Abstract
Male infertility might be clearly associated with aberrant DNA methylation patterns in human spermatozoa. An association between oxidative stress and the global methylation status of the sperm genome has also been suggested. The aim of the present study was to determine whether the global sperm DNA methylation status was affected in the spermatozoa of carriers of chromosome structural aberrations. The relationships between the 5-methylcytosine (m5C) levels in spermatozoa and chromatin integrity status were evaluated. The study patients comprised male carriers of chromosome structural aberrations with reproductive failure (n = 24), and the controls comprised normozoospermic sperm volunteers (n = 23). The global m5C level was measured using thin-layer chromatography (TLC) and immunofluorescence (IF) techniques. The sperm chromatin integrity was assessed using aniline blue (AB) staining and TUNEL assay. The mean m5C levels were similar between the investigated chromosome structural aberrations carriers (P) and controls (K). However, sperm chromatin integrity tests revealed significantly higher values in chromosomal rearrangement carriers than in controls (P < 0.05). Although the potential relationship between sperm chromatin integrity status and sperm DNA fragmentation and the m5C level juxtaposed in both analyzed groups (P vs K) was represented in a clearly opposite manner, the low chromatin integrity might be associated with the high hypomethylation status of the sperm DNA observed in carriers of chromosome structural aberrations.
Keywords: 5-methylcytosine, chromatin integrity, chromosomal rearrangement, chromosome translocation, global DNA methylation, male infertility
INTRODUCTION
The symmetric methylation of 5’ytosine (m5C) in CpG islands is one of the mechanisms involved in the regulation of gene expression. DNA methylation is widely regarded as the most stable epigenetic marker and one of the most informative markers for explaining patterns of gene expression, cell differentiation, and phenotype. Cytosine methylation influences chromatin configuration via histone modification and determines genome-wide DNA methylation patterns, which play a crucial role in gametic imprinting, gene silencing, chromosome X inactivation, and protein conformation changes.1,2 Developmental disturbances might occur in newly formed embryos, reflecting the lack of the activation of genes crucial for normal development and associated with disturbances in proper methylation and demethylation events in gametogenic cells.3,4,5,6,7 In addition, immature gametes without fully established methylation patterns can be used for fertilization in assisted reproductive technologies (ARTs). Thus, a basic knowledge concerning the mechanisms and meaning of the gametic epigenome disturbances is important because of the current relatively high frequency of ART-assisted births (approximately, 1%–3% of all live-births).5,6,8
Male infertility might clearly be associated with the aberrant DNA methylation pattern in human spermatozoa previously confirmed through DNA methylation analyses of selected imprinted or nonimprinted genes.9,10,11,12,13,14 Changes in the methylation patterns were documented for males with decreased semen parameters (i.e., DAZL, H19, NFT3, MT1A, PAX8, and PLAGL1)11,13,15,16 and for infertile men with disturbed protamine ratios (LIT1, SNRPN, MEST, ZAC, and PEG3).10
There are other experimental approaches concerning the status of global DNA methylation. To our knowledge, the global methylation analysis of sperm DNA has only been described in eight studies.17,18,19,20,21,22,23,24 The methodology used for the evaluation of global sperm DNA methylation has been limited to a few techniques, most often based on immunofluorescent or colorimetric measurements, followed by microscopic validation or flow cytometry.17,18,19,20,21,24 Only two studies have described methods based on chromatographic measurement, i.e., ultra-performance liquid chromatography/tandem mass spectrometry (UPLC-MS/MS) or high-performance liquid chromatography (HPLC).22,23
The global methylation status of sperm DNA was also analyzed with respect to sperm apoptosis,17,18,21,23 abnormal P1/P2 ratio,19 IVF,17,18 and male aging.24 Based on the results of previous studies, there is a visible relationship between the global m5C level, sperm quality or IVF outcome, and age of male patient. Thus, it is highly desired to obtain an analysis of various sperm features not only regarding genomic structure but also considering epigenetic changes.
The purpose of the present study was to determine the global DNA methylation level in the spermatozoa of carriers of chromosome structural aberrations. In addition, correlations between the level of global sperm DNA methylation and the sperm chromatin integrity (chromatin deprotamination and sperm DNA fragmentation) were performed. To our knowledge, this study is the first to analyze the methylation status in spermatozoa from a group of infertile males with chromosomal rearrangements. Moreover, this study demonstrated the first application of the TLC method for the examination of the global methylation levels in human sperm cells.
MATERIALS AND METHODS
The study group included patients (P) with reproductive failures, comprising 24 men, 26–37 years of age (mean age: 30.7), and harboring chromosome structural aberrations including 15 reciprocal chromosome translocations (RCTs), 4 Robertsonian translocations, 2 complex chromosome rearrangements (CCRs), and 3 pericentric inversions. The karyotyping of all studied patients was performed using a classical GTG staining method (Giemsa solution; Merck, Darmstadt, Germany). The characteristics of the men in the study group, including reproductive history and semen parameters, are presented in Supplementary Table 1 (1.4MB, tif) . The karyotypes of all female partners were normal (46, XX). The control group (K) comprised 23 healthy males, aged 25–33 years (mean: 28.5), with normozoospermia (according to the WHO criteria, 1999; Supplementary Table 2 (836.3KB, tif) ).25 All men (P and K) were carefully selected to have nonsmoking habits or no stimulants/drug use. Ejaculated sperm samples from all enrolled men were collected after 3–5 days of sexual abstinence. After liquefaction and washing in F-10 medium, the sperm samples were analyzed according to the WHO criteria for seminological evaluation (concentration, volume, motility, and morphology). Next, the semen samples were fixed in fresh fixative solution (methanol: glacial acetic acid, 3:1 v/v, −20°C) and stored at −20°C until further use. All men were notified of the purpose of the planned research, and written consent was obtained according to the guidelines of the Local Bioethical Committee at the Poznan University of Medical Sciences.
Characteristics of individual carriers of chromosomal structural aberrations; the results were obtained from 24 patients
Characteristics of semen parameters in control population group (K) of 23 healthy donors (according to the WHO criteria, 1999)
Sperm chromatin integrity
Chromatin immaturity
The chromatin status was evaluated using aniline blue (AB) staining as previously described.26 Briefly, aniline blue binds to lysine residues (in histones), resulting in dark blue staining for the determination of the protamine: histone proportions. Subsequently, three subpopulations of spermatozoa can be visualized as pink, normal sperm cells with a proper protamine: histone ratio; purple-pink, semi-deprotaminated spermatozoa with a disturbed protamine: histone ratio; and dark blue, deprotaminated sperm cells with a high proportion of the remaining histones (lysine rich). In each examined case (P: n = 24; K: n = 23), at least 1500 spermatozoa were assessed using a light microscope (Olympus BX41, Tokyo, Japan; 100× oil immersion objective).
DNA fragmentation
Sperm DNA fragmentation was evaluated using the TUNEL assay (Flow TACS Apoptosis Detection Kit, R&D Systems, Minneapolis, MN, USA) as previously described.27 The principle of the assay was to identify spermatozoa with fragmented DNA-based complex formation between the biotinylated DNA fragments and streptavidin-conjugated fluorescein (FITC) in the presence of terminal deoxynucleotidyl transferase (TdT). After the reaction, two population of sperm cells were counted: fluorescently (light green) labeled TUNEL-positive cells (fragmented DNA) and TUNEL-negative (nonfragmented DNA) labeled only with DAPI (blue). In each case (P: n = 24; K: n = 23), at least 1100 sperm cells were analyzed using a fluorescent microscope (Olympus BX41, equipped with 100× oil immersion objective and FITC/DAPI filters).
Global methylation analysis
Sperm DNA extraction
For sperm DNA extraction, only samples (P: n = 9; K: n = 14) without other contaminating cells were selected using a phase contrast microscope (Olympus BX41, ×200 magnification). Sperm DNA was extracted using a Maxwell 16® (Promega Corp., Madison, WI, USA) automated instrument for DNA extraction (for the simultaneous extraction of up to 16 samples) using a DNA IQ™ Casework Pro Kit (Promega), according to the manufacturer's instructions and the pretreated samples. Briefly, the fixed sperm samples (methanol: acetic acid, 3:1 v/v, −20°C) were washed twice in 1 × PBS (pH 7.4, Biomed, Lublin, Poland) to remove the fixative solution. Approximately, 2 × 106 sperm cells from each sample were resuspended in 150 μl of 1 × PBS and subsequently used for extraction. Each sample was incubated with 20 μl of Proteinase K (Qiagen GmbH, Hilden, Germany) for 2 h at 56°C with brief vortexing every 30 min. Next, 400 μl of lysis buffer (Promega) and 5 μl of 1 M DTT (Merck, Darmstadt, Germany) were added to the samples, followed by mixing and incubation for 15 min at 56°C. Subsequently, the pretreated samples were transferred to the kit cartridges for DNA extraction. The DNA extraction was performed using a Maxwell 16® instrument with “Forensic” and LEV (low extraction volume) software options (30 min). The purified sperm DNA was subsequently dissolved in distilled sterile nuclease-free water, and the concentration was assessed using a NanoDrop® Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA).
Enzymatic hydrolysis of the DNA and deoxynucleotides labeling using 32P
Approximately, 1–3 μg of the DNA sample was resuspended in a solution containing 20 mM sodium succinate buffer, pH 6.0, with 10 mM CaCl2. The following enzymes were added to the buffer: 0.02 U Micrococcus nuclease (MN; Sigma-Aldrich, St. Louis, MO, USA) and 0.001 U spleen phosphodiesterase (SPD, Sigma-Aldrich). The mixture was subsequently incubated for 5 h at 37°C. A hydrolysate of the DNA containing 3’ deoxynucleoside monophosphates (dNp) was applied for labeling. Next, 3 μl of bicine buffer, pH 9.8 (20 mM bicine, 10 mM MgCl2, 10 mM DTT, 1 mM spermidine; Merck, Darmstadt, Germany), 400 pM ATP, 2 U polynucleotide kinase and 1 μCi [γ-32P] ATP were added to the mixture. The mixture was incubated at 37°C for 30 min. Next, radioactive nucleoside-3’,5’- diphosphates ([32P] Np) were hydrolyzed using nuclease P1 (0.2 μg diluted in 0.5 M ammonium acetate buffer, pH 4.0, 30 min, 37°C) to delete the 3’- phosphate group.
Separation and analysis of the chromatography products
For the analysis of the modified deoxynucleotides in a sample, thin-layer chromatography (TLC) was used. The separation and identification of [32P] dN were performed on cellulose-covered plates (10 cm × 20 cm; Merck) using a mixture of diluents such as (i)first direction (20 cm): isobutyric acid/ammonium/water (66/1/17) and (ii) second direction: 0.1 mmol l-1 sodium phosphate buffer, pH 6.8/ammonium sulfate/propanol-1 (100 ml/60 g/1.5 ml). After resolution, the plates were dried. The chromatographic images were analyzed through mapping using a PhosphorImager Typhoon laser scanner (Molecular Dynamics; GE Healthcare, Sunnyvale, CA, USA) equipped with a FLA-5100 Fluoro Image Analyzer and Multi-Gauge 3.0 software (FujiFilm, Tokyo, Japan). For the quantitative evaluation of m5C, the quantity of m5C decay products (C and T) was assessed. The intensity of individual nucleotide spots was estimated. Quantities of m5C were calculated according to the formula, R = [(m5dC)/(m5dC + dC + dT)] ×100.
Immunofluorescent staining for DNA methylation analysis (IF)
Immunostaining was performed according to previously published methods.18,19,20,21 Slides with fixed spermatozoa were washed as following: 1 × PBS (pH 7.4), 1 × PBST (1 × PBS + 0.5% Triton X-100, Sigma-Aldrich), 1 × PBST, and 1 × PBS for 5 min each. To facilitate the access of methylated DNA, the sperm DNA was incubated and decondensed in 25 mmol l-1 DTT/1 M Tris-HCl (pH 9.5) for 20 min. After double washing in 1 × PBST (5 min) and denaturing in 6N HCl (15 min), followed by incubation in 1 M Tris-HCl (pH 9.0, 15 min), the sperm DNA was subsequently blocked with 1% BSA/1 × PBST for 30 min. The slides were further incubated with primary mouse anti-5-methylcytosine (33D3) antibody (Sigma-Aldrich) diluted 1:200 for 60 min in a humidified chamber, followed by incubation with secondary goat anti-mouse IgG (whole molecule)-FITC antibody (Sigma-Aldrich) diluted 1:400 for 30 min. Next, the slides were washed 3 times with 1 × PBST for 5 min each (light-proof chamber) and 20 μl of DAPI/antifade was applied (MetaSystems, Altlussheim, Germany). A negative control was used in the experiment (no primary antibody incubation). All proceedings were performed at room temperature. The analysis of immunofluorescence was performed within 20 min using an Olympus BX41 fluorescence microscope equipped with an adequate filter set (DAPI/FITC/Triple) and a 100× oil immersion objective. To avoid fading of the fluorescence staining, each field of view was exposed to a fluorescent light for no longer than 3 s. In each experiment (P: n = 24; K: n = 20), at least 200 spermatozoa were documented (ISIS, MetaSystems) and evaluated using ImageJ software (version 1.48; NIH, Bethesda, MD, USA). The intensity of fluorescence (cell fluorescence, CF; arbitrary units, a.u.) was calculated in each case, including the integrated density, area of the sperm cell, and mean fluorescence of the background (measured in 5 areas near the spermatozoa).
Statistical analysis
All tests were performed considering a significance cut-off level at α = 0.05. The normal distribution of obtained results was examined using the Shapiro–Wilk method with log transformation. All obtained results revealed a normal distribution pattern (P > 0.05; range: 0.051–0.920) in both groups of investigated men (P and K), suggesting that the proper statistical methods were used for further significance delineation.
The statistical significance between the individual values and the mean frequencies of sperm seminological parameters, sperm DNA fragmentation, chromatin immaturity, and DNA methylation status were assessed using an unpaired two-tailed t-test. In addition, linear regression analyses among the values for the DNA methylation status versus seminological parameters and chromatin integrity were performed. For all statistical evaluations, Origin (v. 8.1, OriginLab, Northampton, MA, USA) and the Analyse-it Method Evaluation for Microsoft Excel (v. 3.70, Analyse-it Software, Ltd., UK) were applied.
RESULTS
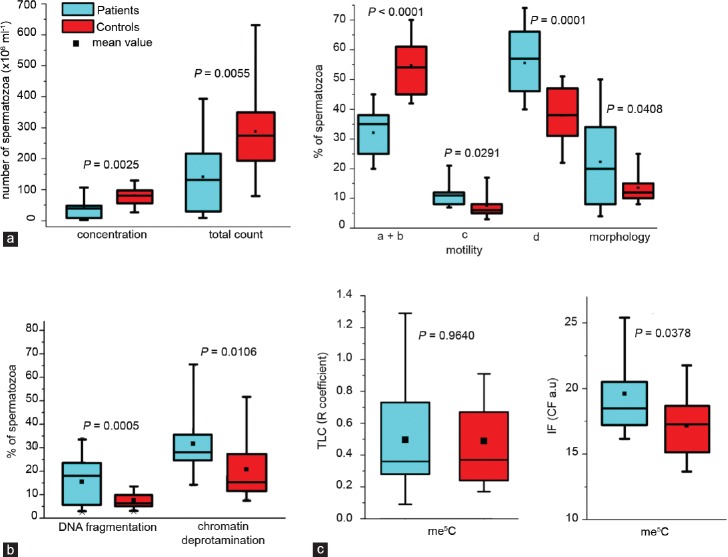
The seminological analysis of a group of males with reproductive failures (P) revealed normozoospermia in 11 individuals, while in 13 other patients, the sperm quality parameters were decreased (Supplementary Table 1 (1.4MB, tif) ). Statistical boxes showing the range, median, and mean of the values obtained in each studied category are presented in Figure 1a. Statistical significance between both studied groups of males was observed after comparing the sperm concentration (P = 0.0025) and total sperm count in the semen samples (P = 0.0055). The mean P seminological values were markedly lower than the K values and showed increased heterogeneity. Statistical significance was observed for the sperm motility (a + b: P <0.0001; c: P = 0.0291; d: P = 0.0001) and sperm morphology (P = 0.0408). The sperm motility was significantly better in control individuals while the sperm morphology was better in the patient group.
Figure 1.
Statistical boxes showing the range (s.d.), median and mean (black square) of the values observed in each performed category of examinations; (a) semen parameters: concentration, total count, motility (a: rapid progressive, b: slow or sluggish progressive, c: nonprogressive, and d: immotile), and morphology; according to the WHO guidelines (1999);25 (b) sperm chromatin integrity: DNA fragmentation and chromatin deprotamination; (c) global m5C level in spermatozoa. The blue color represents Patients (carriers of chromosome structural aberrations with reproductive failures), while the red color corresponds to the Control group (healthy males with normozoospermia). A value of P < 0.05 indicates a statistically significant difference.
Sperm chromatin integrity
Chromatin immaturity
The results of AB staining showed that the mean frequency of spermatozoa with deprotaminated chromatin obtained for the analyzed group of patients (P) was 31.90 ± 14.72% (range: 13.48%–66.29%) was significantly higher (P = 0.0106) compared with the mean control value (K) 21.02 ± 13.21% (range: 7.65%–53.50%) (Tables 1 and 2, Figure 1b).
Table 1.
Results of sperm DNA analysis obtained from a group of 24 patients comprising carriers of chromosome structural aberrations: the data for sperm DNA fragmentation (TUNEL), sperm chromatin deprotamination (AB), and global sperm DNA m5C level (TLC, IF) are presented
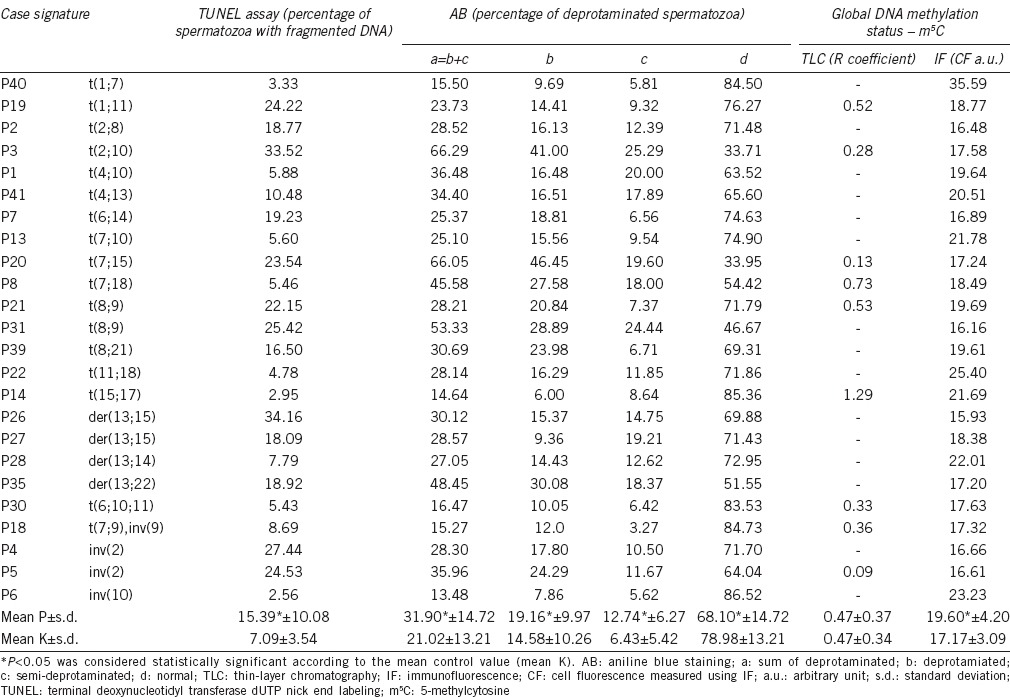
Table 2.
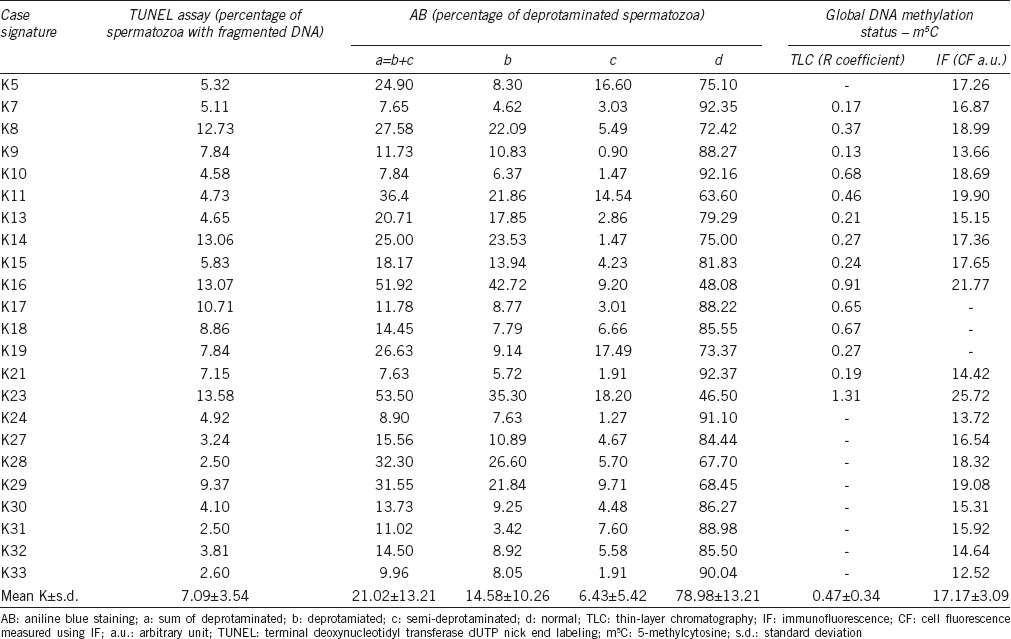
Results of the sperm DNA analysis obtained from a group of 23 normal controls: the data for sperm DNA fragmentation (TUNEL), sperm chromatin deprotamination (AB) and global sperm DNA m5C level (TLC, IF) are presented
No correlations (P > 0.05) were observed in both groups of males when collating global chromatin deprotamination status versus sperm concentration (P: r = −0.3114; K: r = 0.1031), total sperm count (P: r = −0.1459; K: r = −0.0504), sperm morphology (P: r = −0.2161; K: r = 0.0041), and sperm progressive motility (a + b; P: r = −0.2328; K: r = 0.0667). When examining sperm immotility (type d), statistical significance was observed in the control group (K: r = 0.4464, P = 0.0327). No such association was observed in the P group (P: r = 0.4271, P = 0.4732).
DNA fragmentation
The results of the TUNEL assay (Tables 1 and 2, Figure 1b) showed that the mean frequency of sperm DNA fragmentation in the patient group (P) was 15.39 ± 10.08% (range: 2.56%–34.16%), and this value was statistically higher (P = 0.0005) compared with the mean K value 7.09 ± 3.54% (range: 2.50%–13.58%).
A positive correlation between the sperm chromatin deprotamination status and the sperm DNA fragmentation was observed in both groups of males (r = 0.5445, P = 0.0059 for P, r = 0.6260, P = 0.0010 for K), indicating a potential link between the two parameters for chromatin integrity status (Figure 2b).
Figure 2.
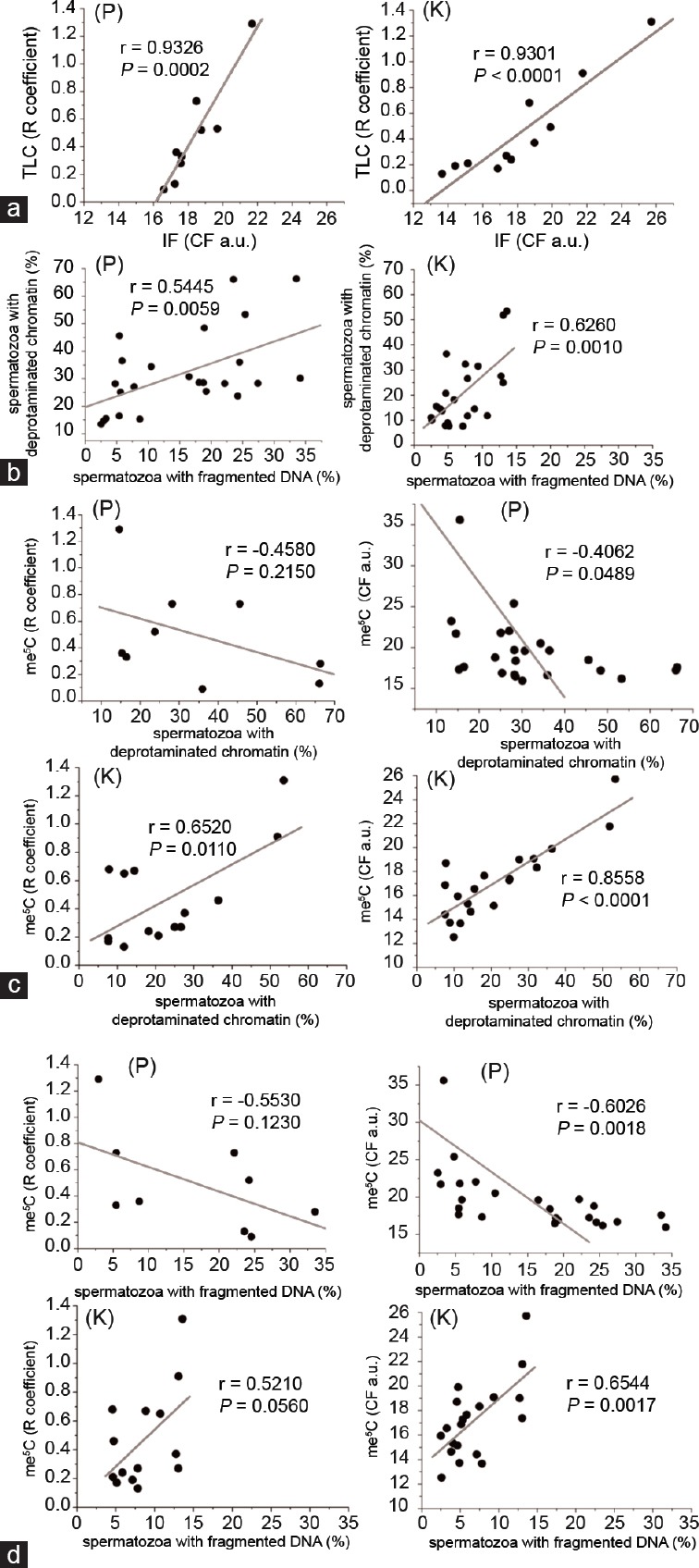
Analysis of the correlations between the global me5C level and chromatin integrity features: (a) correlation between two me5C measurement methods: TLC versus IF; (b) spermatozoa with deprotaminated chromatin versus spermatozoa with fragmented DNA; (c) me5C versus spermatozoa with deprotaminated chromatin; (d) me5C versus fragmented DNA. “P” indicates Patients, while “K” corresponds to the Control. A value of P < 0.05 indicates a statistically significant difference.
No correlations (P > 0.05) were observed in both groups of males when collating the sperm DNA fragmentation level versus the sperm concentration (P: r = −0.0727; K: r = −0.0064), total sperm count (P: r = −0.0898; K: r = 0.0011), sperm progressive motility (a + b; P: r = −0.0284; K: r = −0.1245), sperm immotility (d; P: r = 0.4084; K: r = 0.3723), and sperm morphology (P: r = 0.1114; K: r = −0.1742).
Global methylation of sperm DNA
To quantify the global m5C level in the sperm DNA, TLC and IF methods were used. Using TLC technique, the results obtained for the patient group (P) showed an R coefficient of 0.47 ± 0.37 (range: 0.09–1.29), similar for the controls (K) 0.47 ± 0.34 (range: 0.13–1.31; P = 0.9640; Tables 1 and 2, Figure 1c). Using IF method, the mean control CF result was 17.17 ± 3.09 a.u. (Table 2), and this value was statistically lower (P = 0.0378; Figure 1c) than the primary result obtained for patients (P: 19.60 ± 4.20 a.u.; Table 1). In addition, the results of both methods (TLC and IF) validated each other (r = 0.9326, P = 0.0002 for P, r = 0.9301, P < 0.0001 for K; Figure 2a) and facilitated the interpretation of the obtained results.
An interesting phenomenon was observed from the analysis of the relationship between the parameters of the sperm chromatin status and global sperm methylation (Figure 2c and 2d), and contrasting results were obtained for patients with chromosomal rearrangements versus controls (K). Thus, the analysis of the correlation between the global m5C level and chromatin deprotamination status showed a positive correlation in the control group (K: r = 0.6520, P = 0.0110), and a negative association in the patient group (P: r = −0.4580, P = 0.2150) (Figure 2c). Similar observations were obtained after analyzing the correlation between the global m5C level and the sperm DNA fragmentation (Figure 2d).
No correlations (P > 0.05) were observed in either group of males studied when collating the global me5C levels, measured using both TLC and IF techniques versus all sperm parameters. Representative images of the staining results are presented in Figure 3.
Figure 3.
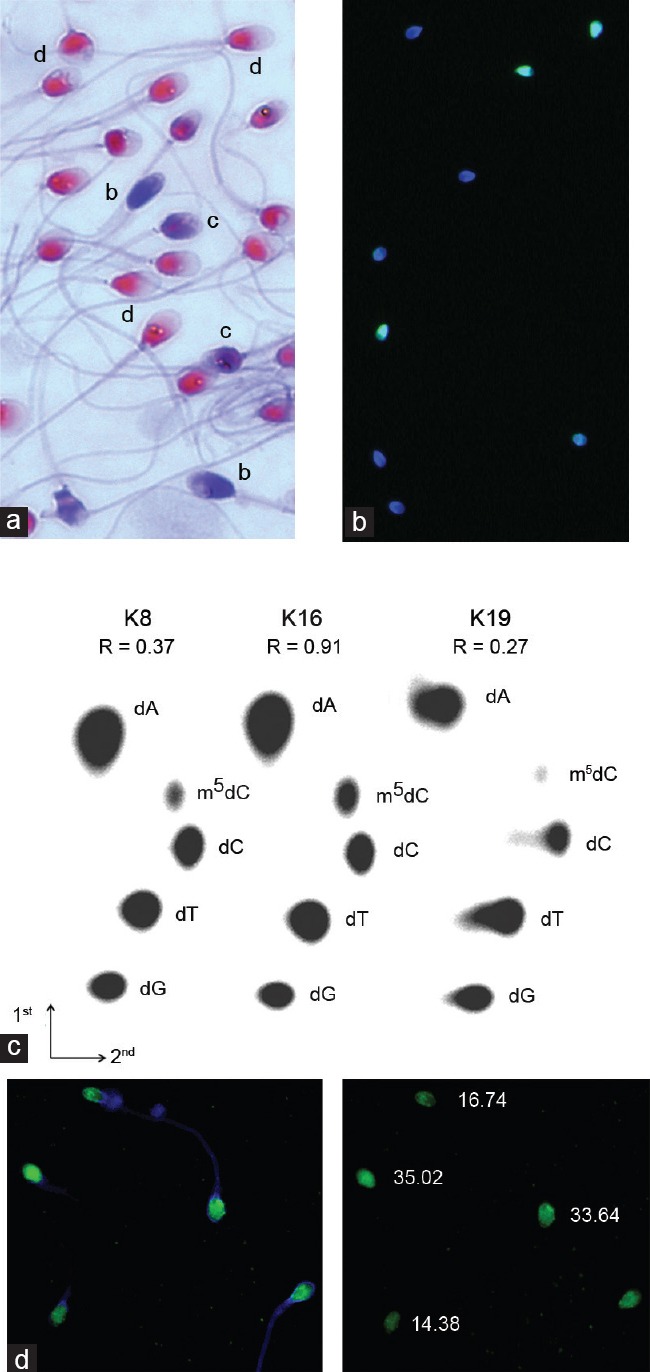
Representative images of the staining results for spermatozoa. The bar represents 5 µm. (a) aniline blue staining of three populations of spermatozoa: b: deprotaminated, c: semi-deprotaminated and d: normal, with a proper protamine: histone ratio; light microscopy, ×1000 magnification; (b) TUNEL assay involving the two populations of spermatozoa: light-green (with fragmented DNA) and blue (only DAPI as a counterstain, without DNA fragmentation); fluorescent microscopy, ×1000 magnification; (c) examples of the two-dimensional thin-layer cellulose chromatography (TLC) analysis of [5’32P]-labeled deoxynucleotides obtained after the enzymatic hydrolysis of DNA. dNp – deoxynucleoside monophosphates: A: adenine; C: cytosine; G: guanine; T: thymine and m5C: 5-methylcytosine. Three different values for the R coefficient are presented; (d) example of the immunofluorescent staining (IF) of spermatozoa. Left panel: merged DAPI and FITC channels; right panel: FITC panel with cell fluorescence (CF) values calculated for five spermatozoa; fluorescent microscopy, ×1000 magnification.
DISCUSSION
To our knowledge, this study is the first to demonstrate the global methylation of sperm DNA from males with chromosome structural rearrangements. Moreover, this study also demonstrated the first application of TLC method for the measurement of the global me5C levels of DNA in human spermatozoa. The results showed that TLC is a good, cheap, and rapid technique for global m5C evaluation. These findings also support the data obtained from previous studies concerning TLC application for the successful measurement of methylation status in different tissue types from patients with a variety of diseases (breast, brain, and colon cancers and blood hypertension).28,29,30,31
Similar mean levels of me5C observed in both groups of males (P vs K) might reflect the fact that the majority of methylation in the genome occurs in areas outside of CpG islands such as repetitive elements and noncoding and nonregulatory regions.9,32 However, it cannot be excluded that the presence of rearranged chromosomes plays an important but indirect role in establishing sperm epigenomic regulation.
The sperm cell is a final product of male spermatogenesis, in which the complexity in cell divisions (mitotic and meiotic) generates a unique haploid cell type with dramatically changed chromatin packaging resulting from the replacement of approximately 85%–90% of the histones with protamines (P1 and P2).33,34 A disruption in expression observed at a proportion of P1:P2, describing the ratio of protamines to the remaining histones, has been implicated in male fertility as a reduction in sperm quality and the induction of sperm DNA damage, leading to the breakdown of embryo implantation.35,36,37,38
Sperm DNA damage can be inflicted (among others) through an oxidative cascade initiated via reactive oxygen species (ROS) directed to unresolved DNA strands. The presence of strand breaks might also result from the remodeling of sperm chromatin during spermiogenesis when high torsion tensions occur.39,40 Such tensions might also accompany chromosomal rearrangements and promote the opening of sperm chromatin prone to apoptosis, leading to a decrease in sperm quality. In the present and previous studies,26 we observed a decrease of chromatin stability in men with chromosomal rearrangements, expressed as both increased sperm DNA fragmentation (2.17× and 2.5×, respectively: present and previous study, P < 0.05) and a significantly increased frequency of deprotaminated spermatozoa (1.52× and 1.97×, P < 0.05). Thus, we hypothesized that chromosome rearrangements not only lead to the disruption of meiotic segregation but also induce disruptions at the chromatin stability level, consistent with the data obtained from previous studies.26,38,39,40,41,42,43 We also proposed that the presence of chromosome aberrations might also disrupt the definitive link between chromatin deprotamination and sperm motility observed in the control group, as this finding was not observed in the patient group, even when the seminal parameters (motility and morphology) in the P group were reduced.
In the control donors (K), the number of spermatozoa with hypermethylated genomes increased with increasing chromatin instability (Figure 2). These results are consistent with those of Barzideh et al.23 who revealed that in normozoospermic males (assuming their normal karyotypes), apoptotic sperm cells were associated with disorders of spermatogenesis, demonstrating the hypermethylation of the genome. In addition, other authors have suggested that global hypermethylation might be an early response to oxidative stress mediated through an increase in DNA methyltransferase (Dnmt) activity.44
In males with chromosomal rearrangements (P), we observed that the global me5C level represented negative association with chromatin deprotamination and sperm DNA fragmentation (Figure 2c and 2d). Tunc and Tremellen21 also reported increased hypomethylation with decreased chromatin integrity, suggesting a potential association between DNA damage and impaired methylation. Moreover, a positive correlation between TUNEL and NBT results (nitroblue tetrazolium staining to evaluate ROS production in semen) and therapy with antioxidants suggested that oxidative damage of the sperm DNA might at least in part be responsible for sperm global DNA methylation.21,45,46 These authors also hypothesized that spermatozoa with hypomethylated DNA might be prone to DNA damage.20,21,45,46 Taken together, these data suggest that oxidative stress, among the other factors, might induce global sperm DNA demethylation, particularly in males with rearranged chromosomes.
A strong hypothesis concerning the role of oxidative stress in establishment of DNA methylation patterns has been presented not only for spermatozoa but also in carcinogenesis. It has been clearly documented that in breast, brain, and colon cancers and in patients with blood hypertension, global DNA hypomethylation is correlated with the severity of the disease stage.28,29,30,31,47 In this scenario, it is reasonable to propose the measurement of global DNA methylation as a potential prognostic and/or diagnostic value biomarker. Indeed, we cannot exclude the fact that the global methylation pattern in different abnormalities/diseases might future determinate the initial symptoms of ailing humans. In addition, the results of the methylation analysis for selected genes showed that the inheritance of affected DNA methylation patterns is a likely etiological candidate for many heritable disorders that were previously considered idiopathic.48,49,50,51
An association between hypomethylation and genomic instability based on the presence of chromosomal breakpoints has also been shown in cancer research.52 Studies have focused on the fact that specified structural genomic elements (such as repetitive GC-rich fragments or regions close to Alu with higher rates of recombination) might be more sensitive to aberrant methylation incidents creating DNA breakpoints. These factors might lead to the epigenetic instability of the genome. Moreover, the inheritance of the affected DNA methylation patterns could serve as a background for many heritable disorders that were previously considered unknown.48,49,50,51 The results of previous studies on spermatozoa have suggested that the affected health of the offspring might reflect epigenetic defects resulting from aberrant sperm DNA methylation.14,21,39,53 Hypomethylation might also alter cell differentiation, and embryonic genomic expression might reveal the disturbed synchronization in development.18 Consequently, the unique epigenetic marks in spermatozoa might be crucial for proper mature gamete function and the activation of specific genes during early embryonic development.3,5,6,7,18 Therefore, understanding the paternal epigenetic “landscape,” particularly in infertile men with an abnormal karyotype, is of paramount importance.
AUTHOR CONTRIBUTIONS
MO designed the study, performed chromatin deprotamination and immunofluorescence experiments, analyzed and interpreted results, drafted the manuscript. MZB and JB performed and interpreted TLC methylation experiments. MF performed sperm DNA fragmentation analysis. NH and VBC collected biological samples and identified structural rearrangements. DZ collected biological samples. MK provided funding, supervised the study and results interpretation, and finalized a manuscript. All authors read and approved the final version of the manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
ACKNOWLEDGMENTS
We would like to thank Prof. Lyubov F Kurilo, PhD, MD (RAMS, Moscow, Russia) and Dr. Ewa Wiland, PhD (IHG PAS, Poznan) for their role in collection of biological samples. This work was supported by National Science Centre in Poland (grant number 2011/01/B/NZ2/04819).
Supplementary information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Black JC, Whetstine JR. Chromatin landscape: methylation beyond transcription. Epigenetics. 2011;6:9–15. doi: 10.4161/epi.6.1.13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schagdarsurengin U, Paradowska A, Steger K. Analysing the sperm epigenome: roles in early embryogenesis and assisted reproduction. Nat Rev Urol. 2012;9:609–19. doi: 10.1038/nrurol.2012.183. [DOI] [PubMed] [Google Scholar]
- 3.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Embryogenesis: demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 4.Allegrucci C, Thurston A, Lucas E, Young L. Epigenetics and the germline. Reproduction. 2005;129:137–49. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- 5.Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23:2826–34. doi: 10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- 6.Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91:305–15. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puri D, Dhawan J, Mishra RK. The paternal hidden agenda: epigenetic inheritance through sperm chromatin. Epigenetics. 2010;5:386–91. doi: 10.4161/epi.5.5.12005. [DOI] [PubMed] [Google Scholar]
- 8.Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, et al. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94:585–94. doi: 10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 9.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 10.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, et al. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod. 2010;10:2647–54. doi: 10.1093/humrep/deq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, et al. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5:60–9. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- 13.Nanassy L, Carrell DT. Analysis of the methylation pattern of six gene promoters in sperm of men with abnormal protamination. Asian J Androl. 2011;13:342–6. doi: 10.1038/aja.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrell DT. Epigenetics of the male gamete. Fertil Steril. 2012;97:267–74. doi: 10.1016/j.fertnstert.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, et al. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aston KI, Punj V, Liu L, Carrell DT. Genome-wide sperm deoxyribonucleic acid methylation is altered in some men with abnormal chromatin packaging or poor in vitro fertilization embryogenesis. Fertil Steril. 2012;97:285–92. doi: 10.1016/j.fertnstert.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Benchaib M, Ajina M, Lornage J, Niveleau A, Durand P, et al. Quantitation by image analysis of global DNA methylation in human spermatozoa and its prognostic value in in vitro fertilization: a preliminary study. Fertil Steril. 2003;80:947–53. doi: 10.1016/s0015-0282(03)01151-8. [DOI] [PubMed] [Google Scholar]
- 18.Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, et al. Influence of global sperm DNA methylation on IVF results. Hum Reprod. 2005;20:768–73. doi: 10.1093/humrep/deh684. [DOI] [PubMed] [Google Scholar]
- 19.Aoki VW, Emery BR, Carrell DT. Global sperm deoxyribonucleic acid methylation is unaffected in protamines-deficient infertile males. Fertil Steril. 2006;86:1541–3. doi: 10.1016/j.fertnstert.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Tavalaee M, Razavi S, Nasr-Esfahani MH. Influence of sperm chromatin anomalies on assisted reproductive technology outcome. Fertil Steril. 2009;91:1119–26. doi: 10.1016/j.fertnstert.2008.01.063. [DOI] [PubMed] [Google Scholar]
- 21.Tunc O, Tremellen K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet. 2009;26:537–44. doi: 10.1007/s10815-009-9346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Suo Y, Yin R, Shen H, Wang H. Ultra-performance liquid chromatography/tandem mass spectrometry for accurate quantification of global DNA methylation in human sperms. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:1647–52. doi: 10.1016/j.jchromb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Barzideh J, Scott RJ, Aitken RJ. Analysis of the global methylation status of human spermatozoa and its association with the tendency of these cells to enter apoptosis. Andrologia. 2013;45:424–9. doi: 10.1111/and.12033. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins TG, Aston KI, Cairns BR, Carrell DT. Paternal aging and associated intraindividual alterations of global sperm 5-methylcytosine and 5-hydroxymethylcytosine levels. Fertil Steril. 2013;100:945–51.e2. doi: 10.1016/j.fertnstert.2013.05.039. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. WHO Laboratory Manual for the Examination of Human Semen and Sperm Cervical Mucus Interaction. 4th ed. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- 26.Olszewska M, Fraczek M, Huleyuk N, Czernikiewicz A, Wiland E, et al. Chromatin structure analysis of spermatozoa from reciprocal chromosome translocation carriers (RCT) with known meiotic segregation patterns. Reprod Biol. 2013;13:209–20. doi: 10.1016/j.repbio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Olszewska M, Huleyuk N, Fraczek M, Zastavna D, Wiland E, et al. Sperm FISH and chromatin integrity in spermatozoa from t(6;10;11) carrier. Reproduction. 2014;147:659–70. doi: 10.1530/REP-13-0533. [DOI] [PubMed] [Google Scholar]
- 28.Zukiel R, Nowak S, Barciszewska AM, Gawronska AM, Gawronska I, et al. A simple epigenetic method for the diagnosis and classification of brain tumors. Mol Cancer Res. 2004;2:196–202. [PubMed] [Google Scholar]
- 29.Barciszewska AM, Murawa D, Gawronska I, Murawa P, Nowak S, et al. Analysis of 5-methylcytosine in DNA of breast and colon cancer tissues. IUBMB Life. 2007;59:765–70. doi: 10.1080/15216540701697412. [DOI] [PubMed] [Google Scholar]
- 30.Barciszewska AM, Nowak S, Naskręt-Barciszewska MZ. The degree of global DNA hypomethylation in peripheral blood correlates with that in matched tumor tissues in several neoplasia. PLoS One. 2014;9:e92599. doi: 10.1371/journal.pone.0092599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smolarek I, Wyszko E, Barciszewska AM, Nowak S, Gawronska I, et al. Global DNA methylation changes in blood of patients with essential hypertension. Med Sci Monit. 2010;16:CR149–55. [PubMed] [Google Scholar]
- 32.Molaro A, Hodges E, Fang F, Song Q, McCombie WR, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–41. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417–35. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- 34.Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- 35.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: what is the link? Hum Reprod Update. 2007;13:313–27. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- 36.Avendaño C, Franchi A, Duran H, Oehninger S. DNA fragmentation of normal spermatozoa negatively impacts embryo quality and intracytoplasmic sperm injection outcome. Fertil Steril. 2010;94:549–57. doi: 10.1016/j.fertnstert.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Peiro A, Martinez-Heredia J, Oliver-Bonet M, Abad C, Amengual MJ, et al. Protamine 1 to protamine 2 ratio correlates with dynamic aspects of DNA fragmentation in human sperm. Fertil Steril. 2011;95:105–9. doi: 10.1016/j.fertnstert.2010.06.053. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Peiro A, Oliver-Bonet M, Navarro J, Abad C, Guitart M, et al. Dynamics of sperm DNA fragmentation in patients carrying structurally rearranged chromosomes. Int J Androl. 2011;34:e546–53. doi: 10.1111/j.1365-2605.2011.01153.x. [DOI] [PubMed] [Google Scholar]
- 39.Brugnon F, Van Assche E, Verheyen G, Sion B, Boucher D, et al. Study of two markers of apoptosis and meiotic segregation in ejaculated sperm of chromosomal translocation carrier patients. Hum Reprod. 2006;21:685–93. doi: 10.1093/humrep/dei401. [DOI] [PubMed] [Google Scholar]
- 40.Perrin A, Caer E, Oliver-Bonet M, Navarro J, Benet J, et al. DNA fragmentation and meiotic segregation in sperm of carriers of a chromosomal structural abnormality. Fertil Steril. 2009;92:583–9. doi: 10.1016/j.fertnstert.2008.06.052. [DOI] [PubMed] [Google Scholar]
- 41.Perrin A, Basinko A, Douet-Guilbert N, Gueganic N, Le Bris MJ, et al. Aneuploidy and DNA fragmentation in sperm of carriers of a constitutional chromosomal abnormality. Cytogenet Genome Res. 2011;133:100–6. doi: 10.1159/000323980. [DOI] [PubMed] [Google Scholar]
- 42.Ferfouri F, Biotrelle F, Tapia S, Gomes DM, Selva J, et al. Sperm FISH analysis of a 46, XY, t(3;6)(p24;p21.2), inv(8)(p11;2q21.2) double chromosomal rearrangement. Reprod Biomed Online. 2012;24:219–23. doi: 10.1016/j.rbmo.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Vozdova M, Kasikova K, Oracova E, Prinosilova P, Rybar R, et al. The effect of the swim-up and hyaluronan-binding methods on the frequency of abnormal spermatozoa detected by FISH and SCSA in carriers of balanced chromosomal translocations. Hum Reprod. 2012;27:930–7. doi: 10.1093/humrep/der445. [DOI] [PubMed] [Google Scholar]
- 44.Doshi T, Mehta SS, Dighe V, Balasinor N, Vanage G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology. 2011;289:74–82. doi: 10.1016/j.tox.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–8. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gharagozloo P, Aitken RJ. The role of sperm oxidative stress in male infertility and the significance of oral antioxidant therapy. Hum Reprod. 2011;7:1628–40. doi: 10.1093/humrep/der132. [DOI] [PubMed] [Google Scholar]
- 47.Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 48.Bennett DA, Yu L, Yang J, Srivastava GP, Aubin C, et al. Epigenomics of Alzheimer's disease. Transl Res. 2015;165:200–20. doi: 10.1016/j.trsl.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Küçükali CI, Kürtüncü M, Çoban A, Çebi M, Tüzün E. Epigenetics of multiple sclerosis: an updated review. Neuromolecular Med. 2015;17:83–96. doi: 10.1007/s12017-014-8298-6. [DOI] [PubMed] [Google Scholar]
- 50.Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–64. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Besaratinia A, Tommasi S. Epigenetics of human melanoma: promises and challenges. J Mol Cell Biol. 2014;6:356–67. doi: 10.1093/jmcb/mju027. [DOI] [PubMed] [Google Scholar]
- 52.Bilke S, Gindin Y. Evidence for a functional link between chromosomal breakpoints and aberrant DNA methylation in cancer. Front Oncol. 2014;4:46. doi: 10.3389/fonc.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milekic MH, Xin Y, O’Donnell A, Kumar KK, Bradley-Moore M, et al. Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry. 2015;20:995–1001. doi: 10.1038/mp.2014.84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of individual carriers of chromosomal structural aberrations; the results were obtained from 24 patients
Characteristics of semen parameters in control population group (K) of 23 healthy donors (according to the WHO criteria, 1999)