Abstract
In the past 10 years, autophagy has emerged as a crucial regulator of T-cell homeostasis, activation and differentiation. Through the ability to adjust the cell’s proteome in response to different stimuli, different forms of autophagy have been shown to control T-cell homeostasis and survival. Autophagic processes can also determine the magnitude of the T-cell response to TCR engagement, by regulating the cellular levels of specific signaling intermediates and modulating the metabolic output in activated T-cells. In this review we will examine the mechanisms that control autophagy activity in T-cells, such as ROS signaling and signaling through common gamma-chain cytokine receptors, and the different aspect of T-cell biology, including, T-cell survival, effector cell function and generation of memory, which can be regulated by autophagy.
Keywords: T-cell, Autophagy, Macroautophagy, Chaperone-mediated Autophagy, Metabolism
Introduction
Autophagy is a highly conserved catabolic cellular process that involves the degradation of intracellular material, which may range from soluble macromolecules to whole organelles, in the cell’s lysosomes. Controlled degradation is crucial to maintaining cell homeostasis by eliminating damaged or non-functional components and recycling the products of that degradation (reviewed in [1]). However, quality control is just one of many functions regulated by autophagy. Autophagy also constitutes an important source of fuel, which can help cells cope with different forms of metabolic stress, including starvation (reviewed in [2]). Autophagy also allows cells to adapt to other stresses, including oxidative stress, hypoxia or ER stress; and may act as a pro-survival mechanism (reviewed in [3]). Furthermore, changes in the proteome which are required to respond to specific stimuli or to move forward in developmental or differentiation pathways, are in many cases also regulated by autophagy (reviewed in [4]).
The material meant to be degraded in autophagy can be delivered to the lysosomes through different mechanisms, which define the three major forms of autophagy in mammalian cells: macroautophagy (generally referred to as autophagy), microautophagy and chaperone-mediated autophagy. Macroautophagy involves the de novo formation of double membrane vesicles, termed autophagosomes, which fuse with lysosomes to form autolysosomes and degrade their cargo (reviewed in [5, 6]). Recognition of different forms of cargo, which are mediated in many cases by specific mechanisms, define specific forms of macroautophagy; some examples include mitophagy – involving the degradation of mitochondria – [7], lipophagy – defined by the degradation of lipid droplets by autophagy –[8] or xenophagy – characterized by the incorporation of pathogens into autophagosomes [9]. Microautophagy involves the incorporation of cytosolic cargo into the lysosome through invaginations in the lysosomal membrane. Better characterized in yeast [10], microautophagy also occurs in mammalian cells, though, at least in murine dendritic cells, this process takes place in late endosomes and has been termed endosomal microautophagy [11]. Finally, chaperone-mediated autophagy (CMA) represents a selective form of autophagy in which cytosolic proteins are transported to the lysosome by a cytosolic chaperone, and translocated into the lysosomal lumen for degradation (reviewed in [12]).
A comprehensive characterization of the mechanisms that control the sequential steps that regulate the different forms of autophagy, and the development of specific tools to detect and quantitate autophagy activity in tissues and cells, have led to the identification of new tissue-specific functions of autophagy. In the past decade, exciting studies have begun to define the involvement of autophagy in many of the key functions of the immune system, including, among others, pathogen clearance, regulation of cell activation and differentiation, or antigen presentation (reviewed in [13]). Conclusive evidence that autophagy could act as an innate defense system for intracellular pathogens was presented in groundbreaking studies, which showed that clearance of intracellular bacteria by non-phagocytic and phagocytic cells was dependent on macroautophagy [9, 14]. These observations were extended to include destruction of viruses and parasites as targets of macroautophagy [15, 16]. It was later described that macroautophagy could also promote cross-presentation of antigens on MHC molecules, allowing for the loading of peptides generated from cytosolic proteins or intracellular pathogens into class II molecules, further implicating autophagy in the regulation of the immune response [17]. Several excellent reviews have been recently published covering the different roles that autophagy plays in the innate and the adaptive immune response [18, 19] and we will not cover most of those topics here. Instead this review will focus on our current knowledge of how different forms of autophagy control specific aspects of T-cell biology.
Regulation of macroautophagy in T-cells
It was not until 10 years ago that the first series of studies describing a physiological role for macroautophagy in T-cells were reported [20, 21]. Although previous studies had already shown that macroautophagy might be activated in HIV-infected CD4+ T-cells [22] or in vitro-aged human CD8+ T-cells [23], it was subsequent work conducted in macroautophagy-deficient genetic models which showed that macroautophagy constituted a prominent mechanism through which T-cells preserved homeostasis and adapted to changes in the intracellular or extracellular environment [20, 21, 24–26]. In mice, ATG5 or ATG7 deficiency was shown not to affect differentiation of CD4+ and CD8+ T-cells per se, but to reduce their numbers in both the thymus and the periphery [21, 24]. Similar results were later reported in studies that analyzed mice carrying Beclin1- or Vps34-deficient T-cells, which confirmed that loss of autophagy affected mature peripheral T-cell homeostasis and survival [27, 28].
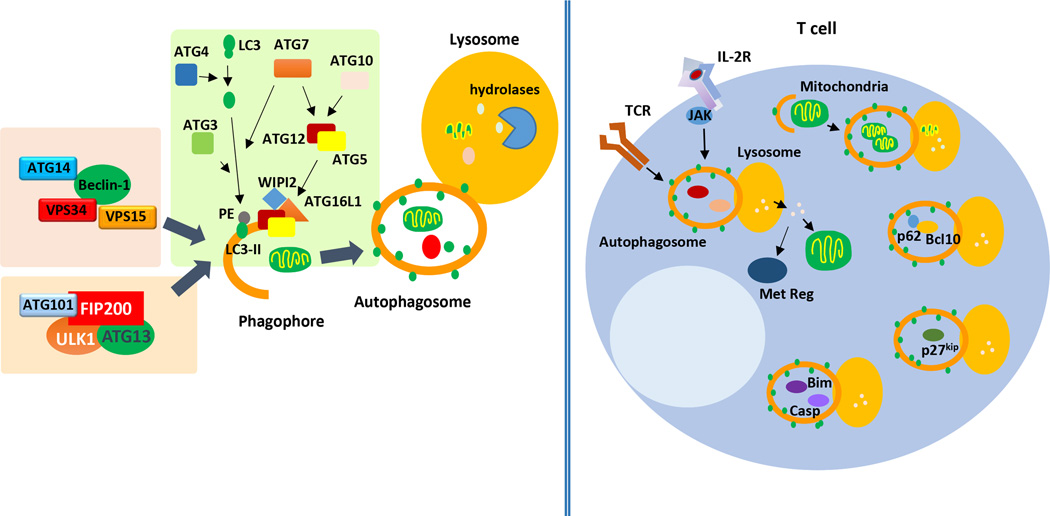
Macroautophagy is primed by the formation of an isolation membrane. This structure elongates, trapping cytosolic content, and ultimately seals to form an autophagosome, (Figure 1). Two major regulatory complexes control the induction of macroautophagy. The UNC-51-like kinases 1 and 2 (ULK1 and ULK2), form part of a complex that contains also the focal adhesion kinase family interacting protein of 200 kD (FIP200), the autophagy related protein (ATG) 101 and ATG13. This complex is required to induce the initial generation of the autophagic vesicle membrane (i.e. autophagosome nucleation), an early stage in autophagosome formation [29] (Figure 1). Activated by the ULK complex, the class III phosphatidylinositol-3-kinase complex (PI3KC3), containing vacuolar protein sorting 34 (Vps34), Vps15, ATG14 and Beclin1, then mediates the recruitment of other ATG proteins to the autophagosome precursor or phagophore to induce elongation [30, 31].
Figure 1.
Macroautophagy in T-cells. Two upstream regulatory complexes: the tetrameric complex formed by ULK1, FIP200, ATG13 and ATG101; and the complex formed by Beclin1, Vps34, Vps15 and ATG14, regulate the initiation of the formation of the autophagosome form the pre-autophagosomal structure or phagophore. Elongation of a double membrane required to form the autophagosome occurs thanks to the activation of two conjugation cascades which are regulated by the E1-like protein ATG7. Processing of LC3 by ATG4 allows for the E2-like protein ATG3 to mediate the conjugation of phosphatidylethanolamine (PE) and LC3 to form LC3-II that integrates into the autophagosomal membrane. ATG7 and the E2-like ATG10 proteins regulate the formation of ATG5/ATG12 conjugates, which are recruited to the autophagosomal membrane by WIPI and ATG16L and also contribute to the elongation of the limiting membrane. Sealing of the elongating autophagosomal to membrane leads to the formation of the autophagosome: a double membrane vesicle that contains the cargo. The autophagosome will fuse with the lysosome, where lysosomal hydrolases will degrade the cargo. In T-cells macroautophagy is activated in response to TCR or IL-2 receptor signaling has been shown to: participate in organelle homeostasis (especially through regulated mitophagy), regulate T-cell proliferation and survival (through the degradation of p27Kip1, Bim and caspases 3 and 8), modulate TCR-induced NFκB activation (though p62/SQSTM1-mediated targeted degradation of Bcl10), and to control T-cell activation, Treg fitness and the generation of CD8+ T-cell memory through the regulation of the T-cell's metabolism (by providing fuel substrates and/or modulating the activity of metabolic regulators).
Elongation of the autophagosomal membrane involves at least two well-known conjugation systems, which function in a similar way as the ubiquitin conjugation system, and are evolutionarily conserved among eukaryotes. The first system conjugates the cytosolic microtubule-associated protein light chain 3 (LC3-I) to phosphatydilethanolamine to generate LC3-II, which incorporates into the autophagosomal membrane and contributes to the subsequent autophagosome expansion and maturation [32]. A cascade of reactions involving the ATG proteins, ATG4, ATG7 and ATG3, is necessary for LC3 processing and conjugation [33]. After the cysteine-protease ATG4 exposes a glycine residue in the LC3 C-terminus, the sequential activities of the E1-like ATG7 and E2-like ATG3 proteins induce the conjugation of LC3 and phosphatydilethanolamine [33] (Figure 1). The dynamic process of LC3-II conjugation and degradation is commonly used as a reliable marker of macroautophagy activity [34, 35]. A second cascade also contributing to autophagosomal membrane elongation involves the E1-like ATG7 and the E2-like ATG10-proteins, which mediate the conjugation of ATG12 to ATG5. ATG5-ATG12 conjugates form a complex with ATG16L, recruited to the pre-autophagosomal membrane by the phosphatidylinositol-3-phosphate-binding protein WIPI [36] (Figure 1). Both conjugation systems require the activity of the ATG7, which represents a regulatory crosstalk between pathways involved in autophagosome elongation [37]. Overall, the current model of macroautophagy regulation suggest that this process is not linear and that the integration of multiple pathways usually takes place to modulate macroautophagy [38].
Several studies have shown that TCR engagement in CD4+ and CD8+ T-cells induces macroautophagy [20, 24, 39, 40]. Full activation of T-cells requires TCR engagement together with costimulatory molecules and cytokine signaling. mTOR is a key molecule activated downstream of these signaling pathways and is indispensable for the fate of the effector T-cell, playing a major role notably in regulating metabolism and protein translation (reviewed in [41]). Inhibition of mTOR signaling by rapamycin during T-cell activation was shown to not only result in defective cellular activation but to also induce anergy [42]. Therefore, T-cell activation introduces a paradigm of concomitant activation of mTOR, a negative regulator of macroautophagy [43], together with macroautophagy induction in human and murine T-cells [39, 40]. A recent study has identified that TNF-α-induced protein 3 (TNFAIP3) restricts mTOR activity and promotes macroautophagy and survival in CD4+ T-cells [44]. However, the fact that activated T-cells activate mTOR and upregulate macroautophagy, and that inhibition of mTOR with rapamycin has little effect on activation-induced macroautophagy in CD4+ T-cells, supports that induction of macroautophagy in response to TCR engagement is likely induced in an mTOR-independent manner [40]. Macroautophagy induction might instead involve Jun kinase (JNK), which in human epithelial cell lines has been shown to phosphorylate B-cell lymphoma 2 (Bcl-2) and cause its dissociation from Beclin1, which becomes then able to interact with Vps34-Vps15-ATG14 in the PI3KC3 complex to induce autophagosome formation [45] (Figure 1). In fact, knocking-down JNK1 or JNK2 in Th1 cells has been shown to reduce the number of LC3-positive vesicles [20]. In line with these results, at least two proteins that form part of the PI3KC3 complex are required for LC3 conjugation in CD4+ T-cells: Vps34- as well as Beclin1-deficient T-cells show reduced macroautophagy activity following TCR and CD28 stimulation [28, 40, 46].
Cytokine signaling plays a major role in the regulation of T-cell function in both naive and effector T-cells. Our laboratory has shown that signaling through common gamma-chain cytokine receptors, such as the IL-2 receptor, induces macroautophagy in CD4+ T-cells; and that this pathway is required to maintain a high macroautophagy flux in activated T helper cells [40]. In CD4+ T-cells, IL-2 is secreted early after TCR engagement and is crucial for sustained proliferation and survival, as well as for effector differentiation and memory programming. It is thus likely that the positive effects that signaling through the IL-2 receptor and other common gamma-chain cytokines has on determining the fate of effector and memory T-cells may stem from their participation in the regulation of macroautophagy. These results highlight the emerging role of cytokines and the JAK-STAT pathway in the regulation of macroautophagy [47, 48].
Regulation of ATG protein expression has also been reported in T-cells. Following TCR engagement, LC3 expression is upregulated in T helper cells through a yet unknown mechanism that appears to act at a posttranscriptional level [40]. Furthermore, the Beclin1 promoter is targeted by NFκB to induce Beclin1 expression in activated Jurkat cells [49]. How changes in the expression of ATG proteins may affect the overall regulation of macroautophagy in T-cells remains to be determined.
Macroautophagy and organelle homeostasis in T-cells
There is mounting evidence that supports that macroautophagy plays an essential role in maintaining organelle homeostasis in T-cells. The reduction in mitochondrial content that occurs in T-cells as they differentiate from an early thymic emigrant to mature peripheral T-cells is controlled by macroautophagy. Consequently, inhibition of macroautophagy in mouse T-cells leads to defective mitochondria turnover, which results in increased ROS generation and altered levels of apoptotic proteins [26, 50]. Accumulation of ER and altered calcium mobilization have also been reported in ATG7-deficient mouse T-cells [51]. Similar defects in mitochondria and ER homeostasis have been confirmed in T-cells lacking ATG3 or Vps34 [25, 46]. Interestingly, aged mice bearing Vps34-deficient T-cell develop an inflammatory syndrome that is likely a consequence of defective Treg function, indicating that macroautophagy is also important for the regulation of this critically important T-cell population for the maintenance of immune homeostasis [46, 52].
Macroautophagy and T-cell survival
Macroautophagy regulates T-cell survival at different levels. Dysregulated organelle accumulation in the cytoplasm may act as an inducer of cell death in macroautophagy-deficient T-cells, possibly as a consequence of increased generation of ROS caused by impaired mitophagy [26, 50, 53]. However, the involvement of macroautophagy in the regulation of apoptosis goes beyond mitochondrial homeostasis, as shown by the fact that Beclin-1 deficient T-cells present increased susceptibility to apoptosis, at least in part, caused by the accumulation of the proapoptotic proteins Bim and caspases 3 and 8 [27]. These results support that the cellular levels of specific pro-apoptotic proteins might be regulated by their rate of degradation through macroautophagy [27]. Interestingly, Vps34-deficient T-cells show disrupted recycling of the alpha chain of the IL-7 receptor, though this defect might be independent of the loss of macroautophagy in those cells [54]. The reduced numbers of T-cells that are observed in mice deficient in Vps34 or ATG proteins results probably from altered regulation of T-cell survival and apoptosis in the absence of macroautophagy [21, 28, 46].
Macroautophagy in the modulation of T-cell metabolism
Following TCR engagement, CD4+ T-cells increase autophagosome formation and degradation, and both ATG5- and ATG7-deficient T-cells show impaired proliferation in response stimulation [21, 24]. The mechanisms that underlie this effect have not been fully defined. ATG7-deficient naïve CD4+ and effector Th1 cells, or cells activated in the presence of either PI3KC3 inhibitors or lysosomal hydrolases inhibitors, show reduced proliferation and cytokine production following TCR and CD28 engagement, which may be a consequence of their inability to generate an efficient energetic output [24]. Macroautophagy-deficient mouse CD4+ T-cells show decreased activation-induced ATP production, which is restored when a cell-permeable substrate able to fuel oxidative phosphorylation is provided [24]. Interestingly, a shift in the nature of the autophagosome cargo occurs in activated effector CD4+ T-cells, which changes from being mainly comprised of organelles in naïve cells, to preferentially excluding organelles following activation [24]. These changes in autophagosome cargo might be important in supporting the ability of macroautophagy to provide substrates required to meet an increased energy demand, while preserving mitochondrial content during activation. The ability of macroautophagy to regulate T-cell metabolism has also been recently reported in memory CD8+ T-cells and Treg. Cells unable to induce macroautophagy show changes in their metabolic profiles when compared with their wild-type counterparts, which in Treg respond to increased c-Myc-induced glycolysis [52, 55]. Consequently, mice bearing macroautophagy-incompetent CD8+ T-cells generate deficient CD8+ T-cell memory, while those where deletion of Atg7 or Atg5 occurs in Foxp3+ T-cells show decreased Treg stability and survival [52, 55]. Interestingly, T-cells lacking Rab7, a small GTPase that promotes fusion between autophagosomes and lysosomes [56], also have a reduced ability to proliferate upon TCR and CD28 engagement [57]. Rab-7 deficient T-cells should still be able to form autophagosomes, though they would fail to effectively fuse with lysosomes and degrade cargo. These data indicate, then, that not only sequestering of cargo into autophagosomes, but also its degradation, are important to support T-cell activation.
Macroautophagy in the regulation of TCR signaling
Though incorporation of cargo into the autophagosome was initially thought to be a merely non-specific in-bulk process, emerging evidence has shown that cargo recognition proteins, such as p62/Sequestosome 1 (SQSTM1) or Neurabin 1 (NRB1), allow macroautophagy to target distinct types of cargo (from selected proteins to specific organelles) for destruction [58]. The ability of macroautophagy to be selective has been recently shown to mediate regulation of specific signaling pathways activated by TCR engagement. In effector Th2 cells and central memory CD8+ T-cells, SQSTM1-mediated targeted degradation of Bcl10 through macroautophagy modulates NF-κB activation downstream of the TCR [59]. Macroautophagy has also been shown to target the cell cycle inhibitor p27Kip1 for degradation in activated T-cells [60], which may account for the reduced activation-induced cell proliferation that has been reported in macroautophagy-deficient T-cells [21, 24]. Interestingly, in the context of HIV infection, selective autophagy in human CD4+ T-cells can act as a protective mechanism by actively inducing the degradation of the viral transactivator protein Tat [61].
Collectively, those studies support that macroautophagy regulates peripheral T-cell activation following in vitro TCR stimulation. In vivo, studies performed using viral infection models in mice with Atg7-deficient CD8+ T-cells, have shown that those mice can mount effective primary anti-viral responses to LCMV, MCMV or Influenza virus, though a small decrease in the magnitude of the expansion phase can still be observed [55, 62]. Interestingly, in an adoptive transfer model using CD8+ T cells in which Atg5 is acutely deleted using a tamoxifen-induced Cre recombinase expression system, infection with influenza virus results in a significant decrease in the magnitude of the primary CD8+ T-cell expansion [63]. This decreased CD8+ T-cell response is however associated with increased apoptosis, which is induced in a ROS and p53-dependent manner [63].
Macroautophagy in the generation of CD8+ T-cell memory
As discussed above, recent studies analyzing CD8+ T-cell responses to viral infections indicate that naïve CD8+ T-cells might not be dependent on macroautophagy for clonal expansion during the acute phase of a viral infection, as the deletion of Atg genes does not significantly limit primary CD8+ T-cell response [55, 62]. Interestingly, it is just before the contraction phase, when cells stop dividing and the pathogen has been cleared, when CD8+ T-cells appear to upregulate macroautophagy, which becomes essential for the efficient generation of memory T-cells [55]. Consequently, macroautophagy-deficient CD8+ T-cells show diminished survival during the contraction phase, which results in a defective generation of memory [55, 62]. This result may respond to the involvement of macroautophagy in the regulation of the memory T-cells’ metabolism, which depend on oxidative phosphorylation much more than activated naïve cells [55]. Interestingly, during MCMV infection, administration of spermidine enhances macroautophagy activation in CD8+ T-cells in old mice, which potentiates the generation of CD8+ T-cell memory in those mice [62]. Taken together, these findings support that modulation of macroautophagy might be used to boost T-cell memory generation and improve vaccine efficacy, anti-pathogen response or even T-cell-based immunotherapy.
Chaperone-mediated autophagy in T-cells
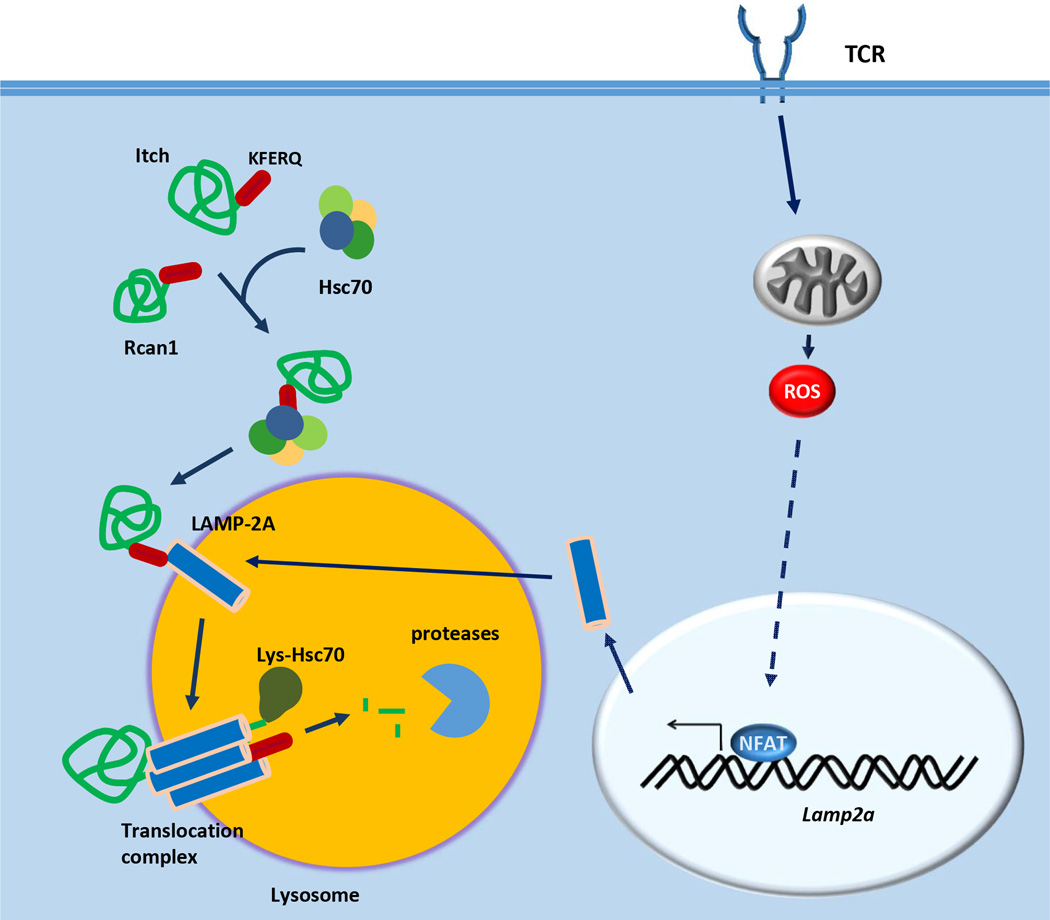
The remarkable specificity of CMA for specific cytosolic proteins is achieved by the cooperative activity of the molecules LAMP-2A and Hsc70 [64, 65]. The CMA-targeted pool of protein substrates, bear one or more consensus motifs biochemically related to the pentapeptide KERFQ, which is recognized by Hsc70 within a particular cytosolic chaperone complex (reviewed in [66], Figure 2). This chaperone complex brings the substrates to the lysosomal membrane, where they are unfolded and transported into the lysosomal lumen for degradation by a translocation complex, which assembles as a multimer of LAMP-2A [67] (Figure 2). The LAMP-2A protein is just one of the three splicing alternative mRNA forms of the Lamp2 gene, which also generates LAMP-2B and LAMP-2C; however only LAMP-2A is involved in CMA [68]. Regulation of the levels of LAMP-2A at the lysosomal membrane usually occurs via modulation of its lysosomal distribution and turnover [69]. However, stressors, such as oxidative stress, may induce transcriptional upregulation of Lamp2a expression, to increase the presence of LAMP-2A at the lysosomal membrane [70].
