Abstract
The prevalence of obesity and type 2 diabetes, two closely linked metabolic disorders, is increasing worldwide. Over the past decade, the connection between these disorders and the microbiota of the gut has become a major focus of biomedical research, with recent studies demonstrating the fundamental role of intestinal microbiota in the regulation and pathogenesis of metabolic disorders. Because of the complexity of the microbiota community, however, the underlying molecular mechanisms by which the gut microbiota is associated with metabolic disorders remain poorly understood. In this review, we summarize recent studies that investigate the role of the microbiota in both human subjects and animal models of disease and discuss relevant therapeutic targets for future research. [BMB Reports 2016; 49(10): 536-541]
Keywords: Gut microbiota, Metabolic disorder, Obesity, Short-chain fatty acids, Type 2 diabetes
THE MICROBIOTA IN THE GUT MUCOSA
The human gut is populated with as many as 100 trillion (1014) cells, including bacteria, fungi, viruses, and other microbial and eukaryotic species (1, 2). These complex, heavily diverse communities provide tremendous enzymatic capability and thus play a fundamental role in manipulating host physiology (3, 4). It is well established that five bacterial phyla, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Verrucomicrobia, are dominant components of the human gut microbiota (5). More than 90% of the bacterial populations are gram-negative anaerobes and include the predominant genera Bacteroides, Eubacterium, Bifidobacterium, and Fusobacterium (6). Additionally, the gut microbiota is indispensable for carbohydrate fermentation and nutrient absorption (1), protection of pathogenic bacteria (7), and regulation of metabolic disorders (8).
The host intestine is unique with respect to its constant exposure to a plethora of antigens from daily food intake and exogenous bacteria. The resident gut microbiota contains a number of components able to activate both innate and adaptive immunity responses (3, 9). For example, the majority of intestinal bacteria are gram-negative anaerobes equipped with diverse agents, such as lipopolysaccharide (LPS) and flagella, allowing for innate signaling to intestinal epithelial cells through toll-like receptors (TLRs) (10). Segmented filamentous bacteria (SFB) embedded in the ileum can also stimulate adaptive, T helper 17 (TH17) responses and induce the production of mucosal immunoglobulin A (IgA) antibody (11). In addition, commensal microbe-derived butyrate is associated with regulatory T (Treg) cell differentiation in the colon (12).
THE GUT MICROBIOTA AND METABOLIC DISORDERS
According to the World Health Organization, more than 1.9 billion adults were overweight in 2014, with over 600 million of those adults classified as obese. Obesity results from energy imbalance and is associated with other metabolic complications such as type 2 diabetes. The relationship between gut microbiota and metabolic diseases was first reported by the laboratory of Jeffrey Gordon at Washington University. Specifically, the Gordon laboratory demonstrated that leptindeficient mice, notable for their excessive appetite and profound obesity, contained fewer Bacteroidetes and more Firmicutes than control mice (13). This study provided the first direct evidence of differences in the microbial communities of lean vs. obese animals. In support of this finding, a follow-up study from the Gordon laboratory observed fewer Bacteroidetes and more Firmicutes in obese human subjects than in lean subjects (14). Furthermore, the proportion of Bacteroidetes increased with either fat- or carbohydrate-restricted diet and subsequent weight loss. In people who lost weight after a gastric bypass procedure, increased levels of Bacteroides and Prevotella were negatively correlated with energy intake and adiposity (15). Other studies, however, have not observed a shift in the ratio of Bacteroidetes and Firmicutes in human subjects with weight loss (16-18). Thus, although it is possible that certain microbial species in the human gut contribute to weight gain and others contribute to weight loss, it is also possible that any observed changes in the gut microbiota are the result of weight shifts.
To address this concern, microbiota transplantation experiments have since been adopted. An initial study conducted by Gordon et al. demonstrated that conventionalization of germ-free mice with a normal microbiota resulted in increased body fat content and insulin resistance within 14 days, despite reduced food intake (19). This study provided the first mechanistic evidence that gut microbes can increase the host’s ability to store body fat. Furthermore, germ-free mice that received gut microbes from an obese twin donor showed an increase of total body and fat mass as well as obesity-associated metabolic phenotypes compared with those that received a lean twin’s microbiota (20). Interestingly, the gut microbiota from a lean mouse could invade the microbiota of an obese mouse and provide protection from weight gain, but this influence was dependent on diet. Other studies have demonstrated that germ-free mice transplanted with obesity-associated microbiota gained weight, but not to an excessive, obese level (21). Thus, the role of diet and other factors need to be considered.
