Abstract
Sumoylation represents a conserved mechanism of post-translational protein modification. We report that Pli1p, the unique fission yeast member of the SP-RING family, is a SUMO E3 ligase in vivo and in vitro. pli1Δ cells display no obvious mitotic growth defects, but are sensitive to the microtubule-destabilizing drug TBZ and exhibit enhanced minichromosome loss. The weakened centromeric function of pli1Δ cells may be related to the defective heterochromatin structure at the central core, as shown by the reduced silencing of an ura4 variegation reporter gene inserted at cnt and imr. Interestingly, pli1Δ cells also exhibit enhanced loss of the ura4 reporter at these loci, likely by gene conversion using homologous sequences as information donors. Moreover, pli1Δ cells exhibit consistent telomere length increase, possibly achieved by a similar process. Point mutations within the RING finger of Pli1p totally or partially reproduce the pli1 deletion phenotypes, thus correlating with their sumoylation activity. Altogether, these results strongly suggest that Pli1p, and by extension sumoylation, is involved in mechanisms that regulate recombination in particular heterochromatic repeated sequences.
Keywords: centromere, genomic instability, SUMO E3 ligase, telomere
Introduction
Covalent protein modification by SUMO (sumoylation) has been shown to play important roles in such diverse processes as nucleo-cytoplasmic transport, chromosome segregation, DNA metabolism, as well as transcriptional regulation (reviewed in Melchior, 2000; Müller et al, 2001; Seeler and Dejean, 2003). Like the modification with ubiquitin and other ubiquitin-like modifiers (Ubls), sumoylation is achieved with a distinct but evolutionarily conserved pathway consisting of E1-activating, E2-conjugating and E3 ligase enzymes. Sumoylation of numerous target proteins can be achieved in vitro using only E1 and E2 enzymes; however, in vivo, it is likely that the substrate specificity and the rate of the reaction depend critically on the activity of E3 ligase-containing complexes, as is also the case for ubiquitylation. To date, three protein families have been implicated as SUMO E3 ligases. The first, or SIZ/protein inhibitor of activated Stat (PIAS) family, was initially shown in budding yeast (Saccharomyces cerevisiae) to mediate the modification of septins (Johnson and Gupta, 2001; Takahashi et al, 2001). Subsequently, members of the mammalian PIAS family were shown to be critical for the modification of transcription factors such as p53, nuclear receptors (e.g. AR, PR), Lef1 and Sp3 (Jackson, 2001; Kotaja et al, 2002; Sapetschnig et al, 2002; Gill, 2003). A second type of E3 ligase is given by the nuclear import factor RanBP2, which has been shown to mediate the modification of the nuclear proteins SP100 and HDAC4 (Kirsh et al, 2002; Pichler et al, 2002), and has provided a further link between sumoylation and nucleo-cytoplasmic transport. Finally, a third SUMO E3 ligase was discovered with the polycomb protein Pc2, which was found to enhance the modification of the transcriptional repressor CtBP1 (Kagey et al, 2003).
Studies of sumoylation in mammalian cells have largely been focused on the effects on transcriptional regulation, largely because the majority of the currently characterized SUMO substrates are either transcription factors or co-factors. A growing number of cases supports a model in which the sumoylation of a transcription factor leads to repression (reviewed in Verger et al, 2003). Consistent with this is the finding that the sumoylation target sites often fall within known repression domains of dual-function transcription factors and that mutation of these sites leads to de-repression. While the precise mechanisms involved remain elusive, it has been shown in some cases (Lef1, Sp3) that sumoylation leads to the sequestration of the transcription factor to specific nuclear domains, such as PML nuclear bodies, and thus to the concomitant attenuation of transcriptional activity. That sumoylation may also play a role at the DNA (or promoter) level has been suggested by Shiio and Eisenman (2003), who showed that the E2 enzyme (Ubc9) fused to a GAL4 DNA-binding domain could lead to transcriptional repression of a reporter gene that is associated with the appearance of sumoylated chromatin, presumably at histone H4 within nucleosomes.
Analysis of the role of sumoylation in genetically tractable organisms has opened new fields of interest such as chromosome condensation and cohesion, and DNA replication and repair. In the budding yeast, the proliferating cell nuclear antigen (PCNA) was shown to be modified by ubiquitin and SUMO on the same lysine residue (K164), sumoylation occurring during S phase and ubiquitylation upon DNA damage, channeling DNA repair towards the RAD6-dependent post-replication repair mechanisms (Hoege et al, 2002; Stelter and Ulrich, 2003). It was proposed that sumoylation of PCNA has an inhibitory function on repair since abrogation of SUMO modification by mutating both K164 and K127 (the other residue subject to sumoylation) partially rescues the DNA damage sensitivity of the PCNA K164R single mutant. Again, in budding yeast, SMT4, a de-sumoylation enzyme, was found as a high copy suppressor of thermosensitive alleles of the SMC2 gene, encoding one of the core condensin subunits (Strunnikov et al, 2001) and of the PDS5 gene, required for proper chromatid cohesion maintenance (Stead et al, 2003). Furthermore, SMT4 mutants exhibit severe defects in chromatin condensation (Strunnikov et al, 2001) and precocious dissociation of sister chromatids during mitosis, which has been linked to the sumoylation of two major proteins TOP2 and PDS5 (Bachant et al, 2002; Stead et al, 2003). In Drosophila melanogaster, the Su(var)2–10 gene, which encodes a member of the Siz/PIAS family, is required for viability, and mutants show defects in both chromosome condensation and inheritance (Hari et al, 2001). In Schizosaccharomyces pombe, mutations in the genes encoding SUMO (Pmt3p; Tanaka et al, 1999), E1 (Rad31p; Shayeghi et al, 1997), E2 (Hus5p; al-Khodairy et al, 1995) in the SUMO pathway produce cells that survive but display severe growth defects, that is, are essentially nonviable, and thus preclude proper genetic analysis.
In this work, we report the characterization of a novel fission yeast nuclear protein, Pli1p, as an E3 ligase in the SUMO pathway. Deletion of the pli1 gene, while exhibiting only mild or no mitotic phenotypes, leads to increased sensitivity to the microtubule spindle poison TBZ and minichromosome loss, indicating improper centromere and/or kinetochore function. Consistent with this, pli1Δ cells display alleviated silencing of an ura4 reporter gene at the central centromeric core required for kinetochore assembly. Furthermore, we present evidence for a role of Pli1p in protecting heterochromatic repeated sequences (i.e. centromeres and telomeres) from illegitimate recombination. The phenotypes described for pli1-deleted cells are totally or partially reproduced by point mutations within the RING finger, in correlation with their E3 ligase activity, thus strongly suggesting that sumoylation via Pli1p is involved in centromeric and telomeric functions in fission yeast.
Results
Isolation of Pli1p, an S. pombe Siz1/Siz2/PIAS homologue
Our previous studies of the biological role of the essential chromatin-associated protein switch-activating protein 1 (Sap1p) (De Lahondes et al, 2003) prompted us to perform a yeast two-hybrid screen of a S. pombe cDNA library with this protein as bait. While the functional relevance of the obtained interactants remains to be elucidated, this screen identified a novel cDNA corresponding to the hypothetical open reading frame (ORF) listed as SPAC1687.05 under the reserved name of pli1 in the Sanger Institute GeneDB database. The pli1+ ORF translates into a putative protein of 727 amino acids with a predicted molecular mass of 80.7 kDa. Amino-acid sequence comparisons revealed significant similarity to the SIZ1 and SIZ2 proteins of budding yeast as well as all known mammalian PