Abstract
The adaptor proteins AP-2 and AP-1/GGAs are essential components of clathrin coats at the plasma membrane and trans-Golgi network, respectively. The adaptors recruit accessory proteins to clathrin-coated pits, which is dependent on the adaptor ear domains engaging short peptide motifs in the accessory proteins. Here, we perform an extensive mutational analysis of a novel WXXF-based motif that functions to mediate the binding of an array of accessory proteins to the α-adaptin ear domain of AP-2. Using nuclear magnetic resonance and mutational studies, we identified WXXF-based motifs as major ligands for a site on the α-ear previously shown to bind the DPW-bearing proteins epsin 1/2. We also defined the determinants that allow for specific binding of the α-ear motif to AP-2 as compared to those that allow a highly related WXXF-based motif to bind to the ear domains of AP-1/GGAs. Intriguingly, placement of acidic residues around the WXXF cores is critical for binding specificity. These studies provide a structural basis for the specific recruitment of accessory proteins to appropriate sites of clathrin-coated vesicle formation.
Keywords: AAK1, clathrin, endocytosis, stonin, synaptojanin
Introduction
Clathrin-mediated vesicle trafficking constitutes a major transport route from the plasma membrane and the trans-Golgi network (TGN) towards the endosomal/lysosomal system and involves the orchestrated recruitment and assembly of proteinaceous coats (Conner and Schmid, 2003; Hinners and Tooze, 2003). Besides clathrin, clathrin adaptors constitute the major protein components of clathrin-coated vesicles (CCVs) and specific subsets of adaptor proteins are used during vesicle formation at specific cellular locations (Ritter and McPherson, 2004; Robinson, 2004). For example, the heterotetrameric AP-2 complex, which is composed of two large subunits, α and β2, a medium-sized μ2 subunit, and the small σ2 subunit, functions in CCV formation at the plasma membrane (Brodsky et al, 2001). CCV formation at the TGN is served by an analogous heterotetrameric complex, AP-1, composed of γ, β1, μ1, and σ1 subunits, alongside the monomeric Golgi-localized, γ-ear-containing, Arf-binding (GGA) proteins (Bonifacino, 2004; Robinson, 2004). The C-terminal domains of the large AP-1 and -2 subunits and the GGA proteins are globular structures, referred to as ears, which extend from the adaptors on flexible hinges (Ritter and McPherson, 2004; Robinson, 2004). The common function of the adaptor ears is the recruitment of accessory proteins to sites of CCV formation. These proteins appear to gain their specificity for different membranes in part through their affinity for the adaptor ears.
The adaptor ear domains bind to short peptide motifs in the accessory proteins. The best characterized example is the AP-2 α-adaptin ear (α-ear), which is subdivided into C-terminal platform and N-terminal sandwich domains and recruits numerous accessory proteins carrying the signature motifs DPF/W and FXDXF (Owen et al, 1999; Traub et al, 1999; Brett et al, 2002). These motifs target overlapping binding sites in the platform domain and a second DPW-specific binding site has been described for the sandwich domain (Brett et al, 2002).
Through proteomics analysis of CCVs, we have identified two previously uncharacterized endocytic accessory proteins, now referred to as NECAP 1 and 2 (Ritter et al, 2003; Blondeau et al, 2004). Their interaction with AP-2 originates solely from a novel α-ear-binding motif, WVQF (Ritter et al, 2003). Variations of the WVQF sequence function in AP-2 binding in several endocytic accessory proteins, demonstrating the global nature of the motif (Ritter et al, 2003; Jha et al, 2004; Walther et al, 2004). However, the determinants within the motif that allow for specific interaction with AP-2 have not been defined in detail. We have thus undertaken a systematic mutational approach to characterize the new motif. Moreover, we have utilized nuclear magnetic resonance (NMR) to identify its binding site on the α-ear, which revealed that the WVQF motif constitutes the major ligand for an α-ear sandwich-binding site formerly shown to bind DPW motifs.
In several studies, we and others have characterized the first γ-adaptin ear (GAE)-binding motif, which is utilized by a large array of accessory proteins, including the NECAPs, for interaction with AP-1 and GGA proteins (Collins et al, 2003; Duncan et al, 2003; Miller et al, 2003; Wasiak et al, 2003; Bai et al, 2004; Mattera et al, 2004). The GAE-binding motifs and the new α-ear-binding motif are both based on ∅XX∅ cores (where ∅ is a large hydrophobic residue) and, in the NECAPs, the sequences WVQF and WGDF enable specific interactions with AP-2 and AP-1, respectively. The striking similarity of the motifs raises the fundamental question of how binding specificity for their appropriate ear domains is achieved. Intriguingly, through a systematic mutational analysis, we have shown that negative charge upstream of the GAE-binding motif and downstream of the WVQF α-ear-binding motif are important determinants for specific interactions with the adaptor ears.
Results
Characterization of the WXXF-based α-ear-binding motif
Previously, we determined that the C-terminal most six amino acids of NECAP 1 and 2 are necessary and sufficient for α-ear binding (Ritter et al, 2003). Alignment of mouse NECAPs with paralogues from frog and fly reveal conservation of the last four amino acids, WVQF, while the two preceding positions are less conserved (Figure 1A). We thus designated the conserved W as position 0 (Figure 1A). Mutations were introduced into the motif in the context of the six amino-acid peptide fused to GST. For both NECAP 1 and 2, substitution of positions −1 and −2 to A did not markedly affect AP-2 binding in pull-down experiments from rat brain protein extracts (Figure 1B). In contrast, substitution of either W0 or F+3 to A abolished AP-2 binding (Figure 1C/D). Moreover, exchange of both hydrophobic positions to A within full-length NECAP 1 eliminates the co-immunoprecipitation of AP-2 seen with the wild-type protein (Figure 1E). We next substituted W0 and F+3 with other aromatic or large hydrophobic residues. Mutation of W0 to F or Y abolishes binding (Figure 1C), whereas F+3 tolerates substitution to W but not Y, L, or M (Figure 1D). These results suggest an initial consensus motif, W–X–X–[FW].
Figure 1.
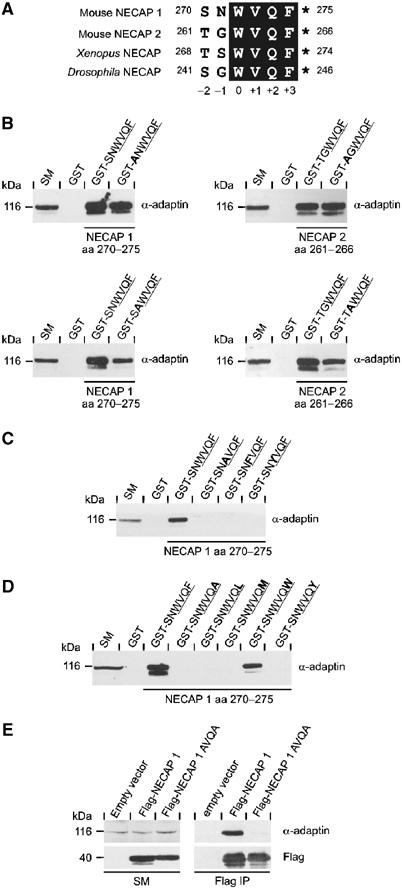
Definition of the α-ear-binding motif in NECAPs. (A) Sequence alignment of the C-terminal most six amino acids of murine NECAP 1 and 2 with NECAP isoforms from Xenopus laevis (Xenopus, gi32484237) and Drosophila melanogaster (Drosophila, gi24642653). The conserved core of the α-ear-binding motif is shaded black. The numbers before and after the sequences indicate amino-acid positions within the protein. The numbering underneath designates positions within the motif. The asterisks indicate free main chain carboxyl groups. (B–D) GST or peptides fused to GST as indicated were precoupled to glutathione–Sepharose and incubated with soluble rat brain extracts. Affinity-selected AP-2 (α-adaptin) was detected by Western blot. An aliquot of brain homogenate (starting material, SM) equal to one-tenth of that added to the fusion proteins was analyzed in parallel. The tetrapeptide cores of the motifs are underlined and bold amino acids indicate the substitutions introduced into the wild-type sequence. (B) Alanine screen for positions −1 and −2 in NECAP 1 and 2. Mutational analysis for W at position 0 (C) and F at position +3 (D). (E) Immunoprecipitation of overexpressed Flag-tagged NECAP 1 variants (Flag) and co-immunoprecipitated AP-2 (α-adaptin) were detected by Western blot. Aliquots of the cell lysates (starting material, SM) equal to one-tenth of that used for the immunoprecipitations were analyzed in parallel.
We next examined the role of positions +1 and +2. V+1 was mutated to residues within its class (small hydrophobic) or to S, a small hydrophilic amino acid. The V+1 to S mutation strongly reduced AP-2 binding (Figure 2A). A and I support binding while exchange to L blocks interaction (Figure 2A). These results suggest that the +1 position contributes to α-ear interactions. The difference in AP-2 binding for peptides bearing L or I substitutions may be explained by the fact that V is structurally closer to I than L. In contrast to the +1 position, substitution of Q+2 to A or to any hydrophilic amino acid tested did not affect binding (Figure 2B), revealing a high degree of flexibility for this position.
Figure 2.
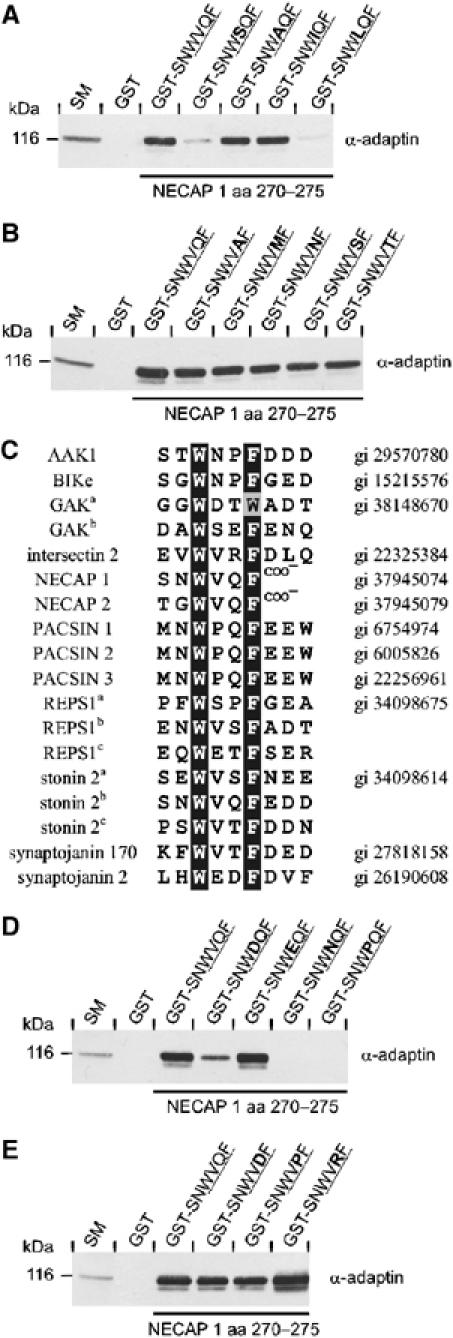
Definition of the inner core positions. (A–E) GST or peptides fused to GST as indicated were precoupled to glutathione–Sepharose and incubated with soluble rat brain extracts. Affinity-selected AP-2 (α-adaptin) was detected by Western blot. An aliquot of brain homogenate (starting material, SM) equal to one-tenth of that added to the fusion proteins was analyzed in parallel. The tetrapeptide cores of the motifs are underlined and bold amino acids indicate the substitutions introduced into the wild-type sequence. (A/D) Mutational analysis for V at position +1 and for Q at position +2 (B/E). (C) Sequence alignment of potential NECAP-like α-ear-binding motifs in proteins connected to vesicle trafficking. The conserved hydrophobic amino acids at the 0 and +3 positions are shaded black and similar residues are shaded gray. Small letters in superscript indicate distinct copies of the motif within the same protein. AAK1, adaptor-associated kinase 1; BIKe, BMP-2 inducible kinase; GAK, cyclin G-associated kinase; PACSIN, PKC and CK2 substrate in neurons; REPS1, RalBP1-associated Eps domain containing protein 1.
To further define the variability allowable at positions +1 and +2, we searched protein databases for additional proteins containing potential NECAP-like α-ear-binding motifs. Given the inflexibility of positions 0 and +3 and the relative flexibility of positions +1 and +2, the search was performed with the short peptide pattern W–X–X–F/W. Within the proteins identified, we recognized several implicated in vesicle trafficking (Figure 2C), including synaptojanin 170 and stonin 2, which have been described recently to interact with AP-2, in part through NECAP-like motifs (Jha et al, 2004; Walther et al, 2004). Strikingly, all but one motif displays an F at position +3, indicating a preference for this residue and leading to the designation of the motif as the WXXF-based α-ear-binding motif.
The alignment revealed amino acids at positions +1 and +2 that we had not tested for AP-2 binding. Introduction of N and P at position +1 within the NECAP 1 sequence abolished AP-2-binding, whereas the charged amino acids D and E were tolerated (Figure 2D). Position +2 tolerates all substitutions introduced based on the alignment (Figure 2E), clearly demonstrating a lack of contribution of this position to α-ear interactions.
The results of the pattern search with W–X–X–[FW] (Figure 2C) revealed an additional interesting detail. In the case of the NECAPs, the WXXF-based motif is found at the very C-terminus, whereas in all other proteins the motif lies within the polypeptide chain, but is followed by at least one and often two or three acidic residues. This raises the possibility that the negative charge provided by these amino acids, or by the free carboxyl group in case of the NECAPs, participates in binding. Consistent with this hypothesis, shifting the free carboxyl group of NECAP 1 three or five positions away from the core through insertion of multiple A residues eliminates AP-2 binding (Figure 3A). The fact that insertion of a single A does not affect binding (Figure 3A) indicates flexibility in the presentation of the negative charge.
Figure 3.
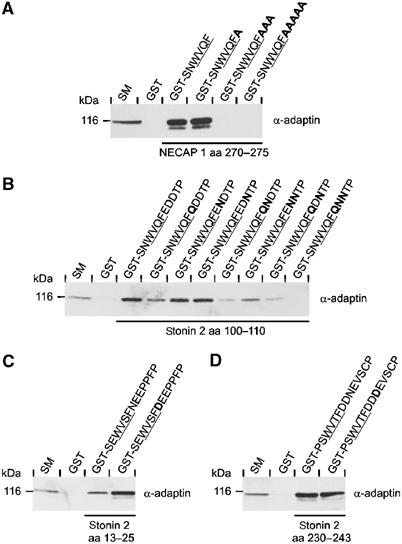
Contribution of downstream negative charge to AP-2 interaction. (A–D) GST or peptides fused to GST as indicated were precoupled to glutathione–Sepharose and incubated with soluble rat brain extracts. Affinity-selected AP-2 (α-adaptin) was detected by Western blot. An aliquot of brain homogenate (starting material, SM) equal to one-tenth of that added to the fusion proteins was analyzed in parallel. The tetrapeptide cores of the motifs are underlined and bold amino acids indicate the substitutions introduced into the wild-type sequence. (A) Contribution of the free carboxyl group in NECAP 1 to AP-2 binding. (B–D) Contribution of the downstream acidic amino acids to AP-2 binding for various motifs in stonin 2 as indicated.
To examine the contribution of acidic amino acids at positions +4 through +6 to AP-2 binding, we focused on stonin 2, which contains three WXXF-based motifs (Figure 2C) (Walther et al, 2004). One of the motifs features the tetrapeptide core WVQF, followed by EDD at positions +4 through +6. We thus introduced single, double, and triple exchanges into the acidic residues (Q for E and N for D). For single exchanges, only the loss of charge at position +4 leads to a reduction in AP-2 interaction (Figure 3B). In all cases, the double exchanges show an obvious reduction, with the strongest effects for combinations in which position +4 is targeted (Figure 3B). Finally, the simultaneous substitution of all the three acidic amino acids eliminates binding to AP-2 (Figure 3B). A second WXXF-based motif in stonin 2, WVSF, is followed by an N at position +4 with E at positions +5 and +6. Conversion of N+4 to D leads to a clear increase in AP-2 binding (Figure 3C). This result is complementary to the decrease in AP-2 interaction seen for the WVQF core upon deletion of the negative charge at position +4 (Figure 3B) and emphasizes the +4 position as having the highest impact on binding. The third core within stonin 2, WVTF, is followed by two D residues at positions +4 and +5 and an N at position +6. Replacing N+6 with D does not affect AP-2 binding (Figure 3D), in agreement with the lack of decrease in AP-2 binding for the complementary loss of charge at position +6 for the WVQF core (Figure 3B). Together with the results on the positioning of the free carboxyl group in the NECAPs, the stonin 2 mutational data demonstrate the importance of a negative charge downstream of the core for interactions with the α-ear.
In summary, two distinct qualities of the WXXF-based motif are necessary for α-ear interactions. First, the core utilizes selected residues with W0–[VAIED]+1–X+2–[FW]+3 (where X is any amino acid) as tested allowable amino acids. Second, the core needs to be conjoint with a source of negative charge at its C-terminus. This charge can be provided by either a free carboxyl group or by acidic amino acids, preferentially at the +4 position.
Binding site for WXXF-based motifs on the α-ear
We next sought to identify the binding site for the WXXF-based motif on the α-ear using NMR analysis. Backbone 1H and 15N resonance assignments demonstrate that the solution structure of the α-ear is very close to the previously determined crystal structure, with the β sandwich domain rigid with respect to the distal α–β platform domain (Owen et al, 1999; Traub et al, 1999; Brett et al, 2002; Denisov et al, 2004). Comparison of 1H–15N heteronuclear single-quantum correlation (HSQC) spectra for 15N-labeled α-ear in the absence or presence of a NECAP 1 peptide containing the WVQF motif (CQAPQPSNWVQF) revealed multiple chemical shift changes in α-ear amino acids caused by peptide interaction (Figure 4A). The magnitude of the amide chemical shift changes was plotted throughout the length of the α-ear and, interestingly, all significant shifts are located within the sandwich domain of the α-ear (Figure 4B, bottom panel). The identified binding surface, which is composed of amino acids located in β strands 2, 3, 4, and 7 (Figure 4C), is identical to a site on the sandwich domain recently described to bind DPW motifs found in epsin (Brett et al, 2002). Since NECAP 1 and epsin fail to compete for α-ear interaction (Ritter et al, 2003), we analyzed the binding behavior of a DPW peptide from epsin (CSDPWGSDPWG). Binding of this peptide to the α-ear induced amide chemical shift changes that revealed a clear preference for a site on the platform domain (Figure 4B (upper panel) and C). In fact, this site is identical to a site originally described to bind to DPF and DPW motifs (Owen et al, 1999; Traub et al, 1999) and to overlap with the site for FXDXF motifs (Brett et al, 2002). The minor shift changes observed in the sandwich domain upon DPW interaction (Figure 4B, upper panel) are consistent with a low-affinity interaction of the peptide with the sandwich domain, or they may reflect structural changes imposed on the sandwich domain upon peptide binding to the platform domain.
Figure 4.

Identification of the α-ear-binding site for WXXF-based motifs. (A) Comparison of 1H-15N HSQC spectra of the 15N-labeled α-ear in the absence (black) or presence (red) of the NECAP 1 peptide (1:1 complex). (B) Magnitude of the amide chemical shift changes ({(Δ1H shift)2+(Δ15N shift × 0.2)2}1/2 in ppm) of the α-ear upon binding CSDPWGSDPWG (epsin, upper panel), CSNWVQFEDDTP (stonin 2, middle panel), and CQAPQPSNWVQF (NECAP 1, lower panel) peptides. The indicated residue numbers correspond to mouse α-adaptin. (C) Amino-acid sequence of the α-ear shown with β-strands in the sandwich domain depicted as green arrows and β-strands and α-helices of the platform domain depicted as blue arrows and rods, respectively. Green shading represents the size of the amide chemical shift changes upon binding of the NECAP1 peptide (dark green: Δδ>0.2; light green: 0.2>Δδ>0.1 ppm). Blue shading represents the size of the amide chemical shift changes upon binding of the epsin peptide (dark blue: Δδ>0.2; light blue: 0.2>Δδ>0.1 ppm). Asterisks mark residues (N713A, Q723A, K727, F740A, and Q782) that, when mutated, affect binding to NECAP 1.
To ensure that the sandwich domain-binding site identified for the NECAP 1 peptide is common for WXXF-based α-ear-binding motifs, independent of their location at the C-terminus or within a protein, we investigated α-ear binding for two peptides derived from stonin 2 (CPSWVTFDDNEV and CSNWVQFEDDTP). Both peptides utilize the sandwich domain-binding surface identified for the NECAP 1 peptide (Figure 4B, middle panel and data not shown). Comparison of the amide chemical shift changes induced by stonin 2 and NECAP 1 peptides revealed only minor variations for positions N713, K727, and S728 of the α-ear (∼0.1 ppm, data not shown). Therefore, the sandwich domain-binding site is the interaction surface for WXXF-based motifs.
A model of the WVQF tetrapeptide bound to the α-ear was generated as described in detail in Materials and methods. The model shows W0 and F+3 binding to Q782 and K727 of the α-ear, respectively (Figure 5A). These residues display among the strongest amide chemical shift changes upon peptide binding. The orientation of the peptide is based in part on a hydrogen bond towards Q782 that was described in the crystallization studies of the epsin DPW peptide on the sandwich domain. In this case, the bond with Q782 can be provided by W but not F (Brett et al, 2002). Moreover, the differences in amide chemical shift changes for the NECAP and stonin 2 peptides involve positions N713, K727, and S728 of the α-ear, and likely reflect the peptide-specific environment for F+3. The hydrophobic side chain of V+1 contacts residues within the α-ear, as confirmed by a broadening of the NMR signals for the side-chain methyl groups of V+1 in 1D spectra of the NECAP 1 peptide (20:1 peptide/protein ratio, data not shown). In contrast, the side chain of Q+2 is oriented into solution, explaining the high degree of flexibility observed in the mutational studies (Figure 2B/E). Finally, the free carboxyl group could stabilize the interaction by forming a hydrogen bond to the positively charged amine group of K727. An electrostatic potential surface analysis of the complexed α-ear reveals a cluster of positively charged amino acids around K727 (labeled blue in Figure 5B), providing a suitable complementary environment for acidic amino acids following the core.
Figure 5.
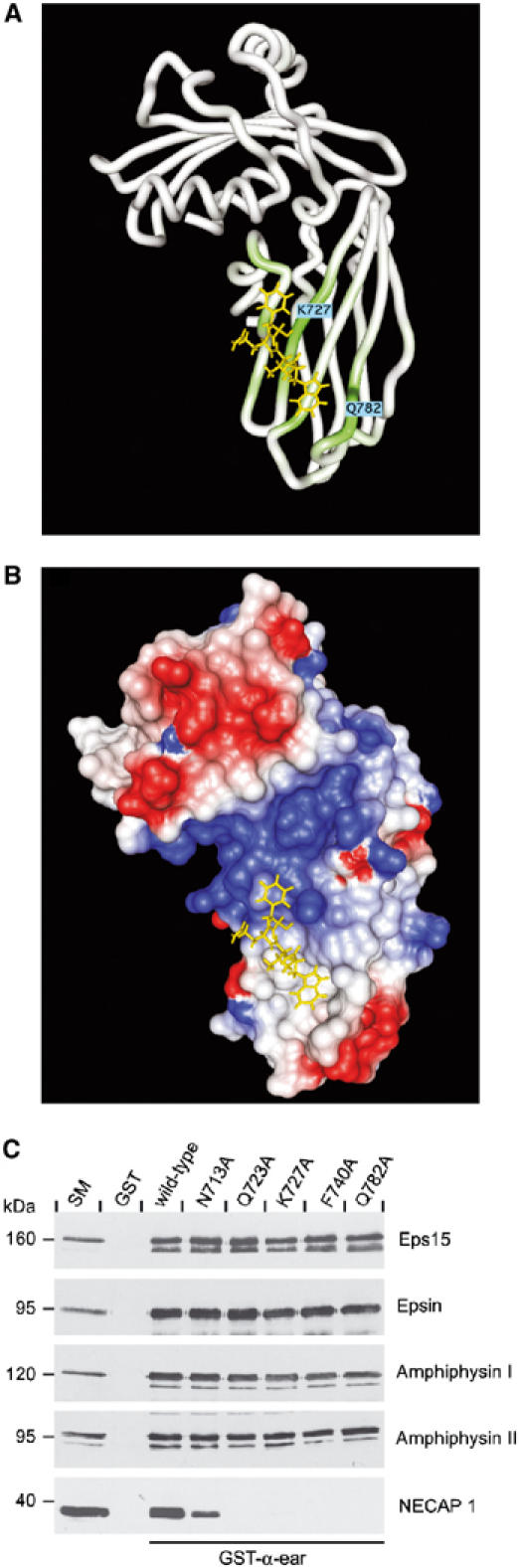
Model of the WVQF core bound to the α-ear. (A) HADDOCK modeled structure of WVQF/α-ear complex. The backbone trace of the α-ear is colored according to the size of the amide chemical shift changes upon binding of the NECAP 1 peptide. (B) Electrostatic potential surface analysis for the WVQF/α-ear complex. The surfaces are color coded, with red indicating negative electrostatic potential (E and D residues) and blue indicating positive potential (K, R, and H residues). (C) GST or GST fusion proteins of wild-type α-ear along with α-ear mutants as indicated were precoupled to glutathione–Sepharose and incubated with soluble rat brain extracts. Affinity-selected Eps15, epsin, amphiphysin I and II, and NECAP 1 were detected by Western blot. An aliquot of brain homogenate (starting material, SM) equal to one-tenth of that added to the fusion proteins was analyzed in parallel.
To test the NMR results, we examined the effect of mutations N713A, Q723A, K727A, F740A, and Q782A on α-ear binding to a series of endocytic accessory proteins. Based on our model, these mutations should affect binding to WXXF-based motifs and, in fact, all mutations reduce or abolish NECAP 1 interaction with full-length α-ear (Figure 5C). In contrast, binding of Eps15 (containing DPF motifs), epsin (containing DPW motifs), and amphiphysin I and II (containing FXDXF motifs) is unaltered (Figure 5C). This further supports that the sandwich-binding site is the interaction interface for WXXF-based motifs and suggests that epsin binding to this site is minimal. 1D-NMR and 15N-1H HSQC spectra of the various α-ear mutants reveal structures that are nearly identical to the wild-type α-ear, demonstrating that the mutants are properly folded. From NMR titration experiments, an epsin-derived peptide containing two DPW motifs and a FXDXF-containing peptide from amphiphysin I, both bind α-ear with an affinity in the range of 40–60 μM, whereas the WXXF motif-harboring peptides from NECAP 1 and stonin 2 display an affinity in the range of 20–30 μM. Thus, in isolation WXXF-based α-ear-binding motifs have a slightly higher affinity than motifs targeting the platform-binding site.
Binding specificity of WXXF-based motifs
Recent studies have characterized ∅XX∅-based motifs that bind to the ear domains of γ-adaptin and the GGA proteins (Collins et al, 2003; Duncan et al, 2003; Miller et al, 2003; Wasiak et al, 2003; Mattera et al, 2004). In addition to the α-ear-binding motif that interacts specifically with AP-2, NECAPs harbor a GAE-binding motif (WGDF) that binds AP-1/GGA specifically (Figure 6A; Ritter et al, 2003; Bai et al, 2004; Mattera et al, 2004), and mutation of aromatic residues within these motifs abolishes co-immunoprecipitation of NECAP 1 with AP-1 and AP-2 (Figure 1E and data not shown). The similarity of these motifs raises the question of how binding specificity to distinct classes of adaptors is achieved.
Figure 6.
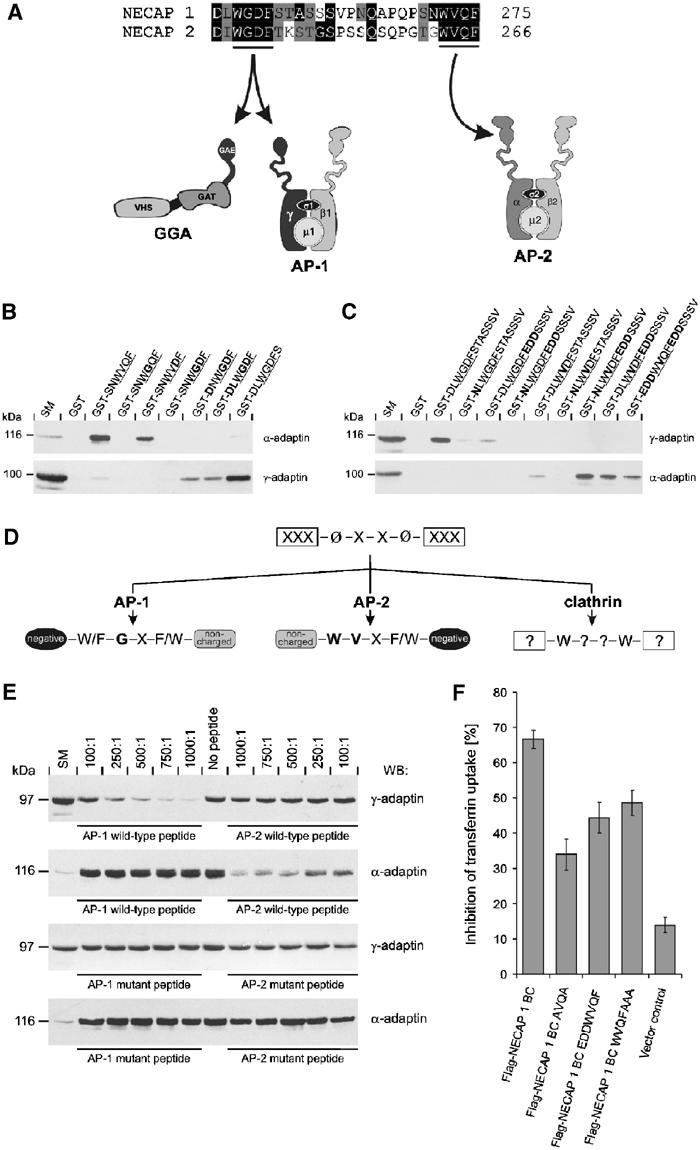
Determinants establishing binding specificity for WXXF-based α-ear- and GAE-binding motifs. (A) Sequence alignment of the C-terminal region of murine NECAP 1 and 2. Conserved amino acids are shaded black, similar residues are shaded gray. The GAE domain- and α-ear-binding motifs are underlined and arrows indicate the specific interaction of these motifs with different adaptor proteins. The numbers after the sequences indicate amino-acid position within the protein. (B/C) GST or peptides fused to GST as indicated were precoupled to glutathione–Sepharose and incubated with soluble rat brain extracts. Affinity-selected AP-2 (α-adaptin) and AP-1 (γ-adaptin) were detected by Western blot. An aliquot of brain homogenate (starting material, SM) equal to one-tenth of that added to the fusion proteins was analyzed in parallel. The tetrapeptide cores of the motifs are underlined and bold amino acids indicate the substitutions introduced into the wild-type sequence. (B) Step-by-step conversion of the α-ear-binding motif into a GAE-binding motif. (C) Step-by-step conversion of the GAE-binding motif into a α-ear-binding motif. (D) Schematic overview on parameters regulating binding specificity of W–X–X–[FW]-based motifs for interaction with GAE domains (AP-1), AP-2, and the clathrin terminal domain (clathrin). The question mark indicates undefined parameters. (E) GST-NECAP 1 was precoupled to glutathione–Sepharose and for each pull-down 100 pmol were incubated with 0.5 mg soluble brain extract without (no peptide) or in the presence of increasing concentrations of various peptides as indicated: AP-1 wild-type peptide, CSNDLWGDFSTAS; AP-1 mutant peptide, CSNNLWVDFSTAS; AP-2 wild-type peptide, CQAPQPSNWVQF; AP-2 mutant peptide, CQAPQPSNWGQFAAA. The molar ratio of peptide to fusion protein is indicated and affinity-selected AP-1 (γ-adaptin) and AP-2 (α-adaptin) were detected by Western blot. Aliquots of brain homogenate (starting material, SM) equal to one-tenth of that added to the fusion proteins were analyzed in parallel. (F) COS-7 cells were transfected with the empty expression vector (vector control) or with Flag-tagged NECAP 1 BC variants as indicated and the percentage of cells that fail to endocytose Cy3-labeled transferrin was quantified (mean±s.e.m., n=5).
A negative charge downstream of WXXF-based α-ear binding motifs appears essential for interaction with AP-2 and all GAE-binding motifs reported thus far are preceded by at least one acidic amino acid (Collins et al, 2003; Bonifacino, 2004). Therefore, the positioning of negative charge on either side of a WXXF-based core may influence its specific binding to ear domains. Another important determinant is likely the +1 position, which within the GAE-binding motif is often occupied by a G (Mattera et al, 2003, 2004; Miller et al, 2003). The +1 position of the NECAP α-ear-binding motif is restricted in its ability to accept mutations (Figure 2) and a V+1 to G exchange abolishes AP-2 interaction (Figure 6A). Thus, the +1 position might function as a switch that allows interaction with one type of ear domain while preventing interaction with the other.
To test these hypotheses, we used a series of point mutations to sequentially vary the three potential determinants (the negative charge in front of the core, the negative charge following the core, and position +1 within the core) in an attempt to convert the AP-2 motif to an AP-1 motif (Figure 6B), and, conversely, to mutate the AP-1 motif to an AP-2 motif (Figure 6C). Full binding to AP-1 could only be achieved when the +1 core position was occupied by G and an acidic residue was located in front of the core. Remarkably, even in this context, the presence of a negative charge immediately following the core was inhibitory to AP-1 binding (Figure 6B/C). To convert the AP-1 site to allow for AP-2 interaction, we first had to mutate the G+1 to V. The addition of negative charge following the core was also required for AP-2 binding. Interestingly and converse to the situation for AP-1 binding, addition of acidic residues preceding the core reduced AP-2 interactions (Figure 6B/C).
In summary, the specificity of WXXF-based motifs for interaction with AP-1 and AP-2 is mainly dependent on the +1 position within the core and the acidic context surrounding the core (Figure 6D). Preceding acidic residues favor AP-1 binding, while downstream negative charges enable AP-2 binding. In both cases, the presence of additional negative charges on the ‘wrong' side of the core creates a less favorable environment for interaction, resulting in reduction or loss of binding. Accordingly, peptides carrying wild-type versions of the α-ear- and GAE domain-binding motifs of NECAP 1 specifically interfere in a concentration-dependent manner with GST-NECAP 1 binding to AP-2 and AP-1, respectively, without influencing NECAP 1 interaction with the other adaptor complex (Figure 6E). Variations of these peptides, in which the +1 position of either motif has been switched and acidic charges have been mispositioned, fail to interfere with GST-NECAP 1 binding to either adaptor complex (Figure 6E).
Overexpression of a Flag-tagged NECAP 1 C-terminal fragment (NECAP 1 BC) interferes with transferrin endocytosis in a dominant-negative manner (Figure 6F; Ritter et al, 2003). Importantly, this endocytic block is significantly reduced for a NECAP 1 BC variant, in which the hydrophobic residues at positions 0 and +3 have been exchanged to A (Figure 6F), a mutation that disrupts AP-2 binding (Figure 1E). Thus, the residual endocytic block must be independent of AP-2 interactions. As for the 0/+3 mutation, introduction of acidic amino acids preceding the core and insertion of multiple A residues following the core reduce the inhibitory effect of the NECAP 1 BC fragment (Figure 6F). These experiments underline the necessity of a functional AP-2-binding motif for NECAP endocytic function.
Discussion
In this study, we have systematically characterized a novel WXXF-based α-ear-binding motif originally identified in NECAPs (Ritter et al, 2003) and also found to function in the endocytic accessory proteins synaptojanin 170, stonin 2, adaptin-associated kinase 1 (AAK1), and cyclin G-associated kinase (GAK) (Jha et al, 2004; Walther et al, 2004). The motif engages a surface on the α-ear sandwich domain composed of β-strands 2, 3, 4, and 7, anchored around residues Q782 and K727. This site was formerly identified as a low-affinity-binding site for DPW motifs within epsin (Brett et al, 2002). However, NMR analysis revealed that a DPW-based peptide from epsin interacts primarily with a site in the platform domain and shows only weak interaction with the sandwich domain-binding site. Moreover, mutations N713A, Q723A, F740A, K727A, and Q782A within the sandwich domain reduce or eliminate NECAP binding, with no detectable effect on epsin interaction. Thus, the sandwich domain-binding site appears to be used primarily for binding to WXXF-based α-ear-binding motifs.
Examination of the residues in WXXF-based motifs that allow α-ear binding, W0–[VAIED]+1–X+2–[FW]+3, reveals a common basic foundation with the core of the GAE-binding motif, which has been systematically determined as [FWY]0–G+1–[PDE]+2–[FWYLM]+3 (Mattera et al, 2004). Most notable are the bulky hydrophobic amino acids at positions 0 and +3. These residues likely mediate the major bond energy for interaction, but they also provide the first degree of specificity. While the GAE-binding motif appears to tolerate any bulky hydrophobic amino acid at either position 0 or +3, the α-ear-binding motif only allows for W at position 0 and F and W at +3. The two motifs also differ in the functions assigned to the inner core positions. For the GAE-binding motif, the inner positions need to shape the peptide into a type I β turn in order to present both hydrophobic positions towards the GAE domain. G is found predominantly at the +1 position as this amino acid provides the conformational flexibility necessary to shape the peptide (Miller et al, 2003; Mattera et al, 2004). In contrast, the side chain of the +1 position in the WXXF-based motif contacts the α-ear and contributes to the interaction, while the +2 position points into solution and is freely interchangeable. These differences, coupled with the observation that the binding sites for the motifs are located on opposite faces of the highly related GAE domains and α-ear sandwich domain (Kent et al, 2002; Nogi et al, 2002; Collins et al, 2003; Miller et al, 2003), suggest that the motifs are a result of co-evolution as opposed to the possibility that one subsequently derived from the other. In this regard, it will be of interest to determine how these motifs compare to the PWXXW motif that binds to the clathrin terminal domain (Figure 6D) (Ramjaun and McPherson, 1998; Slepnev et al, 2000; Drake and Traub, 2001; Miele et al, 2004). Neither NECAP core supports interaction with clathrin and the requirements of the PWXXW motif await further definition.
Our analysis demonstrates that the WXXF cores alone are not sufficient for binding to ear domains. For example, acidic residues upstream of GAE-binding motifs are necessary for AP-1 interaction (Figure 6C). Thus far, different conclusions have been drawn regarding the role of acidic residues preceding the GAE-binding motif. For example, D−1 of the GAE-binding motif in rabaptin-5 directly contacts a positively charged residue in the GAE domain of GGA3, providing secondary stabilization beyond the core (Miller et al, 2003). In contrast, Collins et al (2003) failed to detect stable contacts for the preceding acidic residues in p56 for binding to the GGA1 ear. The authors therefore proposed nonspecific electrostatic attractions towards the GAE domain and/or a function in destabilizing secondary structures within the ligand, ensuring exposure of the motif within the protein chain. In addition to the importance of upstream acidic residues, we have determined that acidic residues C-terminal to the GAE-binding core are inhibitory. The destabilizing effect of negative charge on the ‘wrong' side of the core is likely due to the substitution of positions +4 through +6, which appear to contribute to binding (Miller et al, 2003; Wasiak et al, 2003).
Conversely to the GAE domain-binding motifs, acidic residues upstream of the WXXF-based α-ear-binding motif interfere with binding, which instead requires C-terminal acidic residues or a free carboxyl group (Figure 6D). The contribution of the carboxyl group to binding is not without precedent and has been described for the C-terminal PDZ domain-binding motif S/TXV, where the free carboxyl group of the ligand contacts several positions within the PDZ domain through main-chain amide interactions and also targets an arginine side chain to stabilize the carboxylate ion (Doyle et al, 1996; Kornau et al, 1997). Moreover, in the yeast homologues of epsin and AP180, the clathrin box is located at the very C-terminus and the last acidic residue of the motif is replaced by the free carboxy group of the protein chain (Wendland and Emr, 1998; Wendland et al, 1999). As for NECAPs, shifting the carboxyl group away from the motif through insertion of multiple A residues abolishes binding to the target protein (Wendland et al, 1999). Our NMR studies on the WXXF-based α-ear-binding motifs in NECAP 1 and stonin 2 revealed only small differences in the amide chemical shift changes. We failed to detect any directed bonds between the α-ear and the acidic residues and these amino acids may thus contact the ear only weakly. Even so, the following negative charge is oriented towards a large positively charged surface of the sandwich domain and the residues contributing to this cluster reveal the same high degree of evolutionary conservation as seen for the positions directly involved in interaction (Traub et al, 1999). A secondary contribution to binding with stabilizing effects is thus a likely scenario. The inhibitory effect of upstream acidic residues likely reflects an unfavorable pairing of the negative charge with residues in the α-ear.
The platform domain binds proteins containing DPF/W and FXDXF motifs and the resulting competition between the endocytic accessory proteins that utilize these motifs may necessitate that they function at distinct endocytic steps. Although WXXF-based motifs will compete with each other for the sandwich-binding site, they will not compete with DPF/W and FXDXF-bearing proteins. WXXF-based ligands may therefore contribute an additional regulatory level for AP-2 function. In this regard, it is interesting that several sandwich-binding partners appear to have a direct influence on the function of AP-2 itself. Synaptojanin regulates PI(4,5)P2 levels and this phospholipid docks AP-2 to the plasma membrane (Cremona et al, 1999; Krauss et al, 2003). AAK1 and stonin 2 regulate cargo recognition of the μ2 subunit and the kinases AAK1 and GAK both use AP-2 as substrate (Umeda et al, 2000; Conner and Schmid, 2002; Korolchuk and Banting, 2002; Conner et al, 2003; Walther et al, 2004). It will be of interest to determine if the NECAPs also have a direct regulatory role on AP-2 function. Moreover, the simultaneous presence of α-ear-binding motifs for the platform and sandwich-binding sites as seen in proteins such as synaptojanin 170, AAK1, and GAK could allow these proteins to bridge the α-ear subdomains. This could generate an avidity effect and allow a protein to gain access to the α-ear even in the presence of competing α-ear-binding proteins that harbor only one class of motif. Though the NECAPs lack binding sequences for the platform-binding site, they are the only proteins described thus far to simultaneously harbor WXXF-based motifs designated for GAE domain and α-ear interaction, and might therefore influence clathrin-mediated events at the plasma membrane and the TGN in a common manner.
Materials and methods
Antibodies and peptides
Monoclonal antibodies for α- and γ-adaptin were from BD Transduction Laboratories, and the monoclonal Flag antibody (M2) was from Sigma. Polyclonal serum against Eps15 was from Santa Cruz. Antibodies for amphiphysin I and II were described previously (Ramjaun et al, 1997) and antibody against epsin was a generous gift of Dr L Traub. A polyclonal NECAP 1 serum was raised in rabbits against a synthetic peptide containing amino acids 207–226 of the mouse NECAP 1 sequence (gi27229051) coupled to KLH. Synthetic peptides were purchased from HHMI/Keck Biotechnology Resource Laboratory, Yale University.
Expression constructs
N-terminal GST-tagged peptides for NECAP 1 and 2 and stonin 2 were generated by annealing complementary oligos with subsequent subcloning into pGEX-4T1 (Amersham). For the exact nucleotide sequence of each oligo (see Supplementary data Table I). GST-α-ear was previously described (Ritter et al, 2003) and point mutations were introduced using the megaprimer procedure (Barik, 1993). For NMR analysis, the cDNA encoding amino acids 700–938 of mouse α-adaptin (gi90292) was subcloned into pGEX-2TK (Amersham). For oligos used for all PCR-based manipulations of the α-ear (see Supplementary data Table I). Full-length GST-tagged NECAP 1 was subcloned into pGEX-4T1 as described previously (Ritter et al, 2003). Flag-tagged constructs of full-length NECAP 1 variants and the C-terminal NECAP 1 BC variants were generated by PCR from the full-length cDNA, with subsequent subcloning into pcDNA3 with integrated N-terminal Flag tag. For oligos used to amplify the various NECAP 1 constructs and for generation of the Flag tag (see Supplementary data Table I). All constructs were confirmed by sequence analysis.
Binding studies
Triton X-100-solubilized rat brain extracts were prepared as previously described (Wasiak et al, 2002) in 10 mM HEPES, pH 7.4, supplemented with protease inhibitors and 150 mM NaCl. In each case, aliquots of 2 mg were incubated for 1 h at 4°C with GST fusion protein precoupled to glutathione–Sepharose. For competition pull-down assays, 0.5 mg aliquots of brain extract were incubated for 1 h at 4°C with 100 pmol GST-NECAP 1 precoupled to glutathione–Sepharose in the absence or presence of synthetic peptides at a range of molar ratios relative to the fusion protein as indicated in the figure. For co-immunoprecipitation studies, HEK-293T cells, 48 h post transfection, were resuspended in 10 mM HEPES, pH 7.4, 1% Triton X-100, supplemented with protease inhibitors and 90 mM NaCl, lysed by sonication, and centrifuged at 200 000 g. Aliquots of the supernatant representing 50% of a confluent 10 cm dish were incubated for 3 h at 4°C with 3 μg Flag antibody and 20 μl protein G-agarose.
Endocytosis assay
The assay was performed as described previously (Ritter et al, 2003). Briefly, COS-7 cells were plated on poly-L-lysine-coated coverslips and transfected with Flag-tagged NECAP 1 BC variants. Approximately 48 h after transfection, cells were serum-starved for 30 min, incubated with Cy3-labeled transferrin (25 μg/ml) for 30 min at 37°C and then processed for immunofluorescence with anti-Flag antibody.
NMR spectroscopy
The preparation of 15N-labeled α-ear was described previously (Denisov et al, 2004). The NMR samples contained 0.2 mM protein in 90% H2O/10% D2O, 25 mM sodium phosphate (pH 7.3), 75 mM NaCl, 0.5 mM EDTA, and 5 mM DTT. NMR spectra were acquired at 30°C on a Bruker DRX-600 spectrometer equipped with a triple resonance cryoprobe. Detailed analysis of peptide binding to the α-ear was carried out by comparison of chemical shifts for backbone amide signals in 1H–15N HSQC spectra. HSQC spectra were recorded at 1:20, 1:4, 1:2, 1:1, and 2:1 peptide/protein ratios that were confirmed by UV concentration of both components and intensity of Hɛ1 (W) signals in 1D-NMR.
Determination of dissociation constants
The dissociation constant (Kd) of various motifs was estimated by NMR titrations and analysis of the dependency of chemical shifts of α-ear HSQC signals from peptide concentration. The results for the individual WXXF-based peptides correspond to intermediate exchange on the NMR time scale, and the DPW- or FXDXF-containing peptides correspond to the fast exchange (Hajduk et al, 1999).
α-ear/peptide complex modeling and surface charge analysis
The High Ambiguity Driven protein–protein Docking (HADDOCK) approach (Dominguez et al, 2003) was used to model the binding of a WVQF tetrapeptide to the α-ear domain. We used PDB entry 1KY6 for the structure of the α-ear domain (Brett et al, 2002) and generated a PDB template for the tetrapeptide using standard CNS scripts from ARIA (Nilges et al, 1997). ‘Active' residues were defined as α-ear domain residues with a chemical shift deviation greater than 0.35 ppm (714, 724, 726–728, 743, 781–783) as well as all four residues of the tetrapeptide. Passive residues on the α-ear domain were defined as surface neighbors of active residues. Active and passive residues are used by HADDOCK to define ambiguous intermolecular distance restraints to drive the simulated annealing molecular modeling protocol. To mimic the binding of a DPW peptide to the α-ear domain (Brett et al, 2002), we also introduced a hydrogen bond restraint between the side-chain oxygen of Q782 and the aromatic NH of the tetrapeptide W, and we used standard HADDOCK hydrogen bond energy constants multiplied by 10. The HADDOCK ‘interface' (residues allowed to move during the semi-flexible simulated annealing) was defined as all active and passive residues ±2 sequential residues. A clustering with a 1 Å cutoff was performed among the resulting structural models of the complex and the lowest energy model in the major cluster was selected. Structure figures were prepared with the program MOLMOL (Koradi et al, 1996). The electrostatic surface potential of the protein was also calculated with MOLMOL (Koradi et al, 1996) as described in the software tutorial (example 4). Residue names in the PDB file were corrected to take charges into account and the CalcPot function was used with the default parameters, taking into account the effects of the solvent and appropriate dielectric constants for the protein and solvent.
Note added in Proof
Through a mutational analysis, Mishra et al (J Biol Chem, Epub ahead of print) have independently identified a similar site on the sandwich domain of the α-ear for WXXF-based motifs as that reported here.
Supplementary Material
Supplementary data Table I
Acknowledgments
We thank W Sossin and S Wasiak for discussion, L Traub for epsin antibody, and P Bhagatji for production of NMR samples. This work was supported by Canadian Institutes of Health Research grant MOP-121478 to PSM. Operating grants from the Genome Quebec project Réseau Protéomique de Montréal, Montreal Proteomics Network financially supported this work. BR was supported by a Jeanne Timmins-Costello fellowship from the Montreal Neurological Institute. PSM is a CIHR Investigator, a Killam Scholar, and a McGill University William Dawson Scholar.
References
- Bai H, Doray B, Kornfeld S (2004) GGA1 interacts with the adaptor protein AP-1 through a WNSF sequence in its hinge region. J Biol Chem 279: 17411–17417 [DOI] [PubMed] [Google Scholar]
- Barik S (1993) PCR protocols. In Methods in Molecular Biology, Vol. 15, Tolowa: Humana Press Inc [Google Scholar]
- Blondeau F, Ritter B, Allaire PD, Wasiak S, Girard M, Hussain NK, Angers A, Legendre-Guillemin V, Roy L, Boismenu D, Kearney RE, Bell AW, Bergeron JJM, McPherson PS (2004) Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc Natl Acad Sci USA 101: 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS (2004) The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol 5: 23–32 [DOI] [PubMed] [Google Scholar]
- Brett TJ, Traub LM, Fremont DH (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure (Camb) 10: 797–809 [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol 17: 517–568 [DOI] [PubMed] [Google Scholar]
- Collins BM, Praefcke GJ, Robinson MS, Owen DJ (2003) Structural basis for binding of accessory proteins by the appendage domain of GGAs. Nat Struct Biol 10: 607–613 [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL (2002) Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J Cell Biol 156: 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner SD, Schmid SL (2003) Regulated portals of entry into the cell. Nature 422: 37–44 [DOI] [PubMed] [Google Scholar]
- Conner SD, Schroter T, Schmid SL (2003) AAK1-mediated micro2 phosphorylation is stimulated by assembled clathrin. Traffic 4: 885–890 [DOI] [PubMed] [Google Scholar]
- Cremona O, Di Paolo G, Wenk MR, Luthi A, Kim WT, Takei K, Daniell L, Nemoto Y, Shears SB, Flavell RA, McCormick DA, De Camilli P (1999) Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99: 179–188 [DOI] [PubMed] [Google Scholar]
- Denisov AY, Ritter B, McPherson PS, Gehring K (2004) Letter to the Editor: 1H, 15N and 13C resonance assignments and 15N-1H residual dipolar couplings for the alpha-adaptin ear-domain. J Biomol NMR 29: 441–442 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737 [DOI] [PubMed] [Google Scholar]
- Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R (1996) Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Drake MT, Traub LM (2001) Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J Biol Chem 276: 28700–28709 [DOI] [PubMed] [Google Scholar]
- Duncan MC, Costaguta G, Payne GS (2003) Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat Cell Biol 5: 77–81 [DOI] [PubMed] [Google Scholar]
- Hajduk PJ, Gerfin T, Boehlen JM, Haberli M, Marek D, Fesik S (1999) High-throughput nuclear magnetic resonance-based screening. J Med Chem 42: 2315–2317 [DOI] [PubMed] [Google Scholar]
- Hinners I, Tooze SA (2003) Changing directions: clathrin-mediated transport between the Golgi and endosomes. J Cell Sci 116: 763–771 [DOI] [PubMed] [Google Scholar]
- Jha A, Agostinelli NR, Mishra SK, Keyel PA, Hawryluk MJ, Traub LM (2004) A novel AP-2 adaptor interaction motif initially identified in the long-splice isoform of synaptojanin 1, SJ170. J Biol Chem 279: 2281–2290 [DOI] [PubMed] [Google Scholar]
- Kent HM, McMahon HT, Evans PR, Benmerah A, Owen DJ (2002) Gamma-adaptin appendage domain: structure and binding site for Eps15 and gamma-synergin. Structure (Camb) 10: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- Kornau HC, Seeburg PH, Kennedy MB (1997) Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol 7: 368–373 [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Banting G (2002) CK2 and GAK/auxilin2 are major protein kinases in clathrin-coated vesicles. Traffic 3: 428–439 [DOI] [PubMed] [Google Scholar]
- Krauss M, Kinuta M, Wenk MR, De Camilli P, Takei K, Haucke V (2003) ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J Cell Biol 162: 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Arighi CN, Lodge R, Zerial M, Bonifacino JS (2003) Divalent interaction of the GGAs with the Rabaptin-5–Rabex-5 complex. EMBO J 22: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Ritter B, Sidhu SS, McPherson PS, Bonifacino JS (2004) Definition of the consensus motif recognized by {gamma}-adaptin ear domains. J Biol Chem 279: 8018–8028 [DOI] [PubMed] [Google Scholar]
- Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ (2004) Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat Struct Mol Biol 11: 242–248 [DOI] [PubMed] [Google Scholar]
- Miller GJ, Mattera R, Bonifacino JS, Hurley JH (2003) Recognition of accessory protein motifs by the gamma-adaptin ear domain of GGA3. Nat Struct Biol 10: 599–606 [DOI] [PubMed] [Google Scholar]
- Nilges M, Macias MJ, O'Donoghue SI, Oschkinat H (1997) Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin. J Mol Biol 269: 408–422 [DOI] [PubMed] [Google Scholar]
- Nogi T, Shiba Y, Kawasaki M, Shiba T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Takatsu H, Nakayama K, Wakatsuki S (2002) Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain. Nat Struct Biol 9: 527–531 [DOI] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Noble ME, Hunter JB, Dafforn TR, Evans PR, McMahon HT (1999) A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell 97: 805–815 [DOI] [PubMed] [Google Scholar]
- Ramjaun AR, McPherson PS (1998) Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J Neurochem 70: 2369–2376 [DOI] [PubMed] [Google Scholar]
- Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS (1997) Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem 272: 16700–16706 [DOI] [PubMed] [Google Scholar]
- Ritter B, McPherson PS (2004) Molecular mechanisms in clathrin-mediated membrane budding. Topics in Current Genetics, Keränen S, Jantti J (eds), Regulatory Mechanisms of Intracellular Membrane Transport. Springer-Verlag: Berlin, Heidelberg [Google Scholar]
- Ritter B, Philie J, Girard M, Tung EC, Blondeau F, McPherson PS (2003) Identification of a family of endocytic proteins that define a new alpha-adaptin ear-binding motif. EMBO Rep 4: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS (2004) Adaptable adaptors for coated vesicles. Trends Cell Biol 14: 167–174 [DOI] [PubMed] [Google Scholar]
- Slepnev VI, Ochoa GC, Butler MH, De Camilli P (2000) Tandem arrangement of the clathrin and AP-2 binding domains in amphiphysin 1 and disruption of clathrin coat function by amphiphysin fragments comprising these sites. J Biol Chem 275: 17583–17589 [DOI] [PubMed] [Google Scholar]
- Traub LM, Downs MA, Westrich JL, Fremont DH (1999) Crystal structure of the alpha appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc Natl Acad Sci USA 96: 8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda A, Meyerholz A, Ungewickell E (2000) Identification of the universal cofactor (auxilin 2) in clathrin coat dissociation. Eur J Cell Biol 79: 336–342 [DOI] [PubMed] [Google Scholar]
- Walther K, Diril MK, Jung N, Haucke V (2004) Functional dissection of the interactions of stonin 2 with the adaptor complex AP-2 and synaptotagmin. Proc Natl Acad Sci USA 101: 964–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S, Denisov AY, Han Z, Leventis PA, de Heuvel E, Boulianne GL, Kay BK, Gehring K, McPherson PS (2003) Characterization of a gamma-adaptin ear-binding motif in enthoprotin. FEBS Lett 555: 437–442 [DOI] [PubMed] [Google Scholar]
- Wasiak S, Legendre-Guillemin V, Puertollano R, Blondeau F, Girard M, de Heuvel E, Boismenu D, Bell AW, Bonifacino JS, McPherson PS (2002) Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J Cell Biol 158: 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, Emr SD (1998) Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein–protein interactions essential for endocytosis. J Cell Biol 141: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, Steece KE, Emr SD (1999) Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J 18: 4383–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data Table I