DIET ALTERS THE GUT MICROBIOTA
Diet is a major factor in obesity, and it also helps shape the gut microbiota. Human studies from the past decade have revealed that the gut microbiota responds rapidly to large changes in diet; in many cases, the composition and function of the gut microbiota shifts within 1-2 days (22, 23). Despite these rapid dynamics, long-term dietary habits are still critical in determining the gut composition of an individual (24), and the effectiveness of a specific diet largely relies on the initial composition of the gut microbiota (25).
Extensive research has shown that the gut microbiota of a traditional rural population (i.e., high-fiber, low-fat diet) is more diverse and contains distinct taxa than the microbiota of Western populations (i.e., low-fiber, high-fat diet) (26). Preservation of microbial diversity by a high-fiber, low-fat diet allows individuals to maximize energy intake from fiber while also protecting them from inflammation and noninfectious colonic diseases. Although it is unclear whether increased microbial diversity contributes to protection from metabolic diseases, several metagenomics studies indicate that improved outcomes in metabolic diseases are associated with increased microbial diversity (27, 28). For example, a team of researchers sequenced the microbiomes of 169 obese and 123 non-obese individuals and observed that individuals fell into two groups: a group with a low amount of microbial gene diversity and another group with high diversity (27). Those with fewer genes tended to have more pronounced adiposity, insulin resistance, and dyslipidemia than individuals containing more diverse gut microbiotas. Furthermore, obese individuals with lower bacterial diversity showed more weight gain over time. These data imply that manipulation of microbial diversity in the gut could be a promising avenue for amelioration of metabolic disorders.
MICROBIAL REGULATION OF METABOLITES
The gut microbiota produces numerous amounts of metabolites. For example, the microbiota contributes to host metabolic efficiency by increasing energy availability via the production of short-chain fatty acids (SCFAs), such as acetate, butyrate and propionate (29). Previous studies demonstrated that SCFA levels were elevated in obese human subjects and animal models, consistent with the fact that SCFAs provide extra calories to the host (8, 16). Most recently, researchers observed that amplified production of acetate increased the likelihood of obesity by triggering secretion of ghrelin, the appetite-inducing hormone (30). By contrast, other studies have demonstrated that SCFAs directly contribute to host protection from metabolic diseases. For example, SCFAs are an important energy source for gut epithelial cells through activation of G-protein-coupled receptors, such as GPR41 and GPR43, which influence enteroendocrine regulation (31, 32). Butyrate acts as a main energy source for colonocytes (33), and propionate can induce intestinal gluconeogenesis, thus protecting the host from diet-induced obesity via the gut-brain neural axis (34). Interestingly, consumption of a complex diet resulted in increased levels of SCFAs and increased diversity within the gut microbiota (26). Reduced dietary intake of carbohydrates for obese humans, however, resulted in decreased butyrate levels in their feces and correlated with a reduced abundance of butyrate-producing bacteria (i.e., Roseburia spp. and Eubacterium rectale) (17). Furthermore, levels of propionate specifically correlated with the amount of Bacteroidetes in the gut (35).
In addition to the production of SCFAs, the microbiota forms bile acids from host cholesterol. Bile acids are a family of steroid acids synthesized from cholesterol in the liver and mostly secreted into the lumen of the intestine to control the digestion and absorption of dietary fat and fat-soluble vitamins (36). Commensal microbiota plays a pivotal role in the conversion of primary bile acids in the lower part of the small intestine and the colon to generate secondary bile acids (37, 38). Although it has become clear that bile acids act as signaling molecules for metabolic pathways, fundamental questions remain concerning whether additional administration of specific commensal bacteria can regulate bile acid metabolism and the potential role of these altered bile acids in metabolic diseases, such as obesity and type 2 diabetes (39, 40). A recent study demonstrated that the bile acid profiles in the gut and serum of control mice were quite distinct from those of germ-free mice (41), while another animal study reported that taurine-conjugated bile acids were dominant in germ-free and antibiotic-treated mice (42). Furthermore, administration of probiotics altered gut microbiota composition and enhanced bile acid deconjugation and fecal excretion (43). Notably, the increased probiotic bacteria were associated with the induction of hepatic bile acid synthesis via the farnesoid X receptor (FXR) (43). The gut microbiota also regulates the G-protein-coupled bile acid receptor 1 (known as TGR5) (44). Our group has detected increased levels of bile acids in fecal tissue as well as TGR5 in adipose tissue from B. acidifaciens-fed mice (45). Additional studies have suggested that bile acid-TGR5-cAMP signaling pathways enhance energy expenditure in adipose tissue (46, 47). In total, specific commensal microbiota may regulate host metabolic actions through modulation of bile acid synthesis or reabsorption and by interaction with receptors such as FXR and TGR5.
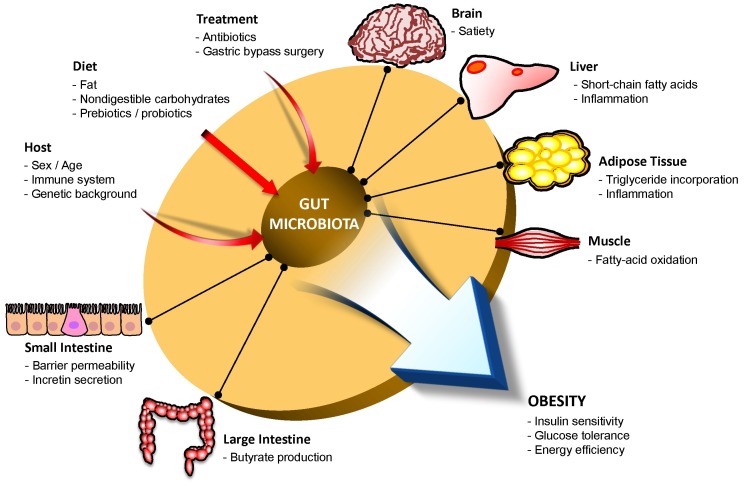
Gut microbiota ultimately contribute to the regulation of incretin hormone secretion through the interaction between the aforementioned metabolites (SCFAs, bile acids) and their receptors (GPR1 and GPR43, TGR5), which are expressed on enteroendocrine L cells (48-52). The stimulated enteroendocrine L cells secrete incretin hormone peptides, such as glucagon-like peptide-1 (GLP-1), glucagon-like peptide-2 (GLP-2), and peptide YY (PYY), which in turn stimulate insulin release and decrease blood glucose levels (48). These secreted peptides affect a wide range of organs and tissues to improve insulin sensitivity, glucose tolerance, and energy homeostasis (Fig. 1), thereby contributing to protection in metabolic disorders such as obesity and type 2 diabetes.
Fig. 1. Interactions between the gut microbiota and host metabolism. The gut microbiota can be influenced by a number of external factors, including host background, diet type, and medical treatments. Imbalance of the intestinal microbiota can lead to severe metabolic disorders (e.g., obesity) by altering host insulin sensitivity or energy homeostasis.
MICROBIAL REGULATION OF INFLAMATION
Several lines of evidence point to a role for gut microbiota in the induction of systemic and adipose tissue inflammation (49, 50). For example, the gut microbiota produces substantial amounts of inflammation-inducing factors, including LPS and peptidoglycan. Additionally, monocolonization of Escherichia coli into germ-free mice promoted macrophage accumulation in white adipose tissues and polarized macrophages toward inflammation, suggesting that gut microbiota-derived endotoxins, such as LPS, play a role in obesity regulation (10). Furthermore, food-derived microbiota exacerbated C-C motif chemokine ligand 2 (CCL2)-dependent macrophage infiltration in white adipose tissues through TLR signaling (51). In mouse models, obesity is associated with increased numbers of effector T cells and decreased numbers of Treg cells (52, 53). Because many studies have demonstrated that microbiotagenerated SCFAs promote anti-inflammatory responses in mucosal and systemic tissues through Treg cells (12, 54, 55), it is tempting to speculate that gut microbiota may control obesity through the generation of Treg cells.
SYMBIONTS IN METABOLIC DISEASES
There are several strategies to control metabolic diseases by a single species of the gut microbiota (Table 1). One such strategy is the consumption of beneficial bacteria as prebiotics or probiotics. For example, Akkermansia (A.) muciniphila, known as the mucin-degrading bacterium (56), can be administered as a prebiotic to reduce the likelihood of obesity and diabetes. This microbe commonly constitutes 3-5% of the human gut microbial community and is more abundant in healthy individuals than in obese/diabetic patients or animals (57-59). In one study, treatment of high-fat diet (HFD)-fed mice with metformin, a widely prescribed type 2 diabetes therapeutic agent, improved the glycemic profile and resulted in a higher abundance of A. muciniphila (60). Moreover, oral administration of A. muciniphila to HFD-fed mice without metformin treatment led to antidiabetic effects. Interestingly, exposure to the cold resulted in significant changes to gut microbiota composition, and reconstitution of cold-suppressed A. muciniphila reduced caloric uptake (61).
Table 1. Gut microbiota associated with obesity.
| Microbes | Mechanisms | References |
|---|---|---|
| Bacteroidetes /Firmicutes ratio↓ | Enhance dietary energy recovery | Ley et al (2005, 2006) |
| Methanobrevibacter smithii↓ | Increase energy harvest | Samuel et al (2007) |
| Bacteroidetes /Firmicutes ratio↑ | Increase SCFA levels | Schwiertz et al (2010) |
| Bacteroides (B.) cellulosilyticus↓ | High invasiveness into the gut | Ridaura et al (2013) |
| B. vulgatus↓ B. thetaiotaomicron↓ | ||
| B. caccae↓ B. uniformis↓ | ||
| Akkermansia muciniphila↓ | Improve glucose homeostasis | Santacruz et al (2010) |
| Bacteroides acidifacients↓ | Activate lipid oxidation | Yang et al (2016) |
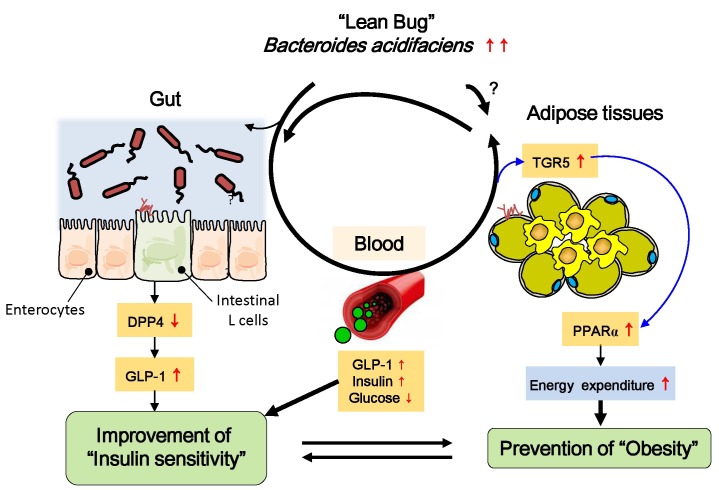
Gordon et al. reported lean phenotypes in mice that were correlated with Bacteroides species, such as B. cellulosilyticus, B. uniformis, B. vulgatus, B. thetaiotaomicron, and B. caccae (20). Oral administration of the B. uniformis CECT 7771 strain ameliorated HFD-induced metabolic dysfunction in obese mice (62). Our group has recently reported that HFD-fed mice given B. acidifaciens for 10 weeks gained less fat mass and body weight than those given PBS alone (45). We have suggested that peroxisome proliferator-activated receptor α (PPARα)-mediated fat oxidation in adipose tissues and an expanded half-life of GLP-1 are involved in the regulation of host adiposity and insulin resistance by B. acidifaciens. Furthermore, metabolites secreted by B. acidifaciens may play a critical role in maintaining low levels of dipeptidyl peptidase-4 (DPP-4) in the gut (45) (Fig. 2). It would be interesting to conduct a follow-up study to identify underlying mechanisms how beneficial symbionts working on and soluble factors produced by those bacterium and their cross-talks.
Fig. 2. Proposed mechanism for modulation of host insulin sensitivity by Bacteroides acidifaciens (BA). The selected commensal bacterium (i.e., BA) causes intestinal epithelial cells to secrete lower amounts of dipeptidyl peptidase-4 (DPP-4) in the gut and increased levels of glucagon-like peptide-1 (GLP-1), which may contribute to glucose homeostasis. At the same time, increased levels of bile acids (i.e., cholate and taurine) may contribute to GLP-1 activation in the intestine and to peroxisome proliferator-activated receptor α (PPARα) activation through TGR5 in adipose tissues, ultimately resulting in fat oxidation and improved insulin sensitivity.
CONCLUSIONS AND PERSPECTIVES
It is likely that nothing is simple when considering the link between diet, gut microbes, and metabolic diseases. Although mechanistic studies in animal models have produced valuable insight and revealed potential therapeutic targets, future studies are challenged with translating these findings into the human patient.
Acknowledgments
This study was supported by the Korean Health Technology R&D Project, Ministry for Health & Welfare, Republic of Korea (Grant HI13C0016).
References
- 1.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010);464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. (2012);486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. (2014);157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. (2016);535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. (2012);489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 6.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. (2003);361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 7.Kamada N, Kim YG, Sham HP, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. (2012);336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho I, Yamanishi S, Cox L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. (2012);488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. (1999);69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 10.Caesar R, Reigstad CS, Backhed HK, et al. Gutderived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. (2012);61:1701–1707. doi: 10.1136/gutjnl-2011-301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009);139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013);504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 13.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. (2005);102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. (2006);444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 15.Furet JP, Kong LC, Tap J, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. (2010);59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwiertz A, Taras D, Schafer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. (2010);18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 17.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. (2007);73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duncan SH, Lobley GE, Holtrop G, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes. (2008);32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- 19.Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. (2004);101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. (2013);341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeWeerdt S. Microbiome: A complicated relationship status. Nature. (2014);508:S61–63. doi: 10.1038/508S61a. [DOI] [PubMed] [Google Scholar]
- 22.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014);505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. (2011);334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. (2011);332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. (2011);5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. (2010);107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. (2013);500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 28.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature. (2013);500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 29.Gao Z, Yin J, Zhang J, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. (2009);58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. (2016);534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge H, Li X, Weiszmann J, et al. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. (2008);149:4519–4526. doi: 10.1210/en.2008-0059. [DOI] [PubMed] [Google Scholar]
- 32.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. (2008);105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donohoe DR, Garge N, Zhang X, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. (2011);13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. (2014);156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Salonen A, Lahti L, Salojarvi J, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. (2014);8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. (2008);7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 37.Midtvedt T. Microbial bile acid transformation. Am J Clin Nutr. (1974);27:1341–1347. doi: 10.1093/ajcn/27.11.1341. [DOI] [PubMed] [Google Scholar]
- 38.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. (2006);47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi M, Ikegami H, Fujisawa T, et al. Prevention and treatment of obesity, insulin resistance, and diabetes by bile acid-binding resin. Diabetes. (2007);56:239–247. doi: 10.2337/db06-0353. [DOI] [PubMed] [Google Scholar]
- 40.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. (2008);57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 41.Sayin SI, Wahlstrom A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Swann JR, Want EJ, Geier FM, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. (2011);108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. (2014);7:12–18. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 44.Kawamata Y, Fujii R, Hosoya M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. (2003);278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 45.Yang JY, Lee YS, Kim Y, et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. (2016) doi: 10.1038/mi.2016.42. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 46.Sato H, Genet C, Strehle A, et al. Anti-hyperglycemic activity of a TGR5 agonist isolated from Olea europaea. Biochem Biophys Res Commun. (2007);362:793–798. doi: 10.1016/j.bbrc.2007.06.130. [DOI] [PubMed] [Google Scholar]
- 47.Tsuboyama-Kasaoka N, Shozawa C, Sano K, et al. Taurine (2-aminoethanesulfonic acid) deficiency creates a vicious circle promoting obesity. Endocrinology. (2006);147:3276–3284. doi: 10.1210/en.2005-1007. [DOI] [PubMed] [Google Scholar]
- 48.Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. (2006);368:1696–1705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- 49.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. (2012);18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 50.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. (2003);112:1796–1808. doi: 10.1172/JCI200319246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. (2015);22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feuerer M, Herrero L, Cipolletta D, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. (2009);15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura S, Manabe I, Nagasaki M, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. (2009);15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 54.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013);504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belzer C, de Vos WM. Microbes inside--from diversity to function: the case of Akkermansia. ISME J. (2012);6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liou AP, Paziuk M, Luevano JM, Jr, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. (2013);5:178ra14. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van den Abbeele P, Gerard P, Rabot S, et al. Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ Microbiol. (2011);13:2667–2680. doi: 10.1111/j.1462-2920.2011.02533.x. [DOI] [PubMed] [Google Scholar]
- 59.Santacruz A, Collado MC, Garcia-Valdes L, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. (2010);104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 60.Shin NR, Lee JC, Lee HY, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. (2014);63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 61.Chevalier C, Stojanovic O, Colin DJ, et al. Gut microbiota orchestrates energy homeostasis during cold. Cell. (2015);163:1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 62.Gauffin Cano P, Santacruz A, Moya A, Sanz Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One. (2012);7:e41079. doi: 10.1371/journal.pone.0041079. [DOI] [PMC free article] [PubMed] [Google Scholar]