Abstract
The short gastrulation (sog) shadow enhancer directs early and late sog expression in the neurogenic ectoderm and the ventral midline of the developing Drosophila embryo, respectively. Here, evidence is presented that the sog primary enhancer also has both activities, with the late enhancer activity dependent on the early activity. Computational analyses showed that the sog primary enhancer contains five Dorsal (Dl)-, four Zelda (Zld)-, three Bicoid (Bcd)-, and no Single-minded (Sim)-binding sites. In contrast to many ventral midline enhancers, the primary enhancer can direct lacZ expression in the ventral midline as well as in the neurogenic ectoderm without a canonical Simbinding site. Intriguingly, the impaired transcriptional synergy between Dl and either Zld or Bcd led to aberrant and abolished lacZ expression in the neurogenic ectoderm and in the ventral midline, respectively. These findings suggest that the two enhancer activities of the sog primary enhancer are functionally consolidated and geographically inseparable. [BMB Reports 2016; 49(10): 572-577]
Keywords: Drosophila, Embryo, Primary enhancer, Shadow enhancer, short gastrulation
INTRODUCTION
The shotgastrulation (sog) gene is one of seven zygotically active genes [decapentaplegic (dpp), zerknüllt (zen), sog, tolloid (tld), twisted gastrulation (tsg), screw (scw) and shrew (srw)] required for dorsal-ventral (DV) patterning in the ectoderm of the Drosophila early embryo (1). Originally, sog was found as one of three X-lined zygotic genes required for specific morphogenetic events of gastrulation (2). The sog gene exhibits dynamic expression during embryonic development (1). sog transcripts are first observed in broad lateral stripes of the neurogenic ectoderm as early as nuclear cleavage cycle 13, the dorsal borders of which abut the ventral limits of the dpp expression domain in the dorsal ectoderm. At least by germ band extension, sog expression is restricted to the ventral midline, which comprises specialized glial cells that secrete signals essential for nerve cord patterning (3).
Early sog expression is dependent on the DV determinants, Dorsal (Dl) and Snail (Sna) (4). Bioinformatics studies of the genome-wide distribution of the Dl recognition sequence have identified a ∼400-bp genomic region that acts as an enhancer to direct the early broad pattern of sog expression in the neurogenic ectoderm (5). Subsequent chromatin immunoprecipitation followed by genomic tiling array (ChIP-chip) analyses indicated that many of the Dl target genes contain two independent and separate enhancers that control the same or similar expression patterns (6). Recently, it was shown that the sog locus has a secondary remote enhancer that directs its expression in the neurogenic ectoderm (7). The two enhancers are referred to as the “primary” and “shadow” enhancers, respectively, according to the chronological order of their identification, rather than functional differences. More recently, the shadow enhancer has been shown to direct late sog expression in the ventral midline after gastrulation (8). The finding that the shadow enhancer has dual enhancer activities in the early neurogenic ectoderm and the late ventral midline raised the possibility that the primary enhancer also has the ability to control late sog expression in the ventral midline.
Here, we demonstrate that the sog primary enhancer also has enhancer activities in the neurogenic ectoderm and the ventral midline, and that the late enhancer activity requires the early enhancer activity. Computational analyses indicated that the primary enhancer contains five Dl-, four Zelda (Zld)-, and three Bicoid (Bcd)-binding sites. Despite its midline enhancer activity, no Sim-binding site was identified within the primary enhancer. Intriguingly, removal of Dl-, Zld-, or Bcd-binding sites abolished lacZ expression in the ventral midline and led to its aberrant expression in the neurogenic ectoderm. These results suggest that late enhancer activity is potentiated by its early transcriptional activation.
