Abstract
Mitochondria import nuclear-encoded precursor proteins to four different subcompartments. Specific import machineries have been identified that direct the precursor proteins to the mitochondrial outer membrane, inner membrane or matrix, respectively. However, a machinery dedicated to the import of mitochondrial intermembrane space (IMS) proteins has not been found so far. We have identified the essential IMS protein Mia40 (encoded by the Saccharomyces cerevisiae open reading frame YKL195w). Mitochondria with a mutant form of Mia40 are selectively inhibited in the import of several small IMS proteins, including the essential proteins Tim9 and Tim10. The import of proteins to the other mitochondrial subcompartments does not depend on functional Mia40. The binding of small Tim proteins to Mia40 is crucial for their transport across the outer membrane and represents an initial step in their assembly into IMS complexes. We conclude that Mia40 is a central component of the protein import and assembly machinery of the mitochondrial IMS.
Keywords: mitochondrial intermembrane space, protein assembly, protein sorting, Saccharomyces cerevisiae, Tim proteins
Introduction
Mitochondria consist of two membranes and two aqueous compartments, the intermembrane space (IMS) and the matrix. Except for a few proteins of the inner membrane and matrix, all mitochondrial proteins are encoded by nuclear genes and synthesized as precursor proteins on cytosolic polysomes (Herrmann and Neupert, 2000; Jensen and Johnson, 2001; Endo et al, 2003; Sickmann et al, 2003). Three main biogenesis pathways of mitochondrial proteins have been characterized so far. Each pathway involves first the general translocase of the outer membrane (TOM complex) and then a specific machinery for sorting to the correct mitochondrial subcompartment.
(i) Most matrix proteins carry positively charged N-terminal presequences that are proteolytically removed after the import. After translocation through the TOM complex, the preproteins are directed to the membrane potential (Δψ)-driven presequence translocase of the inner membrane (TIM23 complex) (Jensen and Johnson, 2001; Truscott et al, 2003). The ATP-powered presequence translocase-associated motor (PAM) in the matrix drives the completion of protein import (Frazier et al, 2004; Kozany et al, 2004). Some inner membrane and IMS proteins use a modified version of the presequence pathway. They carry a hydrophobic sorting sequence behind the cleavable matrix-targeting signal and are thus arrested in the inner membrane (Glick et al, 1992; Hahne et al, 1994). IMS proteins that carry such a bipartite presequence, like cytochrome b2 and NADH-cytochrome b5 reductase, are first inserted into the inner membrane in a Δψ-dependent manner and are then released to the IMS by a proteolytic removal of the sorting sequence. (ii) A majority of inner membrane proteins are synthesized without a cleavable presequence, but contain internal targeting signals. With the help of cytosolic chaperones, the precursors are transferred to the TOM complex in an ATP-dependent manner (Pfanner et al, 1987; Young et al, 2003). After passing through the TOM channel, the precursors do not engage the presequence translocase, but use a separate machinery for transport to and insertion into the inner membrane. Small Tim proteins, including the essential proteins Tim9 and Tim10, guide the hydrophobic precursor proteins through the IMS and transfer them to the Δψ-driven protein insertion complex of the inner membrane (carrier translocase, TIM22 complex) (Sirrenberg et al, 1996, 1998; Kerscher et al, 1997; Koehler et al, 1998a; Davis et al, 2000; Luciano et al, 2001; Curran et al, 2002a, 2002b; Rehling et al, 2003). (iii) The precursors of β-barrel proteins of the outer membrane, like the abundant porin, are imported in an ATP-dependent manner. The precursors are first translocated via the TOM complex to the IMS side. With the help of small Tim proteins, the precursors are then guided to the sorting and assembly machinery (SAM complex) of the outer membrane that promotes their membrane integration and assembly into functional complexes (Kozjak et al, 2003; Paschen et al, 2003; Wiedemann et al, 2003a, 2004a, 2004b; Gentle et al, 2004; Hoppins and Nargang, 2004).
For the majority of IMS proteins, however, no specific import machinery has been identified. Many IMS proteins are small proteins with a size up to 20 kDa and contain conserved cysteine residues that are involved in binding cofactors (metal ions, heme) or formation of disulfide bonds (Lutz et al, 2003). These proteins do not carry cleavable presequences, but are synthesized as mature-sized proteins with internal targeting signals. Typical examples are the five members of the family of small Tim proteins (Tim8, Tim9, Tim10, Tim12 and Tim13) (Koehler, 2004). Like all other mitochondrial precursor proteins, they use the general entry gate, the TOM complex, for translocation across the outer membrane in a loosely folded conformation. The precursor proteins do not engage inner membrane translocases and depend neither on a membrane potential nor on ATP for translocation into the IMS (Lutz et al, 2003). Due to the lack of an IMS protein import machinery for the small Tim proteins, it has been suggested that the refolding of the precursor proteins in the IMS, accompanied by binding of cofactors or formation of disulfide bonds, represents the crucial mechanism for trapping the proteins in the IMS (‘folding-trap model') (Allen et al, 2003; Lutz et al, 2003; Koehler, 2004; Lu et al, 2004a).
We have identified the essential IMS protein Mia40 that binds to incoming precursors of small Tim proteins, facilitating their translocation across the outer membrane. Mia40 directs the precursor proteins onto their assembly pathway into IMS complexes, but is not present in the mature complexes. We conclude that Mia40 is a crucial component of an IMS-specific import and assembly machinery.
Results
Mia40 is an essential protein of the mitochondrial IMS
Since several members of the family of small Tim proteins are essential for cell viability (Koehler et al, 1998a, 1998b; Sirrenberg et al, 1998; Adam et al, 1999), we assumed that a putative IMS import machinery should contain an essential protein. We screened the recently determined mitochondrial proteome of the yeast Saccharomyces cerevisiae (Sickmann et al, 2003) for the presence of essential proteins with a possible bipartite IMS targeting signal (Glick et al, 1992). The protein encoded by the open reading frame YKL195w contained a positively charged N-terminal sequence followed by a hydrophobic segment, that is, a candidate for a bipartite presequence (Figure 1A). Since we show here that the protein is involved in the mitochondrial IMS import and assembly pathway, we term it Mia40. Mia40 possesses a high content of negatively charged residues (pI of 4.51 for the entire precursor and pI of 4.18 for the putative mature region). Mia40 contains a C-terminal domain conserved from yeast to man; however, the function of these proteins has not been known. The domain consists of a cluster of negatively charged residues, followed by a highly conserved sequence with six cysteines (Figure 1B). The four C-terminal cysteines form a twin Cx9C motif. IMS proteins like the copper chaperone Cox17 and the related protein Cox19 also contain this motif, but not the complete conserved Mia40 domain (Beers et al, 1997; Nobrega et al, 2002).
Figure 1.
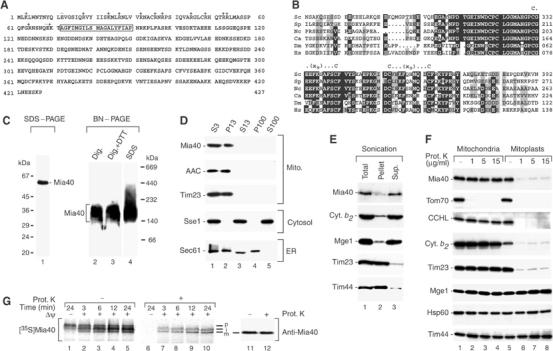
Mia40 is located in the mitochondrial IMS. (A) Predicted primary structure of the translation product of the full open reading frame of S. cerevisiae MIA40 (YKL195w). Box, hydrophobic segment. It has also been proposed that Mia40 starts at the second methionine (residue 25; Kellis et al, 2003). We synthesized both forms in vitro and imported them into isolated mitochondria. The efficiency of in vitro synthesis and the observed import signals were considerably stronger for the protein derived from the full open reading frame. (B) Sequence alignment of S. cerevisiae (Sc) Mia40 (residues 273–392) with homologous protein domains from Schizosaccharomyces pombe (Sp), Neurospora crassa (Nc), Candida albicans (Ca), Drosophila melanogaster (Dm) and Homo sapiens (Hs). Black, identical residues in at least four proteins; gray, similar residues. The conserved twin Cx9C motif is marked. (C) Migration of Mia40 from wild-type S. cerevisiae mitochondria on SDS–PAGE and BN–PAGE, analyzed by Western blotting. Where indicated, 10 mM DTT or 0.2% SDS were added to the digitonin buffer prior to BN–PAGE. The sample containing SDS was boiled for 15 min at 95°C. (D) Cellular fractionation. Equal volumes of the fractions after differential centrifugation steps (numbers refer to 1000 g) were analyzed by Western blot. S, supernatant; P, pellet. (E) Separation of sonicated mitochondria into membrane-bound and soluble fractions by centrifugation. Sup., supernatant. Samples were subjected to Western blot analysis. (F) Treatment of mitochondria and mitoplasts with proteinase K (Prot. K), followed by Western blot analysis. (G) Import and processing of the radiolabeled precursor of Mia40 by isolated mitochondria. The reisolated mitochondria were subjected to SDS–PAGE and digital autoradiography. Mature Mia40 analyzed by Western blotting is shown in samples 11 and 12. p, precursor; i, intermediate; m, mature.
To determine the cellular localization of Mia40, we expressed the protein in Escherichia coli and generated antibodies. Purified recombinant Mia40 as well as the mitochondrial protein decorated with antibodies against Mia40 migrated significantly more slowly on SDS–PAGE than expected according to the predicted molecular mass (Figure 1C, lane 1). This behavior is characteristic for highly acidic proteins due to a reduced binding of SDS. The specificity of the antibody reaction for Mia40 was confirmed by two independent means: the in vitro imported and processed radiolabeled Mia40 showed the same unusual mobility on SDS–PAGE (see below, Figure 1G); and the mobility of the band recognized by anti-Mia40 antibodies was altered in mia40-3 mutant mitochondria (Figure 2B). The mobility of Mia40 on blue native electrophoresis (BN–PAGE) was also highly unusual, probably due to a reduced binding of the negatively charged dye Coomassie blue. Mia40 migrated in a molecular mass range of 150–180 kDa both under mild conditions (digitonin) and under all conditions that were used to dissociate putative oligomers, including reducing conditions and strong denaturants (Figure 1C, lanes 2–4; and data not shown). The comparable mobility of Mia40 on BN–PAGE under denaturing and nondenaturing conditions resembles the behavior of monomeric proteins, while proteins that are present in oligomeric complexes under nondenaturing conditions migrate much faster on BN–PAGE upon dissociation of the complexes by SDS (Dekker et al, 1996, 1997).
Figure 2.
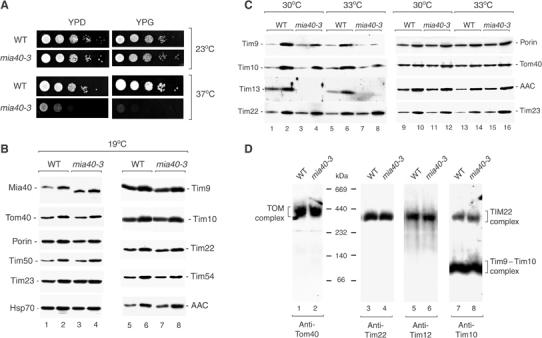
A temperature-sensitive yeast mutant of Mia40 contains reduced levels of small Tim proteins. (A) WT and mia40-3 S. cerevisiae cells were subjected to serial 10-fold dilutions, plated on YPD or YPG medium and incubated for 3 days at the temperatures indicated. (B) Mitochondria from WT and mia40-3 cells grown at 19°C were analyzed by SDS–PAGE and immunodecoration. Mia40, 15 and 45 μg mitochondrial protein, respectively; other proteins, 25 and 50 μg mitochondrial protein, respectively. (C) WT and mia40-3 cells were grown at 30 or 33°C and mitochondria were isolated. Odd-numbered lanes, 15 μg mitochondrial protein; even-numbered lanes, 45 μg mitochondrial protein. (D) Mitochondria (80 μg of protein) from cells grown at 19°C were solubilized by digitonin and subjected to BN–PAGE and immunodecoration.
A fractionation of yeast cells by differential centrifugation demonstrated that Mia40 was selectively present in the mitochondrial fraction like the ADP/ATP carrier (AAC) and Tim23 (Figure 1D). Upon sonication of mitochondria, Mia40 was largely released to the supernatant like the IMS protein cytochrome b2 and the matrix cochaperone Mge1, while Tim23 and Tim44 remained in the membrane fraction (Figure 1E). Mia40 was protected against protease treatment in intact mitochondria (Figure 1F, lanes 2–4), but was degraded by the protease after opening of the IMS space by swelling (formation of mitoplasts), like the IMS proteins cytochrome c heme lyase (CCHL) and cytochrome b2 and the IMS-exposed protein Tim23 (Figure 1F, lanes 6–8). These results demonstrate that Mia40 is located in the mitochondrial IMS.
In order to analyze whether Mia40 contained a cleavable bipartite presequence, the precursor of Mia40 was synthesized and radiolabeled in reticulocyte lysate and imported into isolated yeast mitochondria. Mia40 was processed in two steps (Figure 1G, lanes 2–5) and transported to a protease-protected location in a Δψ-dependent manner (Figure 1G, lanes 7–10). The second processing product showed the same mobility on SDS–PAGE as Mia40 recognized by anti-Mia40 antibodies (Figure 1G, lanes 11 and 12). The biogenesis of Mia40 thus shows the typical characteristics of proteins carrying a bipartite presequence like cytochrome b2 and the IMS form of NADH-cytochrome b5 reductase (Glick et al, 1992; Hahne et al, 1994).
Deletion of the MIA40 gene was lethal to yeast cells, as determined by tetrad analysis and plasmid shuffling (Winzeler et al, 1999; Stevenson et al, 2001; data not shown). To analyze the function of Mia40, we generated a conditional mutant allele of MIA40 that conferred temperature-sensitive growth. The mia40-3 yeast cells grew like wild-type cells at permissive (low) temperature, but were strongly impaired at the nonpermissive temperature of 37°C on fermentable medium and were unable to grow on nonfermentable medium at 37°C (Figure 2A). In summary, Mia40 is an essential protein of the mitochondrial IMS.
Mia40 is required for the import of precursor proteins into the IMS
We grew mia40-3 cells at different temperatures and determined the protein levels of isolated mitochondria by Western blotting. After growth at permissive temperature, mia40-3 mitochondria contained wild-type amounts of the proteins analyzed, including proteins of each of the four mitochondrial subcompartments (Figure 2B). The steady-state level of the mutant protein Mia40-3 was moderately increased. A stop codon in the mia40-3 allele led to a C-terminal truncation of Mia40-3 (see Materials and methods) and a faster mobility on SDS–PAGE than the wild-type protein (Figure 2B, lanes 1–4). When mia40-3 cells were grown at elevated temperature, the steady-state levels of Tim9, Tim10 and Tim13 were significantly reduced (Figure 2C, lanes 3, 4, 7 and 8), raising the possibility that Mia40 was involved in the biogenesis of small Tim proteins.
For a detailed analysis of the function of Mia40, we studied the import of radiolabeled precursor proteins into isolated mitochondria (shown in Figures 3, 4, 5 and 6). To avoid alterations of the steady-state protein levels of mia40-3 mitochondria, the cells were grown at permissive conditions, mitochondria were isolated and then shifted to elevated temperature. BN–PAGE analysis showed that the isolated mitochondria contained intact TOM and TIM complexes (Figure 2D), including the 70 kDa Tim9–Tim10 complex that consists of three Tim9 molecules and three Tim10 molecules; and the 300 kDa TIM22 complex that consists of the integral inner membrane proteins Tim22, Tim54 and Tim18 and small Tim proteins (Tim12 and a fraction of Tim9–Tim10) (Sirrenberg et al, 1996, 1998; Kerscher et al, 1997; Koehler et al, 1998a; Koehler, 2004; Lu et al, 2004b; Wiedemann et al, 2004a).
Figure 3.
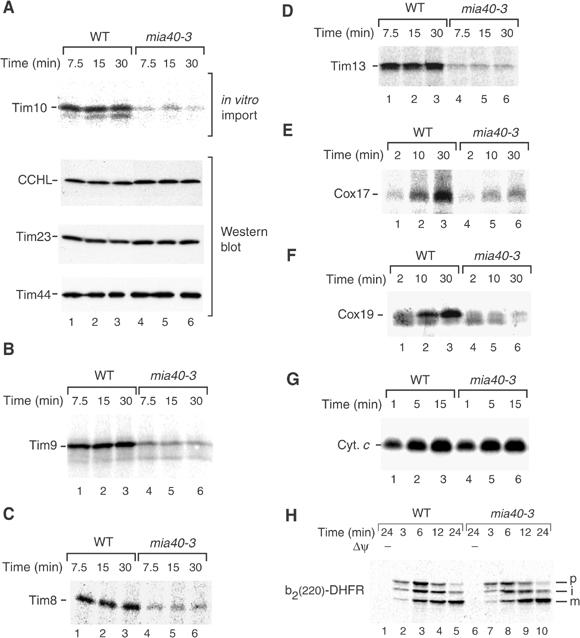
mia40-3 mitochondria are inhibited in import of small Tim proteins, Cox17 and Cox19 into the IMS. (A) Energized mitochondria isolated from WT and mia40-3 cells grown at 19°C were incubated with the radiolabeled precursor of Tim10 at 30°C for the indicated timespans. The import reaction was stopped on ice. Nonimported precursors were removed by treatment with proteinase K. The reisolated mitochondria were subjected to digital autoradiography and Western blot analysis. (B–H) The radiolabeled precursors of Tim9, Tim8, Tim13, Cox17, Cox19, cytochrome c (Cyc1) and b2(220)-DHFR were imported into WT and mia40-3 mitochondria in the presence of a Δψ unless indicated otherwise. The mitochondria were treated with proteinase K.
Figure 4.
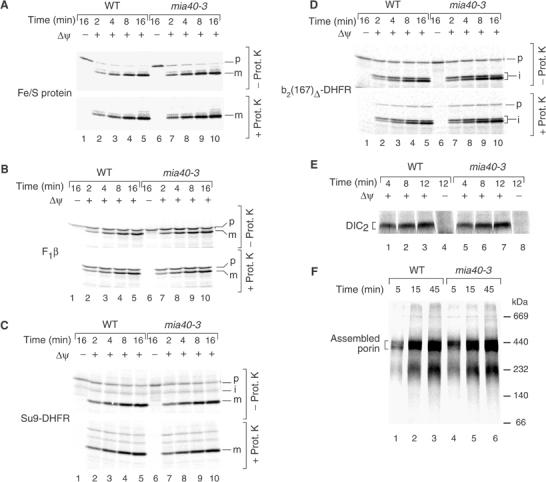
mia40-3 mitochondria are not impaired in protein import to the matrix, inner membrane or outer membrane. (A–D) The radiolabeled precursors of the Rieske Fe/S protein, F1β, Su9-DHFR and b2(167)Δ-DHFR were imported into WT and mia40-3 mitochondria (isolated from cells grown at 19°C) at 30°C in the presence or absence of a Δψ. Where indicated, the mitochondria were treated with proteinase K. The mitochondria were reisolated and analyzed by SDS–PAGE and digital autoradiography. (E) The precursor of DIC was imported into WT and mia40-3 mitochondria. The mitochondria were treated with proteinase K and analyzed by BN–PAGE and digital autoradiography. DIC2, mature dimeric form of DIC. (F) The precursor of porin was imported into WT and mia40-3 mitochondria at 30°C. The mitochondria were analyzed by BN–PAGE.
Figure 5.
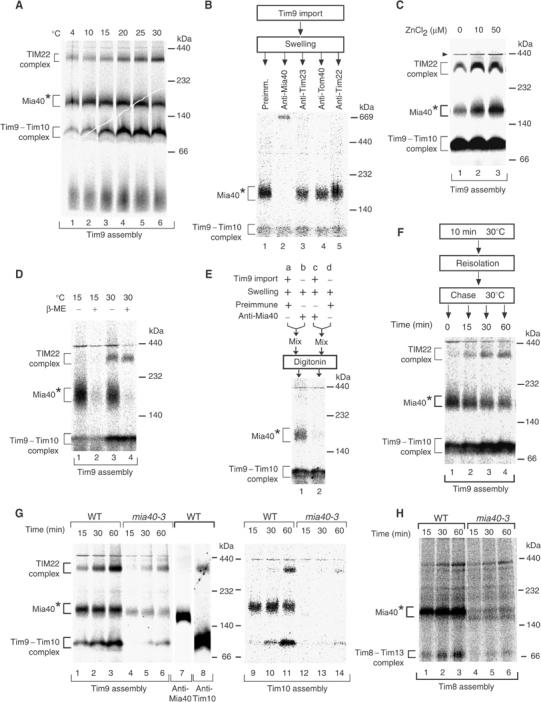
The precursors of small Tim proteins associate with Mia40. (A) The radiolabeled precursor of Tim9 was incubated with energized WT mitochondria for 45 min at the indicated temperatures. The mitochondria were reisolated, solubilized by digitonin and subjected to BN–PAGE and autoradiography. Mia40*, transport intermediate bound to Mia40. (B) Tim9 was imported into WT mitochondria for 10 min at 30°C. The mitochondria were converted to mitoplasts and incubated with antibodies for 30 min on ice. After reisolation and solubilization by digitonin, the samples were analyzed by BN–PAGE and autoradiography. (C) Tim9 was imported into WT mitochondria for 45 min at 30°C in the presence of zinc chloride as indicated. The mitochondria were reisolated, solubilized and subjected to BN–PAGE and autoradiography. Arrowhead, unspecific band. (D) Tim9 was imported into WT mitochondria for 15 min at 15 or 30°C. The mitochondria were lysed in digitonin buffer in the presence or absence of β-mercaptoethanol (β-ME) and analyzed by BN–PAGE. (E) Tim9 was imported into WT mitochondria for 10 min at 30°C where indicated. The mitochondria were subjected to swelling and incubated with antibodies. After reisolation, the mitochondria were mixed as indicated, solubilized with digitonin and analyzed by BN–PAGE and autoradiography. (F) Tim9 was imported into WT mitochondria for 10 min at 30°C. The mitochondria were reisolated, incubated at 30°C as indicated and analyzed by BN–PAGE. (G) Mitochondria isolated from WT and mia40-3 cells grown at 19°C were incubated with the radiolabeled precursors of Tim9 and Tim10 at 30°C. After solubilization with digitonin, the samples were analyzed by BN–PAGE and autoradiography. For comparison, WT mitochondria were analyzed by BN–PAGE and immunodecoration (samples 7 and 8). (H) Tim8 was imported into WT and mia40-3 mitochondria as described in the legend to G.
Figure 6.
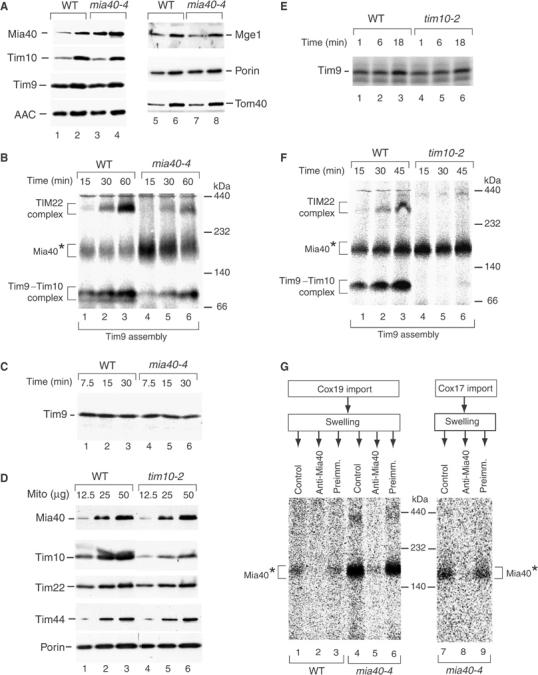
Mia40 is involved in the assembly pathway of small Tim proteins. (A) Mitochondria from WT and mia40-4 cells grown at 19°C were analyzed by SDS–PAGE and immunodecoration. Mia40, Tim10, Mge1 and Tom40, 15 μg mitochondrial protein (odd-numbered lanes), 45 μg mitochondrial protein (even-numbered lanes); other proteins, 25 μg mitochondrial protein (odd-numbered lanes), 50 μg mitochondrial protein (even-numbered lanes). (B) Energized mitochondria isolated from WT and mia40-4 cells grown at 19°C were incubated with the radiolabeled precursor of Tim9 at 30°C. The mitochondria were solubilized with digitonin and analyzed by BN–PAGE and autoradiography. (C) Tim9 was imported into WT and mia40-4 mitochondria at 30°C, followed by treatment with proteinase K. The mitochondria were reisolated and subjected to SDS–PAGE and autoradiography. (D) Mitochondria (μg of protein) from WT and tim10-2 cells grown at 24°C were analyzed by SDS–PAGE and immunodecoration. (E) Tim9 was imported into WT and tim10-2 mitochondria at 30°C, followed by proteinase K treatment and SDS–PAGE. (F) Tim9 was imported into WT and tim10-2 mitochondria at 30°C and analyzed by BN–PAGE and digital autoradiography. (G) Radiolabeled Cox19 and Cox17 were imported into WT or mia40-4 mitochondria in the presence of 10 μM CuCl2. After swelling and incubation with antibodies as indicated, the mitoplasts were reisolated and analyzed by BN–PAGE.
The precursors of IMS proteins were synthesized in reticulocyte lysate in the presence of [35S]methionine/cysteine and incubated with isolated mia40-3 and wild-type mitochondria at 30°C. The mitochondria were then treated with proteinase K to remove nonimported precursor molecules. The transport of Tim10, Tim9, Tim8 and Tim13 to a protease-protected location was strongly inhibited in mia40-3 mitochondria (Figure 3A, upper panel; Figure 3B–D). To exclude that the outer membrane of mia40-3 mitochondria was leaky to the protease, we determined the levels of endogenous CCHL and Tim23 by Western blot analysis. Tim23 and CCHL, which were degraded by a low concentration of proteinase K when the outer membrane was opened (Figure 1F, lane 6), were completely protected in mia40-3 mitochondria (Figure 3A, middle panels), demonstrating the intactness of the outer membrane barrier. Similarly, the import of Cox17 and Cox19 was inhibited in mia40-3 mitochondria in comparison to wild-type mitochondria (Figure 3E and F).
Previous studies showed that the biogenesis pathway of the small IMS protein cytochrome c is different from that of all other mitochondrial proteins studied so far. Although the precursor apocytochrome c employs the TOM complex, the pattern of dependence on individual Tom proteins is unique (Diekert et al, 2001; Wiedemann et al, 2003b). The covalent addition of heme by CCHL is required for the accumulation of cytochrome c in the mitochondrial IMS (Nargang et al, 1988; Dumont et al, 1991). The import of apocytochrome c into mia40-3 mitochondria occurred with similar efficiency as into wild-type mitochondria (Figure 3G). We conclude that, with the exception of cytochrome c, functional Mia40 is required for the mitochondrial import of the small IMS proteins analyzed.
Finally, we asked whether Mia40 was also involved in the biogenesis of an IMS protein with a bipartite presequence that is imported via the TIM23 complex. We used the model protein b2(220)-DHFR that is imported in a Δψ-dependent manner and processed twice (Geissler et al, 2002). mia40-3 mitochondria processed b2(220)-DHFR and transported it to a protease-protected location with the same efficiency as wild-type mitochondria (Figure 3H), indicating that the IMS import pathway via the TIM23 complex does not require functional Mia40.
Protein import into the matrix, inner membrane or outer membrane is not inhibited in mia40-3 mitochondria
We asked how specific the protein import defect of mia40-3 mitochondria was. We synthesized the precursors of four presequence-carrying preproteins in reticulocyte lysate: the inner membrane protein Rieske Fe/S protein (Figure 4A); the matrix protein F1-ATPase subunit β (F1β) (Figure 4B); and two matrix-targeted model proteins carrying N-terminal targeting signals fused to dihydrofolate reductase (Su9-DHFR and b2(167)Δ-DHFR) (Figure 4C and D). All four preproteins were efficiently processed and transported to a protease-protected location by mia40-3 mitochondria in a Δψ-dependent manner.
The import via the carrier import pathway to the TIM22 complex was studied by following the Δψ-dependent assembly of the dicarboxylate carrier (DIC) by BN–PAGE (Figure 4E) (Rehling et al, 2003). The assembly pathway of the outer membrane β-barrel protein porin was similarly analyzed by BN–PAGE (Figure 4F) (Wiedemann et al, 2003a). In both cases, import and assembly were not impaired in mia40-3 mitochondria. We conclude that mia40-3 mitochondria are able to efficiently import outer membrane, inner membrane and matrix proteins.
Small Tim proteins associate with Mia40 on their import pathway
We asked whether Mia40 associated with small Tim proteins on their import pathway. The import of radiolabeled Tim9 into wild-type mitochondria was analyzed by BN–PAGE. Isolated mitochondria were incubated with the Tim9 precursor at different temperatures and then lysed with digitonin. BN–PAGE showed the temperature-dependent formation of the mature 70 kDa Tim9–Tim10 complex and 300 kDa TIM22 complex. In addition, a band migrating at about 180 kDa, designated Mia40*, was observed even at relatively low temperatures (Figure 5A). This band was not found in the mature Tim9–Tim10 or TIM22 complexes (see below, Figure 5G, lane 8), and was slightly larger than the Mia40 band on BN–PAGE (Figure 5G, lane 7).
In order to determine whether this band represented an intermediate of small Tim proteins bound to Mia40, we analyzed its composition by antibody-shift BN–PAGE (Figure 5B) (Truscott et al, 2002; Wiedemann et al, 2003a). Radiolabeled Tim9 was accumulated in Mia40* of wild-type mitochondria by performing the import reaction for a short time. The outer membrane was opened by swelling and antibodies were added (under the swelling conditions, Mia40 remains associated with the mitoplasts like the IMS protein CCHL, see Figure 1F). The mitoplasts were reisolated and radiolabeled complexes were separated by BN–PAGE. Antibodies against Mia40 completely shifted the Mia40* band, but not the Tim9–Tim10 complex (Figure 5B, lane 2). In contrast, neither antibodies against Tom40 nor antibodies against Tim23 or Tim22 shifted the Mia40* band (Figure 5B, lanes 3–5). These results demonstrate that the precursor of Tim9 is associated with Mia40 during its import pathway.
It has been reported that the biogenesis pathway of small Tim proteins involves the binding of zinc ions and/or formation of disulfide bonds (Allen et al, 2003; Lutz et al, 2003; Koehler, 2004; Lu et al, 2004a, 2004b). We noticed that the relative amount of the Mia40* intermediate was variable, depending, for example, on the batch of reticulocyte lysate used. We thus added zinc ions during the import reaction and observed a significant increase in the formation of the Mia40* intermediate (Figure 5C). To determine whether disulfide bonds were also involved in the formation of the Mia40* intermediate, we added β-mercaptoethanol in the lysis buffer after the import of Tim9 into wild-type mitochondria. We used two import conditions: low temperature to mainly generate the Mia40* intermediate (Figure 5D, lanes 1 and 2) and higher temperature to additionally generate the mature TIM complexes (Figure 5D, lanes 3 and 4). In both cases, the Mia40* intermediate was efficiently dissociated by β-mercaptoethanol, while the Tim9–Tim10 complex as well as the TIM22 complex were also present under reducing conditions. Since the reducing conditions do not affect the mobility of the Mia40 protein on BN–PAGE (Figure 1C), we conclude that the interaction between Mia40 and the small Tim proteins also depends on disulfide bonds. These results suggest that the biogenesis of small Tim proteins via Mia40 may involve both metal ions and disulfide bonds.
Of concern was the possibility that the observed interaction between Mia40 and the precursor of Tim9 occurred after lysis of mitochondria. We therefore performed a mixing experiment. Radiolabeled Tim9 was imported into wild-type mitochondria (Figure 5E, reaction a), while in a parallel reaction mitochondria were incubated without radiolabeled precursor (Figure 5E, reaction b). Both mitochondria were subjected to swelling. The mitochondria of reaction b received antibodies directed against Mia40. Then, both mitochondria were reisolated, mixed and lysed by digitonin. In a control reaction, radiolabeled Tim9 was imported into mitochondria that subsequently received anti-Mia40 antibodies (Figure 5E, reaction c) and were then mixed with mitochondria that received neither Tim9 precursor nor antibodies (Figure 5E, reaction d). The BN–PAGE analysis shows that the Mia40* intermediate was only affected by the anti-Mia40 antibodies when the antibodies had been added to the mitochondria importing Tim9 (Figure 5E, lane 2). Thus, the association of radiolabeled Tim9 with Mia40 occurred within mitochondria and not after lysis.
We asked whether a precursor accumulated in Mia40* was chased to the mature TIM complexes. Tim9 was accumulated in Mia40* by a short-term import reaction (Figure 5F, lane 1). Then the reisolated mitochondria were incubated further at 30°C. Thereby, the Tim9 precursor was chased into the mature complexes, the Tim9–Tim10 complex and the TIM22 complex (Figure 5F, lanes 2–4). This finding demonstrates that the association with Mia40 represents a productive intermediate in the biogenesis pathway of the small Tim protein.
We investigated whether the mia40-3 mutant mitochondria were inhibited in formation of the Mia40* intermediate. Radiolabeled Tim9 or Tim10 were imported into wild-type and mia40-3 mitochondria and analyzed by BN–PAGE. In mia40-3 mitochondria, not only was the formation of the Tim9–Tim10 and TIM22 complexes impaired, but also the formation of the Mia40* intermediate (Figure 5G). Similarly, upon import of radiolabeled Tim8 into mia40-3 mitochondria, the formation of Mia40* and of the mature 70 kDa Tim8–Tim13 complex were inhibited compared to wild-type mitochondria (Figure 5H). Therefore, functional Mia40 is required for generation of the intermediate on the import pathway of small Tim proteins.
Small Tim proteins accumulate at Mia40 when the assembly into mature TIM complexes is impaired
The results shown above suggested that the decreased binding of small Tim precursors to Mia40-3 may be the reason for their impaired translocation across the outer membrane. To obtain independent evidence for this view, we selected the temperature-sensitive mia40-4 yeast mutant. Marker proteins were comparable between wild-type and mia40-4 mitochondria when the cells were grown at permissive (low) temperature (Figure 6A). The level of the mutant protein Mia40-4 was increased about two-fold in comparison to wild-type Mia40 (Figure 6A, upper panel, lanes 1–4). When radiolabeled Tim9 was imported into isolated mia40-4 mitochondria, significantly increased amounts of the Mia40* intermediate were observed, while the formation of the Tim9–Tim10 and TIM22 complexes was delayed (Figure 6B, lanes 4–6 versus 1–3). Thus, the mutant protein Mia40-4 is able to bind the precursors of small Tim proteins, but is probably impaired in their release, leading to a prolonged presence of the precursors in the Mia40* intermediate. We then analyzed the translocation of the precursor of Tim9 to a protease-protected location by SDS–PAGE. In mia40-4 mitochondria, the precursor of Tim9 was indeed transported to a protease-protected location, with an efficiency close to that of wild-type mitochondria (Figure 6C), in clear contrast to the import inhibition in mia40-3 mitochondria (Figure 3B). The different ability of the mutant mitochondria mia40-3 and mia40-4 to translocate Tim precursors to a protease-protected location correlates with the ability of the mutant Mia40 to associate with the precursor proteins, indicating that binding to Mia40 is critical for the translocation of small Tims across the outer membrane.
We asked where the precursors of small Tim proteins accumulated when the final assembly into the mature TIM complexes was impaired. We used tim10-2 mutant mitochondria. As reported by Truscott et al (2002) and Wiedemann et al (2004b), tim10-2 mitochondria contain wild-type levels of marker proteins for the different mitochondrial subcompartments, only the levels of Tim10 and Tim9 are reduced. Mia40 was present in wild-type amounts in tim10-2 mitochondria (Figure 6D). The mutant mitochondria efficiently transported the radiolabeled precursor of Tim9 to a protease-protected location (Figure 6E). As expected, due to the mutation and reduced level of the Tim10 partner protein, the assembly of the Tim9 precursor into the mature TIM complexes was strongly impaired in tim10-2 mitochondria (Figure 6F). The radiolabeled Tim9 precursor accumulated as a Mia40* intermediate (Figure 6F). Thus, the final assembly of Tim9 into the mature TIM complexes is not required for translocation of the precursor across the outer membrane, but unassembled Tim9 molecules accumulate at Mia40 in the IMS. Mia40 thus represents a link between translocation and assembly of small Tim proteins.
We investigated whether the precursors of Cox19 and Cox17 interacted with Mia40 on their biogenesis pathway. Radiolabeled Cox19 was imported in the presence of the oxidizing reagent CuCl2, which promotes crosslinking by formation of disulfide bridges (Seedorf and Soll, 1995). BN–PAGE revealed a Mia40* band, which was specifically increased in mia40-4 mitochondria (Figure 6G, lanes 1 and 4). To verify the presence of Mia40 in the intermediate, an antibody-shift BN–PAGE was performed. The mitochondria were subjected to swelling after the import reaction and incubated with antibodies. Anti-Mia40 completely shifted the Mia40* intermediate, whereas preimmune antibodies had no effect (Figure 6G, lanes 2 and 5 versus 3 and 6). A similar Mia40* intermediate was observed for the import of Cox17 into mia40-4 mitochondria (Figure 6G, lanes 7–9). We conclude that Cox17 and Cox19 interact with Mia40 on their import pathway.
Discussion
We report that Mia40 is the first essential protein of an IMS-specific protein import machinery. The precursors of small Tim proteins are transported across the outer membrane in a loosely folded conformation (Lutz et al, 2003; Koehler, 2004; Lu et al, 2004a). The small Tim proteins bind to Mia40 and this interaction is crucial for the complete translocation into the IMS. Mia40 is not present in the mature TIM complexes of the IMS, but is required for formation of an intermediate step in the import and assembly pathway of small Tim proteins.
When the assembly of the Tim9–Tim10 complex is impaired due to a tim10 mutation, radiolabeled precursors of small Tim proteins are completely translocated into the IMS and accumulate at Mia40. The suggestion that Mia40 is important for the import and assembly pathway of small Tims is underscored by the comparison of two different mia40 mutant mitochondria. The mutant protein Mia40-3 is impaired in its interaction with the precursors of small Tim proteins. The translocation of the precursors into the IMS is thus inhibited and, as a consequence, their subsequent assembly into the mature TIM complexes is hindered. The mutant protein Mia40-4, however, efficiently associates with the small Tim proteins and indeed the precursor proteins are translocated into the IMS. However, the assembly of the small Tim proteins into the mature TIM complexes is delayed since the precursors accumulate at this Mia40 mutant protein.
Mia40 is not required for the three previously characterized mitochondrial protein import pathways that direct proteins to the matrix, inner membrane or outer membrane: the presequence pathway via the TIM23 complex, the carrier pathway via the TIM22 complex and the β-barrel pathway via the SAM complex (Herrmann and Neupert, 2000; Jensen and Johnson, 2001; Endo et al, 2003; Wiedemann et al, 2004a). IMS proteins with a bipartite presequence that use the TIM23 complex are also imported independently of functional Mia40. Interestingly, the precursor of Mia40 itself also contains a bipartite presequence and is imported via the TIM23 complex.
Mia40 contains six highly conserved cysteines. Four of these cysteines form a twin Cx9C motif that is also found in other IMS proteins such as the copper protein Cox17 and the related protein Cox19 (Beers et al, 1997; Nobrega et al, 2002). It is currently a matter of debate whether the cysteines of the small Tim proteins are required for coordination of metal ions (zinc) or the formation of disulfide bonds (Sirrenberg et al, 1998; Curran et al, 2002a, 2002b; Allen et al, 2003; Lu et al, 2004a, 2004b). Several studies showed an involvement of the four conserved cysteines of the small Tim proteins in the import and assembly of these proteins (Roesch et al, 2002; Allen et al, 2003; Lutz et al, 2003; Koehler, 2004; Lu et al, 2004a). Recently, it has been suggested that both mechanisms, zinc binding and disulfide bond formation, are involved in the biogenesis of small Tim proteins (Lutz et al, 2003; Koehler, 2004; Lu et al, 2004a, 2004b). The characterization of the role of Mia40 in the import of small Tim proteins is in agreement with this view, since an involvement of both metal ions and disulfide bonds was found. A combination of both mechanisms in protein function or biogenesis has been reported for proteins such as copper/zinc-superoxide dismutase, metallothionein and the heat shock protein Hsp33 (Maret and Vallee, 1998; Jakob et al, 2000; Sturtz Field et al, 2003).
The role of Mia40 in protein import is not restricted to small Tim proteins. The transport of Cox17 and Cox19 into the IMS similarly required functional Mia40. Thus, Mia40 seems to play a general role in the translocation, into the IMS, of small proteins containing conserved cysteine residues. The exception is cytochrome c that contains two cysteines to which heme is covalently attached. The biogenesis of cytochrome c has been addressed in many studies that revealed a unique import mechanism, including an unusual dependence on Tom proteins (Diekert et al, 2001; Wiedemann et al, 2003b). The covalent attachment of heme by CCHL in the IMS is crucial for translocation of the precursor of cytochrome c and its trapping in the IMS (Nargang et al, 1988; Dumont et al, 1991). The ‘folding-trap' model, which predicts a passive diffusion via the TOM complex and trapping in the IMS by refolding (Lutz et al, 2003), may thus apply to the biogenesis of cytochrome c. Other small IMS proteins tested, however, critically depend on the presence of functional Mia40. Since these IMS proteins apparently contain different metal ions or even disulfide bonds, it is unlikely that the role of Mia40 is restricted to the donation of a specific metal ion to an incoming precursor protein.
In summary, Mia40 is a central player of the IMS-import machinery for small IMS proteins. Mia40 binds to the incoming unfolded precursor proteins and promotes the completion of translocation into the IMS. The assembly of imported proteins into functional complexes, like in the case of small Tim proteins, is initiated from the Mia40-bound state. Thus, Mia40 plays an essential function in the mitochondrial IMS protein import and assembly pathway.
Materials and methods
Yeast strains and growth conditions
The S. cerevisiae strains used are derivatives of YPH499 (MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-801) (Sikorski and Hieter, 1989). For the isolation of wild-type mitochondria, the YPH499 strain was grown at 24–28°C in liquid YPG medium (1% (w/v) yeast extract, 2% (w/v) bacto-peptone, 3% (w/v) glycerol).
Conditional mutants of MIA40 (encoded by YKL195w/FMP15) were generated by error-prone PCR and isolated by screening for a temperature-sensitive growth phenotype according to the procedure described in Truscott et al (2002). The mutant strains and the corresponding wild-type strain YPH499-BG-FOMP2-wild type (WT) (MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-801 mia40::ADE2 (pFL39-FOMP2/MIA40-WT)) contained a chromosomal deletion of MIA40 and carried the mutant or wild-type allele of MIA40, respectively, on a centromeric plasmid under the control of its own promoter and terminator. The strains YPH-BG-fomp2-8 (mia40-3; MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-80 mia40::ADE2 (pFL39-FOMP2-8ts/mia40-3)) and YPH-BG-fomp2-7 (mia40-4; MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-80 mia40::ADE2 (pFL39-FOMP2-7ts/mia40-4)) were selected. The mutations led to the following amino-acid changes: Mia40-3: K100R, E142G, E144A, D178G, M361V, E370Stop; Mia40-4: T313A, I316M, F342L.
For the isolation of mitochondria and in vitro import studies, the mia40-3 and mia40-4 strains and the corresponding wild-type strain were grown at 19°C in liquid YPG medium. To study specific defects in vivo, mia40-3 and the corresponding wild-type strain were grown in liquid YPG medium at 30°C or 33°C.
In vitro import of precursor proteins
Mitochondrial precursor proteins were synthesized in rabbit reticulocyte lysate (Amersham) in the presence of 35S-labeled methionine/cysteine. The import of radiolabeled precursors (with or without prior denaturation in urea) into isolated mitochondria was performed at 30°C in the presence of 2 mM ATP, 2 mM NADH and an ATP-regeneration system (5 mM creatine phosphate and 0.1 mg/ml creatine kinase) in the standard import buffer (Ryan et al, 2001). The import reaction was stopped on ice by adding proteinase K (50 μg/ml) for 15 min or by dissipation of the membrane potential (Ryan et al, 2001).
Fractionation procedures
For fractionation of yeast cells, spheroplasts were lysed and subjected to successive differential centrifugation. Pellets and supernatant fractions were analyzed by immunodecoration with antibodies against marker proteins.
To release soluble proteins, mitochondria were sonicated (3 × 30 s with 40% duty cycle in a Branson Sonifier 250) in ice-cold buffer containing 250 mM sucrose, 1 mM EDTA and 10 mM MOPS, pH 7.2. The soluble and membrane-bound material was separated by centrifugation at 100 000 × g for 1 h. Isolated mitochondria were converted to mitoplasts by hypotonic swelling and were treated with proteinase K according to Ryan et al (2001).
BN–PAGE and antibody shift
BN–PAGE was performed essentially as described previously (Dekker et al, 1996, 1997). Briefly, mitochondria (80–100 μg protein) were solubilized in ice-cold 1% digitonin buffer (1% (w/v) digitonin, 20 mM Tris–HCl, pH 7.4, 50 mM NaCl, 10% (w/v) glycerol, 0–0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). Soluble material was separated on a 6–16.5% gradient gel at 4°C and subjected to autoradiography. For immunodetection of protein complexes, the soluble material separated by BN–PAGE was transferred to a PVDF membrane and decorated with specific antibodies. Antibody-shift BN–PAGE was performed as described (Truscott et al, 2002; Wiedemann et al, 2003a), except that the mitochondria were converted to mitoplasts prior to the incubation with antibodies.
Miscellaneous
For generation of antibodies, full-length precursor of Mia40 with a His10-tag at the N-terminus was expressed in E. coli BL21-Codon Plus (DE3)-RIL (Stratagene) after induction by IPTG, purified by Ni-NTA affinity chromatography under denaturing conditions and injected into rabbits. Recombinant Mia40 was used for affinity purification of antibodies.
SDS–PAGE was performed according to standard procedures. For the separation of proteins below 15 kDa, Tris-tricine SDS–PAGE or urea-SDS PAGE were applied (Wiedemann et al, 2003b). In some figures, nonrelevant gel lanes were excised by digital treatment. Western blots were performed on PVDF membranes according to standard procedures.
Acknowledgments
We thank Drs ME Dumont, S Rospert and T Sommer for antisera, and H Müller and B Schönfisch for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388, Max Planck Research Award, Alexander von Humboldt Foundation, Bundesministerium für Bildung und Forschung and the Fonds der Chemischen Industrie.
References
- Adam A, Endres M, Sirrenberg C, Lottspeich F, Neupert W, Brunner M (1999) Tim9, a new component of the TIM22-54 translocase in mitochondria. EMBO J 18: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S, Lu H, Thornton D, Tokatlidis K (2003) Juxtaposition of the two distal Cx3C motifs via intrachain disulfide bonding is essential for the folding of Tim10. J Biol Chem 278: 38505–38513 [DOI] [PubMed] [Google Scholar]
- Beers J, Glerum DM, Tzagoloff A (1997) Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J Biol Chem 272: 33191–33196 [DOI] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Oppliger W, Koehler CM (2002a) The Tim9p–Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J 21: 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Schmidt E, Koehler CM (2002b) The role of the Tim8p–Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J Cell Biol 158: 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Sepuri NB, Holder J, Johnson AE, Jensen RE (2000) Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J Cell Biol 150: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJT, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J 16: 5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJT, Müller H, Rassow J, Pfanner N (1996) Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol Chem 377: 535–538 [PubMed] [Google Scholar]
- Diekert K, de Kroon AI, Ahting U, Niggemeyer B, Neupert W, de Kruijff B, Lill R (2001) Apocytochrome c requires the TOM complex for translocation across the mitochondrial outer membrane. EMBO J 20: 5626–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont ME, Cardillo TS, Hayes MK, Sherman F (1991) Role of cytochrome c heme lyase in mitochondrial import and accumulation of cytochrome c in Saccharomyces cerevisiae. Mol Cell Biol 11: 5487–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M (2003) Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci 116: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Frazier AE, Dudek J, Guiard B, Voos W, Li Y, Lind M, Meisinger C, Geissler A, Sickmann A, Meyer HE, Bilanchone V, Cumsky MG, Truscott KN, Pfanner N, Rehling P (2004) Pam16 has an essential role in the mitochondrial protein import motor. Nat Struct Mol Biol 11: 226–233 [DOI] [PubMed] [Google Scholar]
- Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P (2002) The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111: 507–518 [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164: 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Brandt A, Cunningham K, Müller S, Hallberg RL, Schatz G (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69: 809–822 [DOI] [PubMed] [Google Scholar]
- Hahne K, Haucke V, Ramage L, Schatz G (1994) Incomplete arrest in the outer membrane sorts NADH-cytochrome b5 reductase to two different submitochondrial compartments. Cell 79: 829–839 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Neupert W (2000) Protein transport into mitochondria. Curr Opin Microbiol 3: 210–214 [DOI] [PubMed] [Google Scholar]
- Hoppins SC, Nargang FE (2004) The Tim8–Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem 279: 12396–12405 [DOI] [PubMed] [Google Scholar]
- Jakob U, Eser M, Bardwell JCA (2000) Redox switch of hsp33 has a novel zinc-binding motif. J Biol Chem 275: 38302–38310 [DOI] [PubMed] [Google Scholar]
- Jensen RE, Johnson AE (2001) Opening the door to mitochondrial protein import. Nat Struct Biol 8: 1008–1010 [DOI] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE (1997) The Tim54p–Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol 139: 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM (2004) The small Tim proteins and the twin Cx3C motif. Trends Biochem Sci 29: 1–4 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G (1998a) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279: 369–373 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K (1998b) Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J 17: 6477–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K (2004) The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat Struct Mol Biol 11: 234–241 [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N (2003) An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278: 48520–48523 [DOI] [PubMed] [Google Scholar]
- Luciano P, Vial S, Vergnolle MAS, Dyall SD, Robinson DR, Tokatlidis K (2001) Functional reconstitution of the import of the yeast ADP/ATP carrier mediated by the TIM10 complex. EMBO J 20: 4099–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Allen S, Wardleworth L, Savory P, Tokatlidis K (2004a) Functional TIM10 chaperone assembly is redox-regulated in vivo. J Biol Chem 279: 18952–18958 [DOI] [PubMed] [Google Scholar]
- Lu H, Golovanov AP, Alcock F, Grossmann JG, Allen S, Lian LY, Tokatlidis K (2004b) The structural basis of the TIM10 chaperone assembly. J Biol Chem 279: 18959–18966 [DOI] [PubMed] [Google Scholar]
- Lutz T, Neupert W, Herrmann JM (2003) Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J 22: 4400–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret W, Vallee BL (1998) Thiolate ligands in metallothionein confer redox activity on zinc clusters. Proc Natl Acad Sci USA 95: 3478–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargang FE, Drygas ME, Kwong PL, Nicholson DW, Neupert W (1988) A mutant of Neurospora crassa deficient in cytochrome c heme lyase activity cannot import cytochrome c into mitochondria. J Biol Chem 263: 9388–9394 [PubMed] [Google Scholar]
- Nobrega MP, Bandeira SCB, Beers J, Tzagoloff A (2002) Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J Biol Chem 277: 40206–40211 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426: 862–866 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Tropschug M, Neupert W (1987) Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell 49: 815–823 [DOI] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N (2003) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Roesch K, Curran SP, Tranebjaerg L, Koehler CM (2002) Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a–TIMM13 complex. Hum Mol Genet 11: 477–486 [DOI] [PubMed] [Google Scholar]
- Ryan MT, Voos W, Pfanner N (2001) Assaying protein import into mitochondria. Meth Cell Biol 65: 189–215 [DOI] [PubMed] [Google Scholar]
- Seedorf M, Soll J (1995) Copper chloride, an inhibitor of protein import into chloroplasts. FEBS Lett 367: 19–22 [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M (1996) Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384: 582–585 [DOI] [PubMed] [Google Scholar]
- Sirrenberg C, Endres M, Fölsch H, Stuart RA, Neupert W, Brunner M (1998) Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature 391: 912–915 [DOI] [PubMed] [Google Scholar]
- Stevenson LF, Kennedy BK, Harlow E (2001) A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci USA 98: 3946–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtz Field L, Furukawa Y, O'Halloran TV, Culotta VC (2003) Factors controlling the uptake of yeast copper/zinc superoxide dismutase into mitochondria. J Biol Chem 278: 28052–28059 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Brandner K, Pfanner N (2003) Mechanisms of protein import into mitochondria. Curr Biol 13: R326–R337 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Müller H, Meisinger C, Pfanner N, Guiard B (2002) Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol 22: 7780–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Frazier AE, Pfanner N (2004a) The protein import machinery of mitochondria. J Biol Chem 279: 14473–14476 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C (2003a) Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424: 565–571 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Prinz T, Ryan MT, Meisinger C, Pfanner N, Truscott KN (2003b) Biogenesis of yeast mitochondrial cytochrome c: a unique relationship to the TOM machinery. J Mol Biol 327: 465–474 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N (2004b) Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem 279: 18188–18194 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Whelen Dow S, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Véronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112: 41–50 [DOI] [PubMed] [Google Scholar]


