Abstract
The DNA replication machinery stalls at damaged sites on templates, but normally restarts by switching to a specialized DNA polymerase(s) that carries out translesion DNA synthesis (TLS). In human cells, DNA polymerase η (polη) accumulates at stalling sites as nuclear foci, and is involved in ultraviolet (UV)-induced TLS. Here we show that polη does not form nuclear foci in RAD18−/− cells after UV irradiation. Both Rad18 and Rad6 are required for polη focus formation. In wild-type cells, UV irradiation induces relocalization of Rad18 in the nucleus, thereby stimulating colocalization with proliferating cell nuclear antigen (PCNA), and Rad18/Rad6-dependent PCNA monoubiquitination. Purified Rad18 and Rad6B monoubiquitinate PCNA in vitro. Rad18 associates with polη constitutively through domains on their C-terminal regions, and this complex accumulates at the foci after UV irradiation. Furthermore, polη interacts preferentially with monoubiquitinated PCNA, but polδ does not. These results suggest that Rad18 is crucial for recruitment of polη to the damaged site through protein–protein interaction and PCNA monoubiquitination.
Keywords: PCNA, polymerase η, Rad6, Rad18, ubiquitination
Introduction
Exposure of cells to ultraviolet (UV) light causes several types of DNA damage. Among these, cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts are major DNA lesions. In normal vertebrate cells, 6-4 photoproducts are efficiently repaired by nucleotide-excision repair, but nearly 50% of CPDs remain unrepaired even at 24 h after UV irradiation (Mitchell and Nairn, 1989). In such a situation, the DNA replication machinery often encounters the lesion during the S-phase of the cell cycle, and stalls at the replication fork, resulting in a gap opposite the site of damage in the newly synthesized DNA strand. Cell death may be imminent unless the gap is filled. This gap-filling process is operationally defined as postreplication repair (PRR), which is characterized by restarting of DNA replication without removal of the lesion on a template strand. PRR is observed in diverse species from Escherichia coli to humans. It is hypothesized that PRR is mediated by either translesion DNA synthesis (TLS) or recombination to resolve the stalled replication fork (Broomfield et al, 2001).
In the budding yeast Saccharomyces cerevisiae, genes belonging to the RAD6 epistasis group are involved in the PRR pathway, where Rad18 (a putative ubiquitin ligase) and Rad6 (a ubiquitin-conjugating enzyme, E2) play a pivotal role (Bailly et al, 1994, 1997a). rad6 and rad18 mutants are highly susceptible to various DNA-damaging agents including UV and methylmethanesulfonate (MMS) (Hynes and Kunz, 1981). rad6 and rad18 mutants, however, show reduced mutation frequency following treatments with UV and MMS, possibly because error-prone TLS does not work without Rad18/Rad6. Because Rad18 protein binds to single-stranded DNA and forms a tight complex with Rad6 protein (Bailly et al, 1994, 1997b), it is proposed that Rad18 recruits Rad6 protein at replication stalling sites through binding to gap regions, and that the Rad18 complex ubiquitinates some target molecules on the stalled replication forks. Recently, proliferating cell nuclear antigen (PCNA) was shown to be monoubiquitinated in a Rad18/Rad6-dependent manner, which is necessary for tolerance to DNA damage (Hoege et al, 2002; Stelter and Ulrich, 2003). Interaction with PCNA is essential for the function of Rad30 (Haracska et al, 2001a), a yeast homolog of polymerase η, which is a member of RAD6 epistasis group (McDonald et al, 1997). These results suggest that PCNA might be a major target of Rad18/Rad6 in the PRR process.
In vertebrate cells, thus far only a single homolog of RAD18 has been identified (Tateishi et al, 2000). Human and mouse Rad18 interacts with two forms of the Rad6 homolog, Rad6A and Rad6B, both in vitro and in vivo (Tateishi et al, 2000, 2003; Xin et al, 2000). RAD18 knockout mouse embryonic stem (ES) cells and chicken DT40 cells manifest sensitivity to various DNA-damaging agents and enhanced genomic instability as determined by increased sister-chromatid exchange (SCE) and frequency of stable transformation (Yamashita et al, 2002; Tateishi et al, 2003).
Vertebrate polymerase η (polη), a homolog of the RAD30 gene product of the yeast, is a member of a recently discovered Y-family of novel DNA polymerases including polι and polκ (Burgers et al, 2001; Ohmori et al, 2001). They are shown to be involved in TLS in vitro, and structurally related to each other, but unrelated to the replicative polymerases (polδ and polɛ). Polη has a highly distributive, rather than processive, mode of DNA synthesis on undamaged templates and a relatively low stringency (Johnson et al, 2000; Matsuda et al, 2000). However, polη can insert correct nucleotides opposite CPDs in TLS (Johnson et al, 1999; McCulloch et al, 2004). The gene encoding polη is mutated in a cancer-prone hereditary disorder, xeroderma pigmentosum variant (XPV) (Masutani et al, 1999). It is possible that without normal polη CPD becomes highly mutagenic probably due to TLS by some other error-prone polymerase(s), resulting in skin cancers of sun-exposed areas.
When the replicative machinery encounters unrepaired CPD lesions, it is expected that the replicative polymerase is switched to polη to carry out TLS in normal cells. In UV-irradiated human cells, polη forms discrete nuclear foci in a UV-dose- and time-dependent manner (Kannouche et al, 2001). The sites of these foci are colocalized with PCNA, suggesting that these are sites of stalled replication. Because a polη deletion mutant, which has a polymerase activity but does not show focus formation following UV irradiation, cannot complement the sensitivity of XPV cells to UV irradiation, foci formation is an essential step of polη function. However, molecular mechanisms of how polη forms nuclear foci in UV-irradiated cells are largely unknown.
In the study reported here, in order to understand the role of Rad18 in tolerance to UV-induced DNA damage, we used RAD18−/− mouse fibroblasts from RAD18 knockout mice to show that Rad18 functions as an essential coordinator of the formation of polη foci through PCNA monoubiquitination and physical interaction with polη.
Results
Requirement of Rad18 for PCNA monoubiquitination
To investigate the role of Rad18 in UV-induced TLS, we established cell lines from RAD18 knockout mice (Tateishi et al, 2003). These cells did not express detectable levels of Rad18 protein, but showed normal levels of Rad6A/B (Figure 1A) and normal growth rates (Figure 1B). In wild-type (WT) cells, a band of PCNA corresponding to 44 kDa increased in a UV dose- and time-dependent manner, while in RAD18−/− cells, the band remained at the control level up to 8 h after UV irradiation even at 40 J/m2 (Figure 1C and D). Similar modification of PCNA in MMS-treated HeLa cells was reported (Hoege et al, 2002). We concluded that this band represented a monoubiquitinated form of PCNA for two reasons. (i) Unmodified PCNA was detected at 36 kDa in SDS–PAGE (Figure 1C–E) and, when lysates of UV-irradiated cells expressing transfected HA-tagged ubiquitin were immunoprecipitated, bands of 45 and 44 kDa were detected by immunoblotting with an anti-HA antibody and anti-PCNA antibody, respectively (Figure 1E, lanes 2 and 3). (ii) Unmodified PCNA was converted to a 44 kDa band of monoubiquitinated PCNA in vitro by purified Rad18 and Rad6B of human origin plus ubiquitin (Figure 1E, lanes 7, 11, and 12). When ubiquitin was replaced with FLAG-tagged ubiquitin in this system, a 45 kDa band appeared (Figure 1E, lane 13). These results indicate that Rad18 is a ubiquitin ligase for the monoubiquitination of PCNA.
Figure 1.
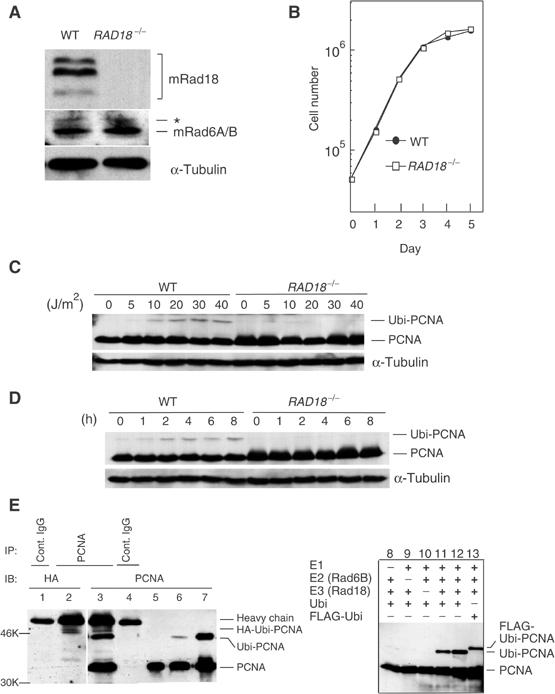
Rad18 dependent monoubiquitination of PCNA by Rad18 and Rad6A/B in vivo and in vitro. (A) Western blot of Rad18 and Rad6A/B in RAD18−/− cells. α-Tubulin was included as a control. An asterisk shows nonspecific bands. (B) Growth curves of RAD18−/− cells. (C, D) Monoubiquitination of PCNA as determined by Western blot. Cells were harvested 5 h later following various doses of UV irradiation (C). In (D), cells were irradiated at 30 J/m2 and harvested at the indicated times. (E) In vivo (left, lanes 1–6) and in vitro (right, lanes 7–13) monoubiquitination of PCNA. GM637 cells were transfected with HA-ubiquitin (lanes 1–4) and irradiated with UV (13 J/m2, 6 h). Lysates were immunoprecipitated and blotted as indicated. In lanes 5 and 6, GM637 cells without transfection were irradiated at 0 and 13 J/m2 (6 h), respectively. Lane 7 represents an in vitro ubiquitination product. In lane 12, two-fold amounts of E2 and E3 were included in the reaction.
In budding yeast, Rad18 binds to Rad6 through its Rad6-binding domain (R6BD) (Bailly et al, 1997b). This domain is highly conserved among various species. To confirm that the putative R6BD in hRad18 (amino-acid residues 340–395; Figure 2A) was a binding site for Rad6A/B, we transfected a Rad18 plasmid lacking R6BD together with a Rad6A/B plasmid into COS-7 cells, and performed co-immunoprecipitation experiments. Rad18 protein lacking R6BD localized in the nuclei like WT Rad18 (data not shown), but did not interact with human Rad6A/B (Figure 2A, lanes 1, 2, 5, and 6). To confirm whether the failure of PCNA ubiquitination in RAD18−/− cells was really due to a defect in Rad18, we established multiple RAD18−/− cell clones stably expressing WT human Rad18 (hRad18) (Figure 2B). PCNA ubiquitination following UV irradiation was restored to the WT level, whereas control RAD18−/− cells transfected with an empty vector did not show such recovery (Figure 2B). To examine whether PCNA ubiquitination required Rad6A/B together with Rad18, we established RAD18−/− cells stably expressing Rad18 but lacking R6BD (hRad18DR6). In these cells, PCNA was not ubiquitinated after DNA damage (Figure 2B). Furthermore, to confirm the requirement of Rad6A/B for PCNA monoubiquitination directly, Rad6 siRNA corresponding to both Rad6A and Rad6B was transfected into human cells, and reduced levels of Rad6A/B protein levels were confirmed by Western blot. In these cells, PCNA monoubiquitination was substantially reduced (Figure 2C). These results clearly indicate that in UV-irradiated mammalian cells, PCNA is monoubiquitinated in a Rad18- and Rad6A/B-dependent manner. To evaluate the significance of the monoubiquitination activity of Rad18, we determined the UV sensitivity of RAD18−/− mouse cells stably expressing hRad18. These cells showed almost normal UV sensitivity, while stable transformants with hRad18 lacking R6BD, or with the vector alone, remained sensitive to UV at the parent cell levels (Figure 2D). These results suggest that the UV sensitivity of RAD18−/− cells is caused at least in part by defects in the monoubiquitination of PCNA and subsequent foci formation of polη.
Figure 2.
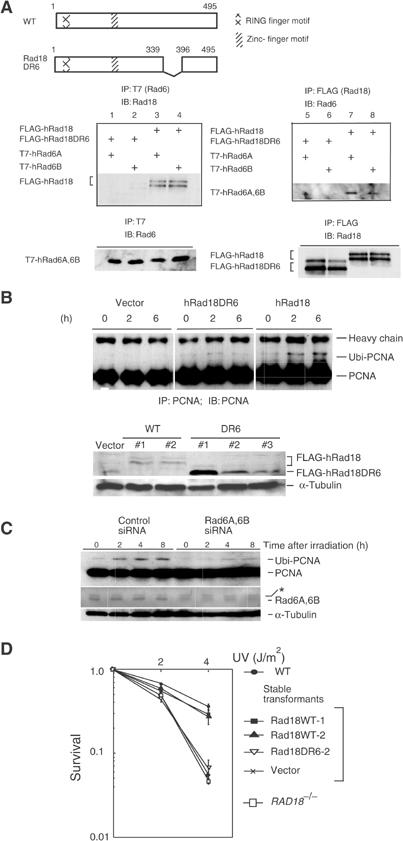
Requirement of Rad6A/B for monoubiquitination of PCNA in UV-irradiated cells. (A) Interaction of WT and mutant hRad18 with hRad6A/B. Full-length and mutant Rad18 proteins are schematically shown on the top panel. Plasmids were transfected into COS-7 cells with different combinations indicated on the left of the middle panels, and immunoprecipitation was performed. Similar levels of expression of hRad18 and hRad6A/B proteins in the transformed cells were confirmed in the lower panel. (B) Restoration of PCNA monoubiquitination in RAD18−/− cells by expression of WT hRad18 but not of mutant hRad18. Cells were incubated for 6 h following UV irradiation at 20 J/m2. Cell lysates were immunoprecipitated and blotted with an anti-PCNA antibody (upper panel). Expression of FLAG-hRad18 or FLAG-Rad18DR6 was confirmed in individual clones of stable transformants of RAD18−/− mouse fibroblasts by Western blot with an anti-Rad18 rabbit antibody (lower panel). α-Tubulin was indicated as a volume control. (C) Inhibition of PCNA monoubiquitination by siRNA for Rad6A/B. WI38VA13 cells were transfected with Rad6A and Rad6B siRNA, incubated for 4 days, and then irradiated with 10 J/m2 of UV light. At the indicated times, protein levels of monoubiquitinated PCNA were determined by Western blot. An asterisk shows a nonspecific band that remained constant following the siRNA treatment. (D) Restoration of UV sensitivity of RAD18−/− mouse cells by introduction of human Rad18 as determined by a colony-forming assay. Two independent clones of stable transformants (WT#1 and WT#2 in (B)) were tested.
Relocalization of Rad18 at stalling sites with PCNA
In mammalian cells fixed with formaldehyde, a substantial fraction of Rad18 was homogeneously localized in the nucleus, while the remaining fraction existed as dots of irregular shapes and sizes (Figure 3A, left). Notably, most of the nuclear dots of Rad18 dispersed throughout the nucleus within 15 min with UV doses as low as 5 J/m2 (Figure 3A, middle). Rad18 dispersion occurred in the presence of cycloheximide (data not shown), suggesting that direct or indirect post-translational modification of Rad18 is involved in this process. Within a few hours after UV irradiation, nuclear foci of Rad18 with uniform sizes appeared (Figure 3A, right). Such dynamic intranuclear translocation of Rad18 was much more clearly detected in cells fixed with methanol (Figure 3B). To investigate the relationship between Rad18 and PCNA, we performed double immunostaining on methanol-fixed cells. Under normal conditions, partial colocalization of Rad18 with PCNA was observed (Figure 3B, upper). Within 1 h after UV irradiation, almost all of Rad18 became colocalized with PCNA (Figure 3B, lower) and such colocalization was observed at least up to 4 h, suggesting that Rad18 translocates to the replication stalling sites. To confirm directly this assumption, UV-irradiated cells were labeled with BrdU and stained for Rad18 and incorporated BrdU. Before UV irradiation, BrdU sites were partially colocalized with Rad18 (Figure 3C, upper), but after UV irradiation most of the BrdU sites were colocalized with translocated Rad18 (Figure 3C, lower). Since colocalization of Rad18 with PCNA was observed in XPV cells with a similar time course, it was inferred that translocation of Rad18 does not require functional polη (data not shown). To determine the subnuclear localization of PCNA, chromatin fractions were separated from UV-irradiated cells. Almost equal amounts of unmodified PCNA were obtained in the soluble and chromatin fractions irrespective of UV irradiation. In contrast, monoubiquitinated PCNA was exclusively recovered in the chromatin fraction of UV-irradiated cells, and it moved to the solubilized nuclear fraction after treatment with micrococcal nuclease (Figure 3D), suggesting that monoubiquitinated PCNA is tightly associated with chromatin. We could not detect any apparent physical interaction between Rad18 and PCNA before or after UV irradiation by co-immunoprecipitation, suggesting that the interaction is weak or transient (data not shown).
Figure 3.
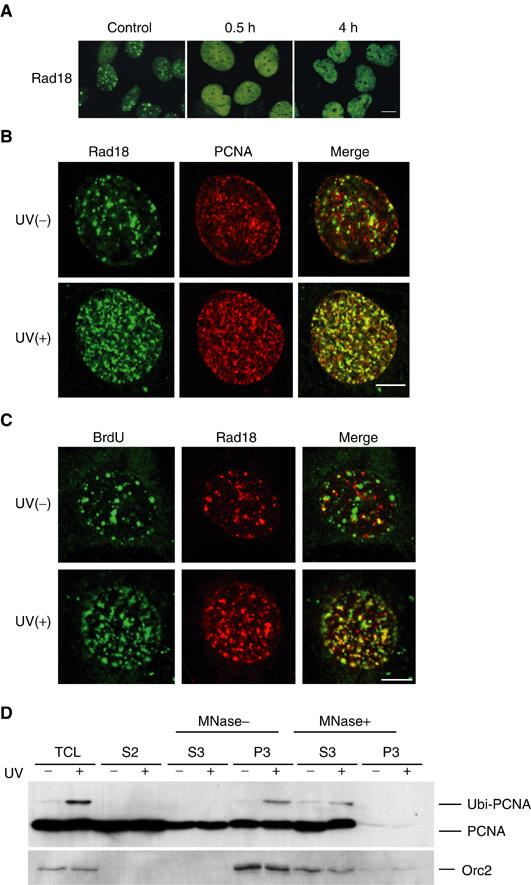
Colocalization of Rad18 with PCNA on chromatin following UV irradiation. (A) Dispersion and relocalization of Rad18. GM637 cells irradiated at 15 J/m2 were fixed with formaldehyde and stained for Rad18. Bar=20 μm. (B) UV-induced colocalization of Rad18 with PCNA. GM637 cells irradiated at 15 J/m2 were fixed with methanol 4 h after UV irradiation and processed for double staining for Rad18 (green) and PCNA (red). Bar=10 μm. (C) Accumulation of Rad18 at the replication stalling sites. UV-irradiated (15 J/m2) GM637 cells were labeled for 2 h with BrdU, fixed with methanol, and processed for double staining for Rad18 (red) and BrdU (green). Bar=10 μm. (D) Binding of monoubiquitinated PCNA to chromatin. Chromatin fractions were isolated from UV-irradiated (15 J/m2, 6 h) or nonirradiated HeLa cells, and then treated with micrococcal nuclease (MNase). The distributions of PCNA in the total cell lysate (TCL), soluble fraction (S2), solubilized nuclear fraction (S3), and chromatin-enriched fraction (P3) are shown. Orc2 is shown as a chromatin fraction marker.
Requirement of Rad18 for polη focus formation
Using polη fused to enhanced green fluorescent protein (eGFP-polη), Kannouche et al (2001) found that polη, which localizes uniformly in the nucleus under normal conditions, formed distinct nuclear foci at the replication stalling sites after treatment with DNA-damaging agents including UV and MMS. This polη focus formation is essential for UV survival, because mutant polη, which is defective in focus formation, could not complement UV survival of XPV cells (Kannouche et al, 2001). To investigate whether Rad18 is required for polη focus formation, we introduced eGFP-hpolη into either RAD18−/− or RAD18+/+ mouse cells. While polη focus formation was clearly observed in the UV-irradiated WT cells, polη remained uniformly dispersed in the nucleus of UV-irradiated RAD18−/− cells (Figure 4A). The formation of polη foci proceeded gradually and reached a plateau at 6 h after UV irradiation at least with dosages ranging 10–20 J/m2 (Figure 4B). Defective focus formation in RAD18−/− cells could be restored by concomitant introduction of WT hRad18 with or without a FLAG tag in its N-terminal region (Figure 4A and C, data not shown). However, hRad18 lacking R6BD did not restore the focus formation (Figure 4C). Furthermore, the formation of polη foci was significantly inhibited in cells treated with Rad6A/B siRNA (Figure 4D). These results indicate that UV-induced polη focus formation is dependent on both Rad18 and Rad6A/B.
Figure 4.
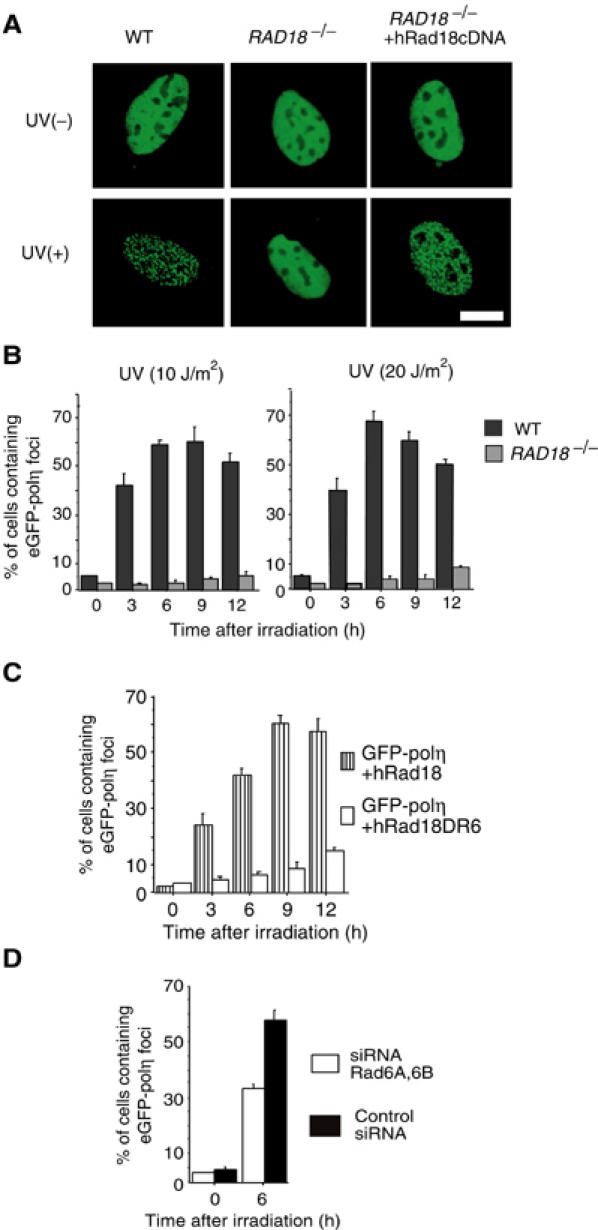
Rad18- and Rad6-dependent formation of polη foci. (A) Focus formation of eGFP-polη following UV irradiation in WT cells but not in RAD18−/− cells. Cells were irradiated at 15 J/m2. After 6 h, the distribution of eGFP-polη was examined after fixation. Defective focus formation of polη was recovered by concomitant expression of Rad18. Bar=10 μm. (B) Time course of eGFP-polη focus formation in UV-irradiated cells. RAD18−/− mouse cells and WT cells were transfected with eGFP-polη. After 20 h, cells were irradiated with UV at the indicated doses. (C) Restoration of eGFP-polη focus formation in UV-irradiated (20 J/m2) RAD18−/− cells by expression of WT hRad18 but not of mutant hRad18 lacking the Rad6-binding domain. (D) Inhibition of polη focus formation by siRNA for Rad6. WI38VA13 cells were transfected with Rad6A and Rad6B siRNA, cultured for 3 days, and then transfected again with an eGFP-polη plasmid. After 20 h, cells were irradiated with UV (10 J/m2), and 6 h later cells containing eGFP-polη foci were counted.
Association of Rad18 with polη
Immunostaining for Rad18 clearly demonstrated that Rad18 colocalized with eGFP-polη at the foci following UV irradiation (Figure 5A, lower). To investigate the interaction between Rad18 and polη, HA-tagged polη was transfected into GM637 cells, and co-immunoprecipitation experiments were performed. Rad18 was consistently associated with polη, irrespective of UV irradiation (Figure 5B). Furthermore, purified polη bound to purified Rad18 in an immunoprecipitation assay, but polδ did not (Figure 5C, lanes 2 and 4), indicating that at least a part of Rad18 is directly associated with polη in a UV-independent manner. To determine the binding site of polη to Rad18, we overexpressed a series of deletion mutants of polη fused with GST at their N-terminal regions (Figure 6A) in insect cells, and purified them with glutathione beads (Figure 6B). GM637 cell lysates were pulled down with these beads. Rad18 interacted with full-length polη and a C-terminal fragment of polη (GST-polη158c) spanning amino-acid residues 556–713 (Figure 6C). We also determined the binding site of Rad18 to polη in a similar way. In this assay, Myc-tagged WT and deleted Rad18 proteins were overexpressed in COS-7 cells (Figure 6D and E), and cell lysates were pulled down with glutathione beads associated with GST-polη158c. Among the deletion mutants, only Rad18 lacking a region spanning amino-acid residues 402–444 could not interact with polη (Figure 6F). To evaluate the biological significance of the interaction between Rad18 and polη, hRad18 lacking the polη-binding domain (hRad18DC2) was transiently expressed in RAD18−/− mouse cells together with eGFP-polη. Formation of eGFP-polη foci was not restored following UV irradiation (Supplementary Figure S1). Furthermore, RAD18−/− cells stably expressing Rad18 lacking the polη-binding domain showed high UV sensitivity like cells transformed with an empty vector (Supplementary Figures S2 and S3), while they had normal levels of monoubiquitination of PCNA after UV irradiation. These results suggest that Rad18 recruits polη to replication stalling sites through direct interaction. Since eGFP-polη localized uniformly in the nucleus with Rad18 under normal conditions (Figure 5A), the nuclear dots of Rad18 in nonirradiated cells might be reservoirs of free Rad18.
Figure 5.
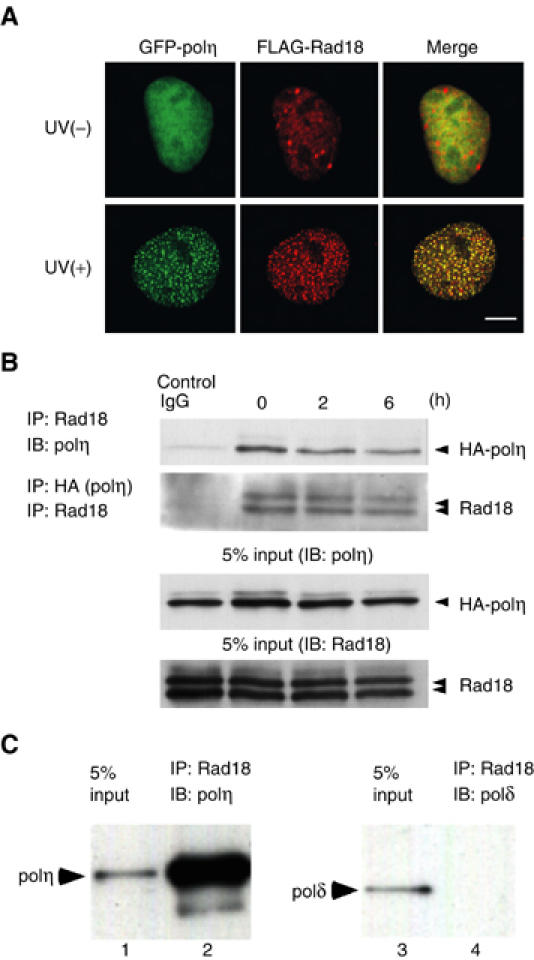
Direct interaction of polη with Rad18. (A) UV-induced colocalization of Rad18 with eGFP-polη in GM637 cells. Cells transfected with eGFP-polη and FLAG-Rad18 plasmids were irradiated at 15 J/m2 and incubated for 6 h. After fixation, cells were stained for Rad18 with an antibody against FLAG. Bar=10 μm. (B) Interaction of Rad18 with polη. HA-polη was transiently expressed in GM637 cells. Immunoprecipitation was performed at various times after UV irradiation (12.5 J/m2). As a control, UV-irradiated cell lysates (6 h) were immunoprecipitated with control IgG. (C) Direct binding of Rad18 with polη. Recombinant Rad18 and polη were purified from insect cells. After incubation of the mixture, Rad18 was immunoprecipitated and polη bound to Rad18 was detected by Western blot. Polδ was used as a control.
Figure 6.
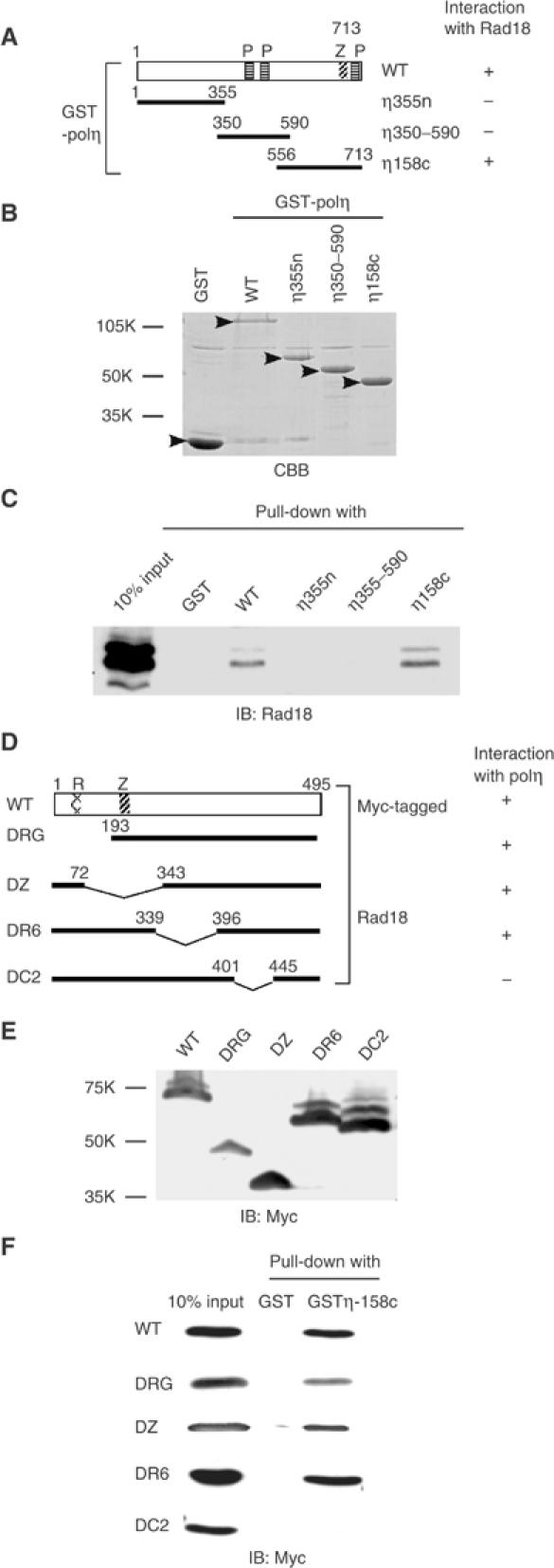
Determination of binding sites. (A) Structural domains of GST-polη fusion proteins. P: putative PCNA-binding domain; Z: zinc-finger domain. (B) Purification of GST- polη fusion proteins by glutathione beads. Proteins bound to the beads were stained with Coomassie brilliant blue (CBB, arrowheads). (C) Pull-down assay. GM637 cell lysates were pulled down with GST-polη fusion proteins bound to glutathione beads. Interaction with Rad18 was analyzed by Western blot. (D) Structural domains of Myc-tagged Rad18 proteins. R: RING finger domain; Z: zinc-finger domain. (E) Deletion mutant proteins were overexpressed in COS-7 cells and their expression was confirmed by Western blot. (F) COS-7 cell lysates containing Myc-tagged mutant Rad18 proteins were pulled down with GST-polη158c bound to glutathione beads. Association of WT and mutant Rad18 proteins with polη was analyzed by Western blot using an anti-Myc antibody.
Preferential binding of polη to monoubiquitinated PCNA
To investigate the molecular mechanism of how UV-induced monoubiquitination of PCNA functions in polymerase switching to polη, the physical interaction between PCNA and polη was determined by a pull-down assay. GST-polη bound to glutathione beads was mixed with lysates prepared from UV-irradiated HeLa cells, and PCNA associated with the GST-polη beads was revealed by Western blot. While monoubiquitinated PCNA was a minor fraction of the total PCNA in the lysates, it was recovered predominantly from the precipitated beads in a time-dependent manner (Figure 7A, right). In contrast, monoubiquitinated PCNA was not associated with GST-polδ in the same assay (Figure 7A, middle). The affinity of monoubiquitinated PCNA for polη was much higher than that of unmodified PCNA, because even at higher salt concentrations, monoubiquitinated PCNA remained bound to polη (Figure 7B, left). Monoubiquitinated PCNA bound to polη was much more refractory to elution by high salt concentrations than unmodified PCNA (Figure 7B, right). To investigate whether polη interacted with monoubiquitinated PCNA in UV-irradiated cells, HA-polη was transiently expressed in GM637 cells, and co-immunoprecipitation assay was performed. In this experiment, cells were treated with 0.1% NP-40 before preparation of cell lysates. This treatment allowed specific crosslinking between chromatin-bound polη and monoubiquitinated PCNA probably by excluding unmodified PCNA and a diffused form of polη from nuclei. Monoubiquitinated PCNA was preferentially immunoprecipitated with HA-polη in the UV-irradiated cells (Figure 7C, lanes 5 and 6). In contrast, monoubiquitinated PCNA was not immunoprecipitated in nonirradiated cells (Figure 7C, lanes 2 and 3). Taken together, these results indicate that polη preferentially binds to monoubiquitinated PCNA both in vitro and in vivo. To prove that the interaction between polη and monoubiquitinated PCNA is direct, PCNA was monoubiquitinated in the in vitro PCNA ubiquitination reaction (Figure 1E). Rad18 and Rad6B were then removed from the in vitro PCNA ubiquitination reaction mixture (Figure 1E) by multiple cycles of immunodepletion with an anti-Rad18 antibody (Figure 7D, upper). Immunodepletion of Rad18 and Rad6B was confirmed by Western blot. Monoubiquitinated PCNA still bound to polη in a pull-down assay (Figure 7D, lane 3). In contrast, polη lacking the three putative PCNA-binding sites on the C-terminus (Kannouche et al, 2001) showed no interaction with PCNA (Figure 7D, lane 2). Immunostaining demonstrated that more than 50% of the transfected eGFP-polη colocalized with endogenous polδ 5 h after UV irradiation (Figure 7E). Furthermore, endogenous polδ colocalized with PCNA in UV-irradiated cells (Figure 7F), suggesting that both polymerases and Rad18 localize at the same stalling sites.
Figure 7.
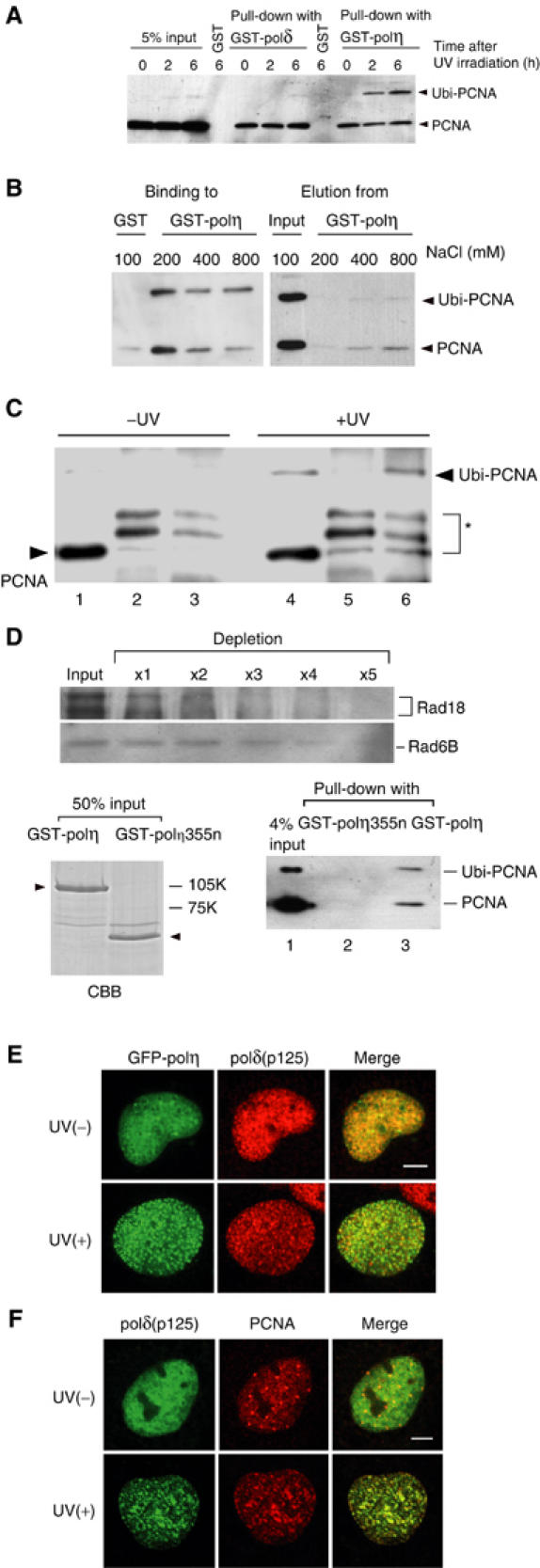
Preferential binding of polη to monoubiquitinated PCNA. (A) Binding of polη to ubiquitinated PCNA. HeLa cells were irradiated with UV at 20 J/m2. PCNA in the cell lysates was pulled down with either GST-polη beads or polδ beads, and analyzed by Western blot using an anti-PCNA antibody. (B) Effects of different salt concentrations on the binding of PCNA to GST-polη (left) and on PCNA elution from GST-polη (right). PCNA pulled down was washed with buffer containing various concentrations of NaCl. PCNA in bound or eluted fractions was analyzed as in (A). (C) Preferential binding of polη to monoubiquitinated PCNA in living cells. GM637 cells were transfected with an HA-polη plasmid. After 2 days, these cells were irradiated with 20 J/m2 of UV (lanes 4–6), or remained untreated (lanes 1–3), and incubated for 5 h. After immunoprecipitation with an anti-polη antibody (lanes 3 and 6) or control IgG (lanes 2 and 5), binding PCNA was detected by Western blot with an anti-PCNA antibody. Lanes 1 and 4 represent 5% samples of the whole-cell lysate. An asterisk shows nonspecific bands. (D) Direct binding of polη to monoubiquitinated PCNA. Rad18 was removed from the in vitro PCNA ubiquitination reaction mixture by immunoprecipitation with an anti-Rad18 antibody (upper panel). Note that Rad6B was also depleted, probably due to direct interaction with Rad18 (upper). Remaining PCNA and monoubiquitinated PCNA were pulled down with purified GST-polη bound to glutathione beads (lower, right). GST-polη355n was used as a control. Purity of the polymerase samples is shown on the left panel (arrowheads) by the Coomassie brilliant blue (CBB) staining. (E) Colocalization of eGFP-polη with polδ in UV-irradiated (10 J/m2, 5 h) GM637 cells. Bar=5 μm. (F) Colocalization of polδ with PCNA in UV-irradiated (10 J/m2, 5 h) WI38VA13 cells. Bar=5 μm.
Discussion
Tolerance to UV-induced DNA damage involves damage-induced PCNA monoubiquitination (Hoege et al, 2002) and the formation of polη foci at the replication stalling sites, thereby inducing TLS by polη. We have addressed two important questions: How is polη recruited to replication stalling sites after UV irradiation? And, what is the function of monoubiquitinated PCNA in the PRR process? In the work presented here, we analyzed these processes using mammalian cells, and found that Rad18 plays pivotal roles as a coordinator in both PCNA monoubiquitination and the formation of polη foci.
Polη synthesizes DNA with a low fidelity, misincorporating nucleotides with a frequency of 10−2–10−3 (Washington et al, 1999). Therefore, to prevent incidental mutagenesis, it is critical to restrict accession of polη to DNA replication sites under normal conditions. This task seemed to be performed by regulation of intranuclear localization of polη. Polη localizes uniformly in the nucleus under normal conditions, but relocalizes at discrete nuclear foci following UV irradiation (Kannouche et al, 2001). We found that Rad18 interacts directly with polη in cells irrespective of genotoxic stresses (Figure 5B). Purified polη bound to Rad18, while polδ did not (Figure 5C). Rad18 relocalized to the replication stalling sites in response to UV irradiation, where polη and PCNA colocalized (Figures 3B and 5A). These results support the view that Rad18 and polη relocate to replication stalling sites simultaneously as a complex and form nuclear foci. Furthermore, polη focus formation requires Rad6A/B, because RAD18−/− cells expressing mutant Rad18 lacking the Rad6A/B-binding domain or normal cells suppressed expression of Rad6A/B by siRNA show little or reduced focus formation, respectively. These results indicate that both Rad18 and Rad6A/B are essential for polη focus formation. When cells are exposed to UV irradiation, CPDs form on both the leading and the lagging strands. In either case, single-stranded gap regions are formed on the 3′ side of the lesion by uncoupling replication (Svoboda and Vos, 1995). Because Rad18 can bind to single-stranded DNA (Bailly et al, 1994), Rad18 together with polη might be located at the gaps where replicative machinery containing PCNA and polδ stalled on the template DNA. In the initial step of this pathway, Rad18 might be activated through a mechanism sensing stalling of replication. We observed that some fractions of Rad18 exist as nuclear dots under normal conditions, and that, following UV irradiation, these dots disperse throughout the nucleoplasm in a short time. Rad18 dispersion is also observed in cells treated with hydroxyurea (data not shown), suggesting that it is triggered not by DNA damage but by stalling of replication. In both cases, Rad18 dispersion takes place even under conditions where protein synthesis is inhibited. It is possible that post-translational modification (e.g. phosphorylation) of Rad18 is involved in the reaction.
The binding site of polη to Rad18 is located on the C-terminal region, which contains one putative PCNA-binding site and a zinc-finger domain (Figure 6A). It is reported that polη lacking a C-terminal region, which overlaps with the Rad18-binding domain, does not show polη focus formation following UV irradiation, and that a fragment of polη containing the C-terminal 120 amino-acid residues is able to form UV-induced foci (Kannouche et al, 2001). Furthermore, clinical manifestations of XPV patients with large deletions in the C-terminal region of polη (class I) are similar to those of XPV patients with small deletions corresponding to the Rad18-binding domain (class III) (Broughton et al, 2002). Probably, inability of polη to form a complex with Rad18 may be a primary cause of dysfunction of polη in these cases.
As in the case of yeast (Hoege et al, 2002), we showed that PCNA is monoubiquitinated in UV-irradiated mammalian cells in a Rad18- and Rad6-dependent manner. The PCNA monoubiquitination reaction is reconstituted in vitro by the presence of human Rad18 and Rad6B proteins (Figure 1E), indicating that Rad18 protein is definitely a ubiquitin ligase (E3) specific for PCNA. At present, it is not clear whether PCNA monoubiquitination occurs at the stalling sites, or to PCNA free in the nucleoplasm. Because Rad6 interacts not only with Rad18 but also other E3s such as Ubr1 and Bre1 under normal conditions (Dohmen et al, 1991; Watkins et al, 1993), dispersion and relocalization of Rad18 might increase the chance of interaction of Rad18 with Rad6A/B, thereby channeling Rad6A/B to a DNA damage tolerance pathway. In the case of polα–polδ switching on regular replication, loading of PCNA onto DNA is critical because polδ binds PCNA while polα does not (Tsurimoto and Stillman, 1991). However, in the case of the switch from polδ to polη, loading of unmodified PCNA would not be sufficient to promote the switch because both polδ and polη interact with PCNA (Tsurimoto and Stillman, 1991; Haracska et al, 2001b). We found that polη preferentially binds to mono-ubiquitinated PCNA while polδ does not (Figure 7A). We assume that the preferential binding is the motive force for the polymerase switch. Immunostaining results revealed that polδ remains associated with the Rad18–polη complex at the foci, suggesting that both of the polymerases are retained at the replication stalling sites.
Rad18 performs two roles in polη foci formation in UV-irradiated cells: targeting Rad6A/B and polη to stalling sites as a guide, and directing monoubiquitination of PCNA as a ubiquitin ligase. Although Rad18 associates constitutively with polη, Rad18 lacking the Rad6A/B-binding domain did not induce the formation of polη foci in UV-irradiated RAD18−/− cells. It is probable that translocation of a Rad18/polη complex to stalled replication sites is not sufficient to form stable polη foci, and this may require monoubiquitinated PCNA. In the budding yeast, monoubiquitinated PCNA is further ubiquitinated by an Mms2/Ubc13/Rad5 complex (Hoege et al, 2002). Following the appearance of monoubiquitinated PCNA, we detected a new band corresponding to approximately 50 kDa by Western blot using an anti-PCNA antibody (data not shown). Judging from its size, this band could be either a multiubiquitinated or sumoylated form of PCNA. The role of multiubiquitination of PCNA is largely unknown, but it probably stimulates polymerase switching in the process of TLS. Our finding that Rad18 interacts with polη and is involved in the monoubiquitination of PCNA accounts well for the requirement for Rad18 for polη focus formation after UV irradiation.
Materials and methods
Establishment of RAD18−/−cells
RAD18+/− ES cells were injected into C57BL/6 blastocysts to generate chimeric mice (Tateishi et al, 2003), which transmitted the mutation into gametes. RAD18+/− mice were intercrossed to generate RAD18−/− mice. RAD18−/− fibroblasts were obtained from the lungs of the mice and immortalized with SV40. RAD18+/+ cells were similarly established from WT mice. Stable transformants of RAD18−/− cells expressing FLAG-hRad18 were selected with hygromycin B, and clones expressing high levels of the transgenes were used.
Protein purification
Human Rad18 and Rad6B proteins were expressed simultaneously in Sf9 insect cells by using a recombinant baculovirus. Rad6B protein had a polyhistidine tag at the N-terminal region. A Rad18 and His-Rad6B protein complex was purified with an Ni ion-loaded HiTrap Chelating HP column (Amersham).
Immunostaining
To examine the colocalization of PCNA with Rad18, GM637 cells were fixed with 100% methanol and processed for indirect double staining. First, cells were stained with an anti-Rad18 rabbit antibody (Tateishi et al, 2000) and an anti-PCNA human serum (Toschi and Bravo, 1988), and then stained with FITC-anti-rabbit IgG (goat, Cappel) and rhodamine-anti-mouse IgG (goat, Cappel). To address the site of Rad18 relocalized after dispersion, UV-irradiated (15 J/m2) GM637 cells were cultured for 30 min, and then labeled with BrdU (10 μM) for 2 h. As a control, nonirradiated cells were labeled with BrdU (80 μM) for 20 min. After fixation with 100% cold methanol, these cells were stained for Rad18 with an anti-Rad18 rabbit antibody and a rhodamine-anti-rabbit antibody (goat, Cappel). After fixing again with 3.7% formaldehyde for 10 min, the cells were treated for 20 min with 2.5 N HCl, and then stained for BrdU with an FITC-anti-BrdU mouse monoclonal antibody (Progen). GM637 purchased from NIGMS (USA) is a normal human fibroblast cell line immortalized with SV40. Colocalization of polδ with PCNA was examined similarly by using WI38VA13 cells and an anti-polδ goat antibody (A-9, Santa Cruz). To examine the colocalization of eGFP-polη with FLAG-Rad18, GM637 cells were cotransfected with peGFP-polη and pcDNA3 FLAG-Rad18 using Fugene 6 (Roche). Cells were pre-fixed with 3.7% formaldehyde, washed with phosphate-buffered saline (PBS), and fixed with 80% methanol. Cells were stained for FLAG with an anti-FLAG antibody (M2, Sigma), and a rhodamine-anti-mouse IgG antibody (Cappel) consecutively. To examine the colocalization of polδ with eGFP-polη, GM637 cells were transfected with eGFP-polη and stained for polδ with an anti-polδ antibody (A-9, Santa Cruz). Stained cells were observed with a confocal laser-scanning microscope (FV300, Olympus).
Immunoprecipitation
COS-7 cells were transfected with plasmids using Fugene 6, and incubated for 48 h. Cells were harvested in a lysis buffer (50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 0.5% NP-40, 8% glycerol, 0.5 mM dithiothreitol (DTT), 50 mM NaF, and protease inhibitors). Cell lysates were mixed with an anti-T7 monoclonal antibody (Novagen) or an anti-FLAG antibody for 1 h at 4°C, and immunoprecipitated with Protein G Sepharose beads (Amersham Pharmacia) for another 1 h at 4°C. Precipitated proteins were separated by SDS–PAGE and analyzed by Western blot. To determine the interaction of HA-polη and Rad18 in vivo, GM637 cells were transfected with pCAGGS HA-polη, incubated for 48 h, and irradiated with UV at 12.5 J/m2. These cells were treated with 5 mM dimethyl 3,3′-dithiobispropionimidate.2HCl (DTBP (Pierce)) for protein crosslinking before preparation of cell lysates (Pearson et al, 2000). Cell lysates were prepared in a buffer (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1.0% NP-40, 50 mM NaF, and protease inhibitors). After sonication, the supernatants were immunoprecipitated with either a monoclonal anti-HA antibody (HA.11, BAbCO) or an anti-hRad18 rabbit antibody. Precipitates were analyzed by SDS–PAGE and Western blot. To investigate the interaction between HA-polη and monoubiquitinated PCNA in vivo, GM637 cells were transfected with pCAGGS HA-polη and incubated for 48 h. After UV irradiation at 20 J/m2, cells were cultured for 5 h and treated with 0.1% NP-40 for 5 min on ice. These cells were washed once with PBS, and then treated with DTBP for crosslinking. Cells were collected in a lysis buffer (50 mM Tris–HCl (pH 7.5), 200 mM NaCl, 1.0% NP-40, 0.1% SDS, 0.1% Na-deoxycholic acid, and protease inhibitors) and disrupted by sonication. After centrifugation, cleared supernatants were immunoprecipitated with an anti-polη antibody. PCNA binding to polη was detected by SDS–PAGE and Western blot using an anti-PCNA antibody.
To examine the interaction between Rad18 and polη in vitro, His-Rad18 and His-polη were overexpressed in Sf9 insect cells and purified with Ni-NTA agarose beads (Invitrogen). Purified Rad18 protein was incubated in buffer A (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.2% Triton X-100, and 0.5 mM DTT) for 30 min at 25°C either with purified polη or with polδ. Final concentrations of the proteins were set at 0.1 μM. Rad18 protein was then immunoprecipitated with an anti-Rad18 antibody. Polymerases bound to Rad18 were detected by Western blot using an anti-polη antibody (B-7, Santa Cruz) or an anti-polδ antibody.
Analysis of eGFP-polη foci after UV irradiation
Cells were transfected with eGFP-polη, or cotransfected with eGFP-polη and RAD18 cDNA, and cultured for 20 h before UV irradiation. For observation, cells were fixed with 3.7% formaldehyde. At least 200 cells were counted for each time point, and the experiments were repeated three times.
In vitro ubiquitination of PCNA
HHR6B (hRad6B) and hRad18 were prepared by using a baculovirus, and purified with Ni-NTA agarose beads. E1 enzyme was purchased from Boston Biochem. Recombinant PCNA purified from E. coli (Tsurimoto and Stillman, 1991) was incubated for 1 h at 25°C with other components in a buffer containing 50 mM HEPES (pH 7.6), 0.05 mM DTT, 1 mM MgCl2, 1 mM ATP, and an ATP-regenerating system together with either 5 mM ubiquitin (Boston Biochem) or FLAG-ubiquitin (Sigma). Samples were analyzed for PCNA by Western blot.
Physical interaction between Rad18 and polη
GST-polη cDNA was generated by PCR by using peGFP-polη as a template with the following primers: gcgaattcATGGCTACTGGACAGGATCG and gcgaattcCTAATGTGTTAATGGCTTAAAAAATGATTC. A fragment digested with EcoRI was ligated into the EcoRI site of pAcGHLT-A vector (PharMingen). GST-polη mutant cDNA was subcloned by PCR by using peGFP-polη as a template. To generate polη355n, gcgaattcATGGCTACTGGACAGGATCG and cgggatccTTAGTCTTTAGTCAGTCTCTCCTC were used as primers. After digestion with EcoRI and BamHI, the product was inserted into the pAcGHLT-A vector digested with EcoRI–BglII. To obtain a fragment of polη350–590, ggaattcGAGAGACTGACTAAAGACCG and gcagatctGCTTTAGAGGATTCTTCTAGC were used as primers. The product was digested with EcoRI and BglII and inserted into the pAcGHLT-A vector digested with EcoRI–BglII. To generate polη158c, pAcGHLT-A-polη was digested with NcoI, and the 3′ 477 bp fragment was inserted into the pAcGHLT-A vector. Full-length Rad18 cDNA tagged with Myc at its C-terminal region was generated by PCR with the following primers: cggaattcATGGACTCCCTGGCC and cggaattCTTACAAGTCCTCTTCAGAAATGAGCTT TTGCTCATTCCTATTACGCT. A fragment digested with EcoRI was ligated into the EcoRI site of the pCAGGS vector. Myc-tagged Rad18 DRG cDNA containing amino-acid residues 193–495 was subcloned by PCR. Myc-tagged Rad18 DZ cDNA was produced by ligation of the two fragments corresponding to amino-acid residues 1–72 and 343–495. The product was inserted into the EcoRI site of the pCAGGS vector. DR6 and DC2 plasmids were generated by inverse PCR using pCAGGS containing Myc-tagged full-length Rad18 as a template by using the primers GTGGATTTCATCTATTTCCTTTTCTG/ACCTCAG TAACAAACCAC and GTGGTTTGTTACTGAGGTC/GACATCATAAGAGAT CTTTTAGAAG, respectively. Final plasmids were obtained by self-ligation of the PCR products. To investigate the binding domain of Rad18 to polη, COS-7 cells were transfected with pCAGGS plasmids containing Myc-tagged full-length Rad18 or deletion mutant Rad18, and cultured for 48 h. Cells were harvested in a lysis buffer (50 mM Tris (pH 7.5), 100 mM NaCl, 1 mM DTT, 0.25% NP-40, and protease inhibitors). Cell lysates were mixed with GST-polη158c beads for 1 h at 4°C. Precipitated proteins were separated by SDS–PAGE and analyzed by Western blot using an anti-c-Myc antibody (9E10, Santa Cruz). To investigate the binding domain of polη with Rad18, GM637 cell lysates were mixed with GST-polη or deletion mutants of polη beads for 1 h at 4°C. Precipitated proteins were separated by SDS–PAGE and analyzed by Western blot using an anti-Rad18 antibody.
Pull-down assay
GST-polη overexpressed in Sf9 insect cells was harvested with glutathione Sepharose 4B (Amersham). His-polδ (p125) overexpressed in insect cells together with GST-p66 and His-p50 was harvested with glutathione beads as a complex (Shikata et al, 2001). After washing with PBS, the beads were suspended in a lysis buffer (20 mM sodium phosphate pH 7.3, 10% glycerol, 10 μM β-mercaptoethanol, 300 mM NaCl, 1% NP-40, and protease inhibitors). Cell lysates were prepared from UV-irradiated (20 J/m2) HeLa cells with a lysis buffer (50 mM Tris–HCl pH 7.5, 1 mM DTT, 100–800 mM NaCl, 1% NP-40, 50 mM NaF, and protease inhibitors), and mixed with either GST-polη beads or GST-polδ beads for 1 h at 4°C. As a control, GST beads were used. Precipitated proteins were analyzed for PCNA by Western blot. To examine elution of PCNA from GST-polη beads, the beads were washed with a lysis buffer containing 200–800 mM NaCl, and eluted PCNA was analyzed by Western blot.
Immunodepletion
PCNA was monoubiquitinated in vitro. Rad18 was removed from the reaction mixture by five cycles of immunodepletion using an anti-Rad18 antibody and protein G Sepharose. After immunodepletion, PCNA was pulled down with either GST-polη or GST-polη355n in a buffer containing 50 mM NaCl, 20 mM HEPES–KOH (pH 7.4), 1 mM MgCl2, 2 mM DTT, and 0.005% NP-40. Binding was revealed by Western blot using an anti-PCNA monoclonal antibody (PC10, Santa Cruz). GST-polη355n is an N-terminal fragment of polη (amino-acid residues 1–355) fused with GST at the N-terminus.
Chromatin isolation
Chromatin fractions were isolated from UV-irradiated (15 J/m2, 6 h culture) or nonirradiated HeLa cells as described elsewhere (Mendez and Stillman, 2000). A part of chromatin fractions was further treated with 0.2 U micrococcal nuclease (MNase) for 1 min at 37°C, and separated into solubilized fractions as described (Mendez and Stillman, 2000).
Transfection of siRNA duplexes
WI38VA13 is a normal human lung fibroblast cell line immortalized with SV40 (purchased from the American Type Culture Collection). WI38VA13 cells were transfected with Oligofectamine (Invitrogen) with a mixture of siRNA specific for HHR6A and HHR6B, the coding strands of which were CGGGAAUAUAUGAAAAGCGU(TT) and GAGUUUCGGCCAUUGUUGA(TT), respectively. These cells were cultured for 72–96 h before the second transfection. For the control transfection, the negative control siRNA (QIAGEN) was used.
UV survival assay
Appropriate numbers of cells were inoculated onto 60-mm dishes and left to attach for 8–9 h. Subsequently, cells were washed with PBS, exposed to UV light (254 nm) at a fluence rate of 0.63 J/m2/s, and cultured for 6–7 days. Colonies were fixed with 80% methanol and stained with 5% Giemsa solution. For each UV dose, at least three dishes were used.
Supplementary Material
Supplementary Figures
Acknowledgments
This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan, and by a grant of the Princess Takamatsu Cancer Research Fund. We thank A Lehmann for eGFP-polη, Y Takasaki for anti-PCNA human serum, S Watanabe for fluorescent pictures, H Niwa, J Miyazaki, K Araki, and K Yamamura for establishing RAD18−/− knockout mice, and D Catcheside for critical reading of the manuscript.
References
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev 8: 811–820 [DOI] [PubMed] [Google Scholar]
- Bailly V, Lauder S, Prakash S, Prakash L (1997a) Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem 272: 23360–23365 [DOI] [PubMed] [Google Scholar]
- Bailly V, Prakash S, Prakash L (1997b) Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol Cell Biol 17: 4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield S, Hryciw T, Xiao W (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res 486: 167–184 [DOI] [PubMed] [Google Scholar]
- Broughton BC, Cordonnier A, Kleijer WJ, Jaspers NGJ, Fawcett H, Raams A, Garritsen VH, Stary A, Avril MF, Boudsocq F, Masutani C, Hanaoka F, Fuchs RP, Sarasin A, Lehmann AR (2002) Molecular analysis of mutation in DNA polymerase η in xeroderma pigmentosum-variant patients. Proc Natl Acad Sci USA 99: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R (2001) Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem 276: 43487–43490 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Madura K, Bartel B, Varshavsky A (1991) The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci USA 88: 7351–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S (2001b) Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol 21: 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L (2001a) Interaction with PCNA is essential for yeast DNA polymerase η function. Mol Cell 8: 407–415 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hynes RH, Kunz BA (1981), In The Molecular Biology of the Yeast Saccharomyces, Strathern JN, Jones EW, Broach JR (eds) pp 371–414. Plainview, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Johnson RE, Prakash S, Prakash L (1999) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Pol η. Science 283: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L (2000) Fidelity of human DNA polymerase η. J Biol Chem 275: 7447–7450 [DOI] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR (2001) Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev 15: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA (2000) Low fidelity DNA synthesis by human DNA polymerase-η. Nature 404: 1011–1013 [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA (2004) Preferential cis–sin thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature 428: 97–100 [DOI] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DL, Nairn RS (1989) The biology of the (6-4) photoproduct. Photochem Photobiol 49: 805–819 [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R (2001) The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406: 207–210 [DOI] [PubMed] [Google Scholar]
- Shikata K, Ohta S, Yamada K, Obuse C, Yoshikawa H, Tsurimoto T (2001) The human homologue of fission Yeast cdc27, p66, is a component of active human DNA polymerase η. J Biochem (Tokyo) 129: 699–708 [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Svoboda DL, Vos JM (1995) Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc Natl Acad Sci USA 92: 11975–11979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S, Niwa H, Miyazaki J, Fujimoto S, Inoue H, Yamaizumi M (2003) Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol Cell Biol 23: 474–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S, Sakuraba Y, Masuyama S, Inoue H, Yamaizumi M (2000) Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc Natl Acad Sci USA 97: 7927–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschi L, Bravo R (1988) Changes in cyclin/proliferating cell nuclear antigen distribution during DNA repair synthesis. J Cell Biol 107: 1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B (1991) Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer–template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem 266: 1950–1960 [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash S, Prakash L (1999) Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J Biol Chem 274: 36835–36838 [DOI] [PubMed] [Google Scholar]
- Watkins JF, Sung P, Prakash S, Prakash L (1993) The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev 7: 250–261 [DOI] [PubMed] [Google Scholar]
- Xin H, Lin W, Sumanasekera W, Zhang Y, Wu X, Wang Z (2000) The human RAD18 gene product interacts with HHR6A and HHR6B. Nucleic Acids Res 28: 2847–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Okada T, Matsusaka T, Sonoda E, Zhao GY, Araki K, Tateishi S, Yamaizumi M, Takeda S (2002) RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J 21: 5558–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