RESULTS
The sog primary enhancer directs lacZ expression in the ventral midline of the late embryo
Sim is a master regulatory gene that directly regulates the expression of many ventral midline genes (9). sim transcripts are first observed in a single row of cells, called the mesectoderm, found on along either side of the presumptive mesoderm (Fig. 1A). The symmetric lines of mesectodermal cells converge at the ventral midline during gastrulation (Fig. 1B). Once induced, sim expression is maintained via autoregulation during germ band elongation (Fig. 1C and D) and later stages of embryogenesis. The sog gene has a broad expression pattern in the neurogenic ectoderm as early as nuclear cleavage cycle 13 (Fig. 1E). Through gastrulation (Fig. 1F), sog expression is restricted to the ventral midline (Fig. 1G and H), which is comparable with the sim expression pattern in the corresponding developmental stages. Endogenous sog midline expression is thought to be controlled directly by the sim gene product. To test if sog is a target of the Sim protein, sog expression was examined in mutant embryos homozygous for a sim null allele (simH9) (Fig. 1I-L). Early sog expression was not affected by lack of Sim (Fig. 1I and J), while sog transcripts were not detected in the ventral midline, at least after completion of gastrulation (Fig. 1K and L). These results suggest that sog expression is directly controlled by Sim in the ventral midline of the developing embryo.
Fig. 1. The sog primary enhancer directs expression in the ventral midline of the late embryo. Approximately 2-10 hours (h) after egg deposition (AED), embryos were collected, dechorionated, and fixed. Whole-mount in situ hybridization was performed with fixed embryos and digoxigenin (DIG)-UTP labeled antisense RNA probes complementary to sim, sog, and lacZ. Each probe used in the individual in situ hybridization is shown on the top of each column. Expression patterns of sim (A-D) were visualized in wild-type (yw) Drosophila embryos. An antisense sog RNA probe was used to target endogenous sog transcripts in both wild-type (yw) (E-H) and sim mutant (sim−/−) (I-L) embryos. The sim mutant embryo was homozygous for the sim2/H9 allele. (M-P) Expression of a lacZ fusion gene directed by a ∼0.4-kb sog primary enhancer in a transgenic embryo recapitulated the endogenous pattern of sog expression in the neurogenic ectoderm and the ventral midline (compare with E-H). ‘st’ indicates the developmental stage of Drosophila embryogenesis. Developmental stages were defined according to previously established criteria (23).
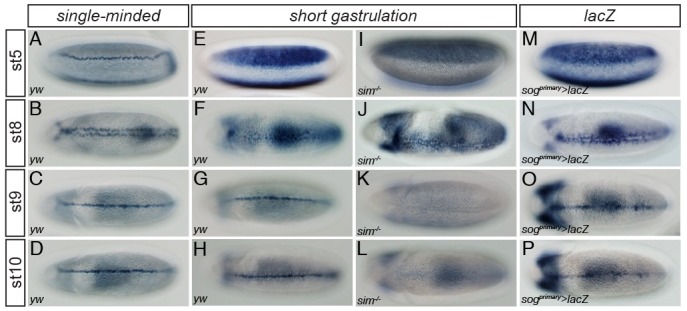
The previous finding that the sog shadow enhancer can direct its midline expression (8) prompted us to test if the sog primary enhancer has similar activity. An approximately 400-bp primary enhancer directed broad stripes of lacZ expression on either side of the neurogenic ectoderm in transgenic embryos (Fig. 1M) (5). The early broad domain of lacZ expression narrowed to the ventral regions of the neurogenic ectoderm during gastrulation (Fig. 1N). From the onset of germ band elongation, lacZ transcripts were observed in the ventral midline (Fig. 1O and P), recapitulating endogenous sim and sog patterns in the ventral midline (compare with Fig. 1C, D, G, and H, respectively). These results suggested that, in addition to the sog shadow enhancer, the primary enhancer also contains ventral midline enhancer activity, and thus that the sog midline expression pattern is determined by both enhancers. The finding that both enhancers can direct both early and late sog expression explains why efforts to identify a sog ventral midline enhancer have long been unsuccessful.
The sog primary enhancer does not contain Sim-binding sites
The loss of sog expression in the ventral midline of a sim mutant embryo together with the ability of the sog primary enhancer to direct lacZ expression in the ventral midline raised the possibility that the primary enhancer contains Sim-binding sites. To test this possibility, ClusterDraw analyses were performed with position frequency matrices (PFMs) (Fig. S1 and S2 in Supplementary Material) for Dl-, Zld-, Sna-, Bcd-, and Sim-binding sites (Fig. 2). ClusterDraw is an r-scan-based program that has been used to identify binding motifs and binding clusters of specific combinations of transcription factors (10). To increase the statistical power of the computational analyses, an identical ClusterDraw analysis was performed twice with two different sets of PFMs for the five transcription factors. One set of PFMs was generated by motif alignments obtained from the in vitro binding data (10) (Fig. 2A) and the other from the in vivo binding data (11) (Fig. 2B). Zld- and Bcd-binding sites were included in those searches because lack of Zld and Bcd proteins led to impaired endogenous sog expression in the neurogenic ectoderm, suggesting that the primary enhancer contains at least one binding site for each transcription factor.
Fig. 2. ClusterDraw analyses across a ∼62-kb genomic region encompassing the sog locus. ClusterDraw analyses were performed with two different types of PFMs for the Dorsal (Dl)-, Zelda (Zld)-, Snail (Sna)-, Bicoid (Bcd)- and Single-minded (Sim)-binding sequences. One was built by motif alignments obtained from in vitro binding data (10) (A) and the other from in vivo binding data (11) (B). Each analysis yielded two statistically significant best cluster P values. Although the patterns of the best cluster P values along the axis of match probability cutoff (−logP) (x axis) differed slightly between the two independent analyses, the two best clusters in each analysis coincided with the primary and shadow enhancers (dotted boxes, Table S6). No Sim-binding sites were identified in the sog primary enhancer. Gene models over the 62-kb genomic region are depicted below panel B. (C) ClusterDraw analyses also indicated the location and cumulative match probability (−logP) value of each motif found in the primary enhancer (Tables S2-S5). The thick line denotes ∼400-bp of the sog primary enhancer (Table S6) (5). Triangles and squares shown above and below the line represent motifs identified in the sense and antisense strands relative to the transcription start sites of the sog gene, respectively.
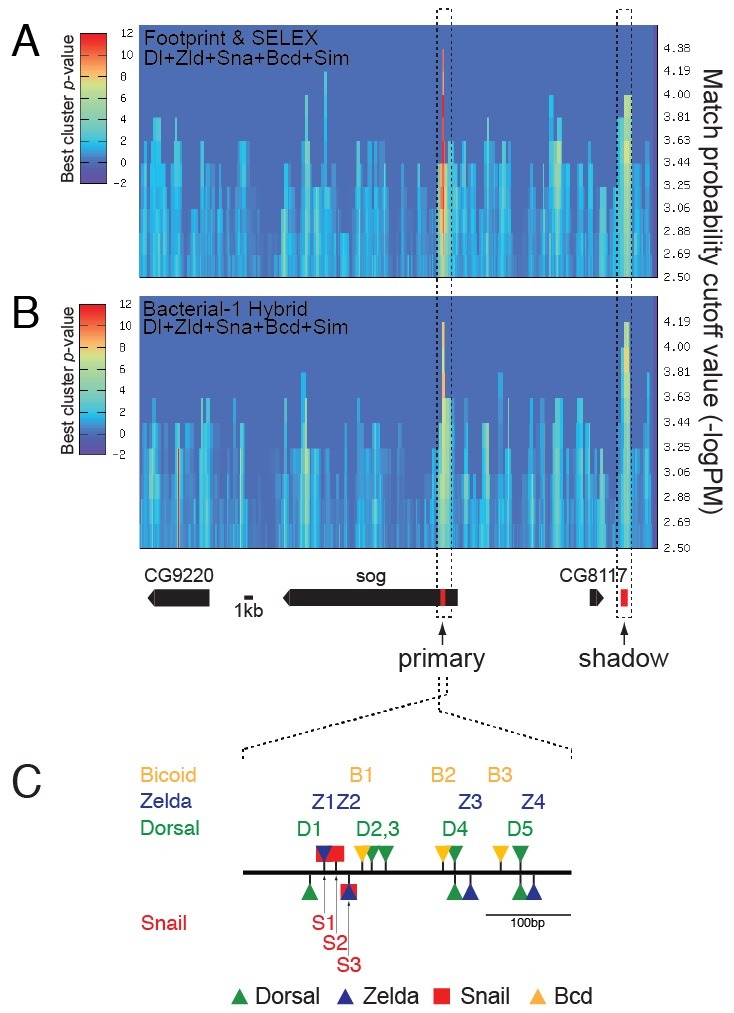
ClusterDraw analyses over a ∼62-kb genomic region encompassing the sog locus identified two clusters repeatedly (Fig. 2A and B). Although the patterns of the best cluster P values along the axis of match probability cutoff (−logP) were slightly different in the two independent analyses, the two best clusters in each analysis coincided with the primary and shadow enhancers (Fig. 2A and B, dotted boxes, Table S6). These results suggest that the primary and shadow enhancers of sog contain the most significant clusters of Dl-, Zld-, Sna-, and Bcd-binding motifs across the ∼62-kb genomic region. The ClusterDraw algorithm also displays the location and quality [cumulative match probability (−logP)] of each binding site found in the clusters that it identifies. Binding sites commonly identified by the two repeated analyses and whose cumulative match probability values were higher than the match probability cutoff value were defined as functional binding motifs (Tables S2-S5, see Supplementary Material for more details). The cluster corresponding to the primary enhancer contained five Dl-, four Zld-, three Sna-, and three Bcd-binding sites, whereas no Sim-binding site was identified in either of the repeated analyses (Fig. 2C). The absence of Sim-binding sites within the ∼400-bp primary enhancer is consistent with the previous observation that the sog shadow enhancer does not include the ‘5-ACGTG-3’ Sim-binding site (8), which has been found in all ventral midline enhancers tested to date. These results strongly suggest that, like the shadow enhancer, the primary enhancer may also function as a HOT region (12) to control sog ventral midline expression.
Early activity of the primary enhancer in the neurogenic ectoderm is required for its late activity in the ventral midline
Recently, it was shown that synergistic interactions between Dl and Zld and between Dl and Bcd in the shadow enhancer play a critical role in generating broad lacZ expression in the neurogenic ectoderm (13). Thus, the close proximities of Dl to both Zld and Bcd in the primary enhancer (Fig. 2C) raised the possibility that the early broad stripes of lacZ expression directed by the primary enhancer (Fig. 3A) also require transcriptional synergy between D1 and Zld or Bcd. To examine this possibility, consensus sequences of Dl-, Zld-, and Bcd-binding sites were changed by site-directed mutagenesis (Fig. 3E, I, M and Table S1). Removal of four Dl-binding sites in the primary enhancer led to complete failure of lacZ expression in the neurogenic ectoderm (Fig. 3E). In addition, loss of either Zld- or Bcd-binding sites resulted in severe reduction in lacZ expression width (compare Fig. 3I and M with A). These lacZ expression patterns are reminiscent of those mediated by the mutant shadow enhancers where synergistic interaction between Dl and either Zld or Bcd is hampered (13). In addition, removal of only the first Zld-binding site (Z1) (Fig. 2C) created a narrow pattern of lacZ expression similar to that mediated by the mutant construct containing no Zld-binding site (Fig. S3). Furthermore, mutation of either the second (B2) or third (B3) Bcd-binding site (Fig. 2C) also resulted in a dramatic reduction in lacZ expression width (Fig. S3). Intriguingly, the remaining narrow lacZ expression patterns directed by constructs containing mutant Zld- and Bcd-binding sites (Z1, B2 and B3) were completely abolished by removal of the linked Dl-binding sites (Fig. S3). These results suggest that transcriptional synergy between Dl and either Zld or Bcd is required for creating the early broad pattern of sog expression throughout the neurogenic ectoderm.
Fig. 3. Intact activity of the sog primary enhancer in the presumptive neurogenic ectoderm is required for its late activity in the ventral midline of the developing Drosophila embryo. Approximately 2-10 h AED embryos were collected, dechorionated, fixed, and hybridized with DIG-UTP labeled antisense lacZ RNA. Binding sites of Dl (green), Zld (blue), Sna (red), and Bcd (yellow) in the sog primary enhancer are depicted on top of each column. Triangles and squares represent binding sites for a transcriptional activator and repressor, respectively. Mutagenized Dl-, Zld-, Sna-, and Bcd-binding sites are marked with asterisks (*). (A-D) The wild-type (WT) sog primary enhancer directs lacZ expression in the early neurogenic ectoderm and the late ventral midline of Drosophila embryo. (E-H) Mutations in four Dl-binding sites abolished lacZ expression both early in the neurogenic ectoderm and late in the ventral midline. (I-L) Lack of Zld-binding sites in the primary enhancer led not only to severe reduction in lacZ expression width in the neurogenic ectoderm, but also to complete loss of lacZ expression in the ventral midline. Note that the remaining lacZ expression also gradually diminished along the anterior-posterior (AP) axis (I). (M-P) Removal of Bcd-binding sites in the primary enhancer produced similar lacZ expression patterns to those of the primary enhancer containing no Zld-binding site, except that narrow lacZ expression gradually increased from the anterior to posterior pole. This lacZ pattern appears to be a mirror image of that produced by the △Zld1234 construct (compare M with I).
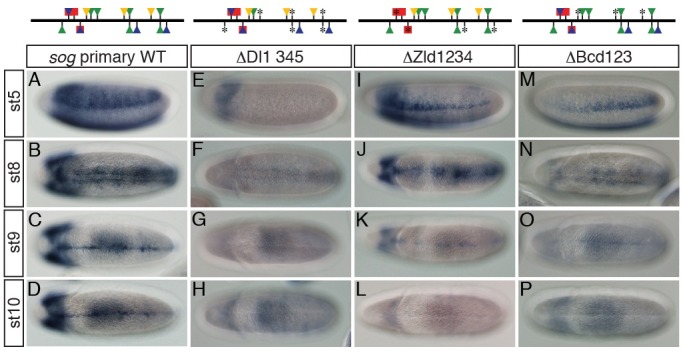
One of the most intriguing features of the shadow enhancer is that linked Dl- and Zld-binding sites are required for its late ventral midline enhancer activity (8). Lack of those linked sites in the shadow enhancer results in catastrophic reduction in late enhancer activity in the ventral midline. Because sog expression is controlled by primary and shadow enhancers at the same time and in a identical space (7), we reasoned that the late activity of the primary enhancer may depend on early enhancer activity. To test this hypothesis, lacZ expression patterns directed by the wild-type and mutant versions of the primary enhancers in the ventral midline were investigated. During late gastrulation, residual lacZ transcripts were still observed in wild-type and mutant embryos (Fig. 3B, F, J, and N). However, after gastrulation, none of the mutant primary enhancers activated lacZ in the ventral midline (compare Fig. 3G, H, K, L, O, and P with C and D). In contrast to the Dl mutant enhancer (Fig. 3E), the primary enhancers containing either mutant Zld or Bcd binding sites could still direct lacZ expression, even though there was severe reduction in lacZ expression width and strength. Nevertheless, the early impaired lacZ expression in the neurogenic ectoderm led to serious failure of late lacZ expression in the ventral midline.