Figure 2.
Chaperone-mediated autophagy in T-cells. In CD4+ T-cells, TCR engagement induces ROS generation, which in turn leads to the activation of NFAT. NFAT induces expression of the lysosomal protein LAMP-2A. Recognition of exposed KFERQ motifs in cytosolic proteins such as Itch and Rcan1 by an Hsc70-containg chaperone complex translocates those proteins through the lysosomal membrane. Interaction of the Hsc70-substrate complex with LAMP-2A induces the formation of a multimeric translocation complex that, with the help of an intralysosomal form of Hsc70, will unfold and transport the substrate to the lysosomal lumen, where it is degraded by lysosomal proteases.
Our laboratory has recently has recently shown that CMA regulates CD4+ T-cell responses through the targeted degradation of negative regulators of TCR signaling, such as Rcan1 and Itch [71]. CMA is activated in response to engagement of the TCR, which induces upregulation of the transcription of Lamp2a in both naive and effector human and mouse CD4+ T-cells [71] (Figure 2). ROS generated in activated T-cells controls calcium signaling [72, 73] and are essential for the induction of the NFAT-dependent expression of Lamp2a [71] (Figure 2). The relevance of CMA for CD4+ T-cell activation is supported by the finding that specific inhibitors of TCR signaling, including the E3 ubiquitin ligase Itch and the calcineurin inhibitor Rcan1, are selectively degraded by CMA [71] (Figure 2). Accordingly, T-cells that lack LAMP-2A, and are therefore CMA-deficient, show a marked reduction in activation-induced proliferation and cytokine secretion; whereas mice bearing LAMP-2A-deficient T-cells respond poorly to immunization and infection with Listeria [71].
Further research will be required to characterize the full breadth of processes that might be controlled by CMA in T-cells. For instance, as reported for macroautophagy, CMA can also contribute to the regulation of cellular energetics, due to its capacity to modulate the levels of key metabolic enzymes, including pyruvate kinase or glyceraldehyde-3-phosphate dehydrogenase [74]. Whether this also occurs in T-cells and how CMA and macroautophagy may coordinate common activities, remain to be determined.
Autophagy and autoimmune disease
Dysfunction of autophagy has been associated with a number of inflammatory diseases (reviewed in [19, 75]). Different mechanisms have been proposed to account for those associations. For instance, basal macroautophagy suppresses inflammasome activation and prevents excessive tissue-damage by directly degrading NACHT, LRR and PYD domains-containing protein 3 (NALP3), or clearing NALP3 agonists, such as ROS, through mitophagy [76, 77]. Consequently, cells with defective macroautophagy have hyperactivation of the inflammasome and increased secretion of inflammatory cytokines [77–79].
The link between macroautophagy and inflammation is, however, not limited to the regulation of the innate inflammatory response, as it also extends to the direct control of T-cell-mediated immunity. Given the important roles of macroautophagy in cross-presentation of antigens [80], in shaping the T-cell repertoire through modulation of thymocyte selection [81] and in altering the nature of the peptides presented by dendritic cells [82], macroautophagy may contribute to the maintenance of immune tolerance. In the experimental autoimmune encephalomyelitis mouse model of multiple sclerosis, macroautophagy has been shown to degrade proapoptotic proteins in T-cells and to function as a prosurvival mechanism, allowing the rapid expansion of autoreactive T-cells [27]. Furthermore, in patients suffering from multiple sclerosis, an increase in the expression of ATG5 in T-cells has been described at inflammatory sites [83]. A similar scenario occurs in systemic lupus erythematosus, where both mouse models and human patients exhibit higher autophagic activity in T-cells, which promotes aberrant activation and extended T-cell survival, which translates into persistent inflammation (reviewed in [84]).
On the other hand, a hypoautophagic phenotype in T-cells has been linked to. T-cells from patients with rheumatoid arthritis have lower levels of the glycolytic enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, which not only prevents efficient utilization of glucose, but also limits macroautophagy activity, increasing susceptibility to apoptosis. [85].
As mentioned before, analysis of mice lacking Vsp34 exclusively in T-cells have shown that these mice develop intestinal inflammation which correlates with impaired Treg function [46], while macroautophagy has recently been reported to maintain Treg stability and function [52]. These studies indicate that regulation of Treg function might constitute another mechanism through which macroautophagy may contribute to the maintenance of immune tolerance.
Autophagy and T-cell immunosenescence
Age is the single largest risk factor for many major diseases in humans. As with other tissues and organs, the immune system’s function also declines with age. The age-associated changes in the T-cell compartment are primarily responsible for the defective adaptive immune responses that are characteristic of the elderly, and result in decreased efficiency of vaccination, increased severity of infectious diseases and diminished immune surveillance [86, 87].
Presently, there is increasing evidence of a strong connection between autophagy and aging, and a clear decrease in the activity of different forms of autophagy is observed with age [88]. Moreover, experimental evidence supports that restoring autophagic function in old or aged organisms leads to improved tissue homeostasis and enhanced function [89]. The immune system is not different from other tissues in this respect, and age-associated dysfunction of autophagy might also condition the magnitude and quality of the immune response in the elderly (reviewed in [90]). In the T-cell compartment, decreased levels of basal macroautophagy have been observed on CD8+ T-cells isolated from old healthy humans [91], while enhancing macroautophagy activity causes a more robust generation of CD8+ T-cell memory in mice in response to viral infection [62]. Activation-induced CMA activity is also diminished with age in mouse and human CD4+ T-cells and restoration of CMA activity in old mouse CD4+ T-cells improves proliferation and cytokine production in response to TCR engagement [71].
A deeper understanding of the biology behind the loss of autophagy associated with age will be essential for the development of therapeutic interventions that may allow for the modulation of autophagy in T-cells. Approaches aimed at restoring autophagy activity in T-cells from older individuals might prove powerful tools to boost T-cell function and improve responses to vaccination and pathogens in the elderly.
Conclusions
In T-cells, autophagy has emerged as an important regulatory mechanism that controls numerous aspects of T-cell biology. Whereas basal macroautophagy is essential to maintain T-cell homeostasis, activation-induced macroautophagy has been characterized as a key regulator of TCR-induced cell proliferation and metabolism, and is key for the generation of effector CD8+ T-cell memory. Both CMA and selective forms of macroautophagy can also modulate the levels of specific proteins to adjust the activity of defined signaling pathways in activated T-cells. However, there is still much left to learn about the regulation of these two forms of autophagy in T-cells. For instance, the full range of functions they may control, or the existence in T-cells of different levels of functional cooperation and cross-talk between them, remain yet to be determined. Moreover, further research is still necessary to better understand the mechanisms that are responsible for the alterations in the regulation of autophagy in T-cells in different pathologies. This knowledge should prove very valuable for the design of therapeutic approaches that might target autophagy and modulate its activity in those diseases or situations where autophagy dysregulation leads to loss of cell function.
Acknowledgments
This work was supported by grants from the National Institutes of Health, the Glenn Foundation, the Hirschl-Caulier Trust (to FM) and the American Federation for Aging Research (to IG-R)
Footnotes
Conflict of Interest
The authors declare no financial or commercial conflict of interest
References
- 1.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 6.Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, et al. Mammalian Autophagy: How Does It Work? Annu Rev Biochem. 2016 doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 7.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 10.Muller O, Sattler T, Flotenmeyer M, Schwarz H, Plattner H, Mayer A. Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J Cell Biol. 2000;151:519–528. doi: 10.1083/jcb.151.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deretic V. Autophagy: an emerging immunological paradigm. J Immunol. 2012;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 15.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 16.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 21.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerland LM, Genestier L, Peyrol S, Michallet MC, Hayette S, Urbanowicz I, Ffrench P, et al. Autolysosomes accumulate during in vitro CD8+ T-lymphocyte aging and may participate in induced death sensitization of senescent cells. Exp Gerontol. 2004;39:789–800. doi: 10.1016/j.exger.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 26.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 27.Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, Roodman DG, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012;19:144–152. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109:8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 31.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 33.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 34.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 35.Botbol Y, Macian F. Assays for Monitoring Macroautophagy Activity in T cells. Methods Mol Biol. 2015;1343:143–153. doi: 10.1007/978-1-4939-2963-4_12. [DOI] [PubMed] [Google Scholar]
- 36.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nazarko VY, Zhong Q. ULK1 targets Beclin-1 in autophagy. Nat Cell Biol. 2013;15:727–728. doi: 10.1038/ncb2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnold CR, Pritz T, Brunner S, Knabb C, Salvenmoser W, Holzwarth B, Thedieck K, et al. T cell receptor-mediated activation is a potent inducer of macroautophagy in human CD8(+)CD28(+) T cells but not in CD8(+)CD28(−) T cells. Exp Gerontol. 2014;54:75–83. doi: 10.1016/j.exger.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Botbol Y, Patel B, Macian F. Common gamma-chain cytokine signaling is required for macroautophagy induction during CD4 T cell activation. Autophagy. 2015:0. doi: 10.1080/15548627.2015.1089374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. J Immunol. 1999;162:2775–2784. [PubMed] [Google Scholar]
- 43.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuzawa Y, Oshima S, Takahara M, Maeyashiki C, Nemoto Y, Kobayashi M, Nibe Y, et al. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy. 2015;11:1052–1062. doi: 10.1080/15548627.2015.1055439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagomez D, Cover TL, et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol. 2013;190:5086–5101. doi: 10.4049/jimmunol.1202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris J. Autophagy and cytokines. Cytokine. 2011;56:140–144. doi: 10.1016/j.cyto.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 48.Jonchere B, Belanger A, Guette C, Barre B, Coqueret O. STAT3 as a new autophagy regulator. JAKSTAT. 2013;2:e24353. doi: 10.4161/jkst.24353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe R, Fujii H, Shirai T, Saito S, Ishii T, Harigae H. Autophagy plays a protective role as an anti-oxidant system in human T cells and represents a novel strategy for induction of T-cell apoptosis. Eur J Immunol. 2014;44:2508–2520. doi: 10.1002/eji.201344248. [DOI] [PubMed] [Google Scholar]
- 51.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, Wu C, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol. 2016;17:277–285. doi: 10.1038/ni.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stephenson LM, Miller BC, Ng A, Eisenberg J, Zhao Z, Cadwell K, Graham DB, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–635. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLeod IX, Zhou X, Li QJ, Wang F, He YW. The Class III Kinase Vps34 Promotes T Lymphocyte Survival through Regulating IL-7Ralpha Surface Expression. J Immunol. 2011;187:5051–5061. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, Konieczny BT, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 57.Roy SG, Stevens MW, So L, Edinger AL. Reciprocal effects of rab7 deletion in activated and neglected T cells. Autophagy. 2013;9:1009–1023. doi: 10.4161/auto.24468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reggiori F, Komatsu M, Finley K, Simonsen A. Autophagy: more than a nonselective pathway. Int J Cell Biol. 2012;2012:219625. doi: 10.1155/2012/219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia W, He MX, McLeod IX, Guo J, Ji D, He YW. Autophagy regulates T lymphocyte proliferation through selective degradation of the cell-cycle inhibitor CDKN1B/p27Kip1. Autophagy. 2015;11:2335–2345. doi: 10.1080/15548627.2015.1110666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sagnier S, Daussy CF, Borel S, Robert-Hebmann V, Faure M, Blanchet FP, Beaumelle B, et al. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol. 2015;89:615–625. doi: 10.1128/JVI.02174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, Watson AS, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014:3. doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlie K, Westerback A, DeVorkin L, Hughson LR, Brandon JM, MacPherson S, Gadawski I, et al. Survival of effector CD8+ T cells during influenza infection is dependent on autophagy. J Immunol. 2015;194:4277–4286. doi: 10.4049/jimmunol.1402571. [DOI] [PubMed] [Google Scholar]
- 64.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 66.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 67.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113(Pt 24):4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 69.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, et al. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15:1046–1054. doi: 10.1038/ni.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwon J, Shatynski KE, Chen H, Morand S, de Deken X, Miot F, Leto TL, et al. The nonphagocytic NADPH oxidase Duox1 mediates a positive feedback loop during T cell receptor signaling. Sci Signal. 2010;3:ra59. doi: 10.1126/scisignal.2000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–432. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kernbauer E, Cadwell K. Autophagy, viruses, and intestinal immunity. Curr Opin Gastroenterol. 2014;30:539–546. doi: 10.1097/MOG.0000000000000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chuang SY, Yang CH, Chou CC, Chiang YP, Chuang TH, Hsu LC. TLR-induced PAI-2 expression suppresses IL-1beta processing via increasing autophagy and NLRP3 degradation. Proc Natl Acad Sci U S A. 2013;110:16079–16084. doi: 10.1073/pnas.1306556110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 78.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 79.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klein L, Munz C, Lunemann JD. Autophagy-mediated antigen processing in CD4(+) T cell tolerance and immunity. FEBS Lett. 2010;584:1405–1410. doi: 10.1016/j.febslet.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 81.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 82.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med. 2011;208:2625–2632. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb AL, Frausto RF, Bhan V, et al. Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy. 2009;5:152–158. doi: 10.4161/auto.5.2.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gros F, Arnold J, Page N, Decossas M, Korganow AS, Martin T, Muller S. Macroautophagy is deregulated in murine and human lupus T lymphocytes. Autophagy. 2012;8:1113–1123. doi: 10.4161/auto.20275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Z, Matteson EL, Goronzy JJ, Weyand CM. T-cell metabolism in autoimmune disease. Arthritis Res Ther. 2015;17:29. doi: 10.1186/s13075-015-0542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goronzy JJ, Li G, Yu M, Weyand CM. Signaling pathways in aged T cells - a reflection of T cell differentiation, cell senescence and host environment. Semin Immunol. 2012;24:365–372. doi: 10.1016/j.smim.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haynes L, Maue AC. Effects of aging on T cell function. Curr Opin Immunol. 2009;21:414–417. doi: 10.1016/j.coi.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev. 2011;10:205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cuervo AM, Macian F. Autophagy and the immune function in aging. Curr Opin Immunol. 2014;29:97–104. doi: 10.1016/j.coi.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phadwal K, Alegre-Abarrategui J, Watson AS, Pike L, Anbalagan S, Hammond EM, Wade-Martins R, et al. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8:677–689. doi: 10.4161/auto.18935. [DOI] [PMC free article] [PubMed] [Google Scholar]