb.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li N, Ragheb K, Lawler G, et al. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. (2003);278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. (2013);153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells. (2010);15:923–933. doi: 10.1111/j.1365-2443.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- 52.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. (2011);7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito T, Sadoshima J. Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circ Res. (2015);116:1477–1490. doi: 10.1161/CIRCRESAHA.116.303790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abounit K, Scarabelli TM, McCauley RB. Autophagy in mammalian cells. World J Biol Chem. (2012);3:1–6. doi: 10.4331/wjbc.v3.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. (2013);13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris J, Hartman M, Roche C, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. (2011);286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. (2011);469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 58.Shi CS, Shenderov K, Huang NN, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. (2012);13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle KB, Randow F. The role of 'eat-me' signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol. (2013);16:339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 60.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. (2012);482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huett A, Heath RJ, Begun J, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. (2012);12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manzanillo PS, Ayres JS, Watson RO, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. (2013);501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. (2010);8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. (2010);107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. (2010);191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarten M, Mohrluder J, Ma P, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. (2009);5:690–698. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 67.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. (2012);287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. (2010);11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. (2012);14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 70.Zhong Z, Umemura A, Sanchez-Lopez E, et al. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. (2016);164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim MJ, Bae SH, Ryu JC, et al. SESN2/sestrin2 suppresses sepsis by inducing mitophagy and inhibiting NLRP3 activation in macrophages. Autophagy. (2016);12:1272–1291. doi: 10.1080/15548627.2016.1183081. [DOI] [PMC free article] [PubMed] [Google Scholar]