DISCUSSION
Almost all of the developmental enhancers tested so far only control the transcription of their target genes at a particular time and in a defined space (14). However, most genes involved in development are repeatedly used at different times and in diverse spaces during an entire process of differentiation (15). Thus, each versatile gene needs various developmental enhancers that switch its transcription on and off at the correct time and in the proper location (16). Expression of rhomboid (rho) that is regulated by at least two discrete enhancers is one good example (17). rho, a DV patterning gene involved in the development of the central nervous system (CNS), is expressed in the neurogenic ectoderm and the ventral midline of the developing Drosophila embryo. The sequential expression in these two different locations is directed by two separate enhancers, the neurogenic ectoderm enhancer (NEE) and the ventral midline enhancer (VME). Although these enhancers are located close to each other, they are functionally autonomous and geographically separable. In contrast to this property of the rho enhancers, however, recent studies performed with transgenic embryos showed that the sog shadow enhancer can also direct sog expression in the ventral midline as well as in the presumptive neurogenic ectoderm (18). As far as we know, the sog shadow enhancer is the first developmental enhancer reported to direct its target gene expression at two different times and in two discrete spaces during Drosophila embryogenesis. The current study presents evidence that the sog primary enhancer shares three functional similarities with the shadow enhancer. First, the primary enhancer is also able to direct sog expression in the ventral midline of a developing embryo (Fig. 1). All of the ventral midline genes that have been examined so far have expression that is controlled by the ventral midline enhancers that are not able to allow expression of midline genes in locations other than the ventral midline in a developing embryo. Second, like the sog shadow enhancer, the primary enhancer also does not have a canonical Sim-binding site (Fig. 2 and Table S6). All of the ventral midline enhancers have been shown to have at least one Sim-binding consensus sequence containing a ‘5’-ACGTG-3’ core motif (19). However, both the primary and shadow sog enhancer do not have such a Sim-binding site (Table S6). These findings suggest that the two sog enhancers may direct ventral midline expression with an unknown mechanism to bypass involvement of Sim. This may be the reason why intense efforts to identify the sog midline enhancer with the canonical Sim-binding consensus sequences have not been successful. Third, the midline enhancer activity of the primary enhancer also requires its neurogenic ectoderm activity (Fig. 3). The early broad sog expression in the neurogenic ectoderm depends on transcriptional synergy between Dl, Zld and Bcd in the two sog enhancers (Fig. 3 and S3) (13). The impaired synergistic interaction between them led to severe reduction in the early neurogenic ectoderm enhancer activity, which in turn abolished the late midline enhancer activity in both enhancers (Fig. 3) (8). It is conceivable that in contrast to the two separate rho enhancers, the two enhancer activities embedded in the primary enhancer are functionally consolidated and geographically inseparable.
The early and late enhancer activities of the sog primary are determined by independent transcription factors in the neurogenic ectoderm and the ventral part of the developing embryo, respectively. For example, early and late sog expression was directed by Dl, Zld, and Sna in the neurogenic ectoderm and Sim in the ventral midline, respectively. Although their transcriptional activities are not coincidental in time or space during embryogenesis, changes in early transcriptional input of the enhancers catastrophically interfered with their late transcriptional outcome. The simplest interpretation for this paradoxical observation is that the early determinants may potentiate the enhancers for the subsequent transcriptional burst by late transcription factors. For example, the preceding interaction between the early transcription factors and enhancers may make the chromatin structure more accessible for the late factors. Indeed, the maternal transcription factor Zld, which is involved in creating the broad sog expression in the neurogenic ectoderm (Fig. 3I), functions as a pioneer factor to increase chromatic accessibility (20). A pioneer factor is a transcription factor that confers transcriptional competency to inactive target enhancers by binding condensed chromatin prior to the binding of other transcription factors (21). It is plausible that early binding of Zld to the primary enhancer establishes an open chromatin environment in the neurogenic ectoderm and causes transcriptional synergy between Dl and Zld by facilitating their cooperative site occupancy. However, the functional dependency of the late enhancer activity on the early one observed in the primary enhancer does not seem to rely entirely on the pioneering activity of Zld, because Zld still binds the primary enhancer containing either mutant Dl- or Bcd-binding sites (Fig. 3). It is possible that there may exist unidentified pioneering factors that prime the developmental enhancers in addition to the zinc-finger transcription factor Zld. Pioneering activity has mostly been observed in transcription factors containing a forkhead box (FOX) or zinc-finger domain (22), which strongly supports the possibility that the fly genome encodes more than one pioneering factor involved in the regulation of early DV gene expression.
MATERIALS AND METHODS
Detailed information is provided in the online Supplementary Material.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (MSIP and MOE) (NRF-2010-0002792 and NRF-2012R1A1 A2038502).
References
- 1.Francois V, Solloway M, O'Neill JW, Emery J, Bier E. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. (1994);8:2602–2616. doi: 10.1101/gad.8.21.2602. [DOI] [PubMed] [Google Scholar]
- 2.Wieschaus E, Nüsslein-Volhard C, Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster III. Zygotic loci on the X-chromosome and fourth chromosome. Roux's Arch Dev Biol. (1984);193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 3.Menne TV, Luer K, Technau GM, Klambt C. CNS midline cells in Drosophila induce the differentiation of lateral neural cells. Development. (1997);124:4949–4958. doi: 10.1242/dev.124.24.4949. [DOI] [PubMed] [Google Scholar]
- 4.Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. (2005);9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. (2002);99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeitlinger J, Zinzen RP, Stark A, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. (2007);21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. (2008);321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin DH, Hong JW. Midline enhancer activity of the short gastrulation shadow enhancer is characterized by three unusual features for cis-regulatory DNA. BMB Rep. (2015);48:589–594. doi: 10.5483/BMBRep.2015.48.10.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nambu JR, Franks RG, Hu S, Crews ST. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell. (1990);63:63–75. doi: 10.1016/0092-8674(90)90288-P. [DOI] [PubMed] [Google Scholar]
- 10.Papatsenko D. ClusterDraw web server: a tool to identify and visualize clusters of binding motifs for transcription factors. Bioinformatics. (2007);23:1032–1034. doi: 10.1093/bioinformatics/btm047. [DOI] [PubMed] [Google Scholar]
- 11.Zhu LJ, Christensen RG, Kazemian M, et al. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. (2011);39:D111–D117. doi: 10.1093/nar/gkq858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kvon EZ, Stampfel G, Yanez-Cuna JO, Dickson BJ, Stark A. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. (2012);26:908–913. doi: 10.1101/gad.188052.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin DH, Hong JW. The short gastrulation shadow enhancer employs dual modes of transcriptional synergy. Int J Dev Biol. (2016) doi: 10.1387/ijdb.160165jh. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. (2010);20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Placzek M, Briscoe J. The floor plate: multiple cells, multiple signals. Nat Rev Neurosci. (2005);6:230–240. doi: 10.1038/nrn1628. [DOI] [PubMed] [Google Scholar]
- 16.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. (2006);133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 17.Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. (1992);6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 18.Shin DH, Hong JW. The shadow enhancer of short gastrulation also directs its expression in the ventral midline of the Drosophila embryo. Genes Genom. (2015);37:743–750. doi: 10.1007/s13258-015-0302-5. [DOI] [Google Scholar]
- 19.Hong JW, Park KW, Levine MS. Temporal regulation of single-minded target genes in the ventral midline of the Drosophila central nervous system. Dev Biol. (2013);380:335–343. doi: 10.1016/j.ydbio.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Foo SM, Sun Y, Lim B, et al. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol. (2014);24:1341–1346. doi: 10.1016/j.cub.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. (2011);25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. (2011);27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer-Verlag; Berlin; Heidelberg: (1985). [Google Scholar]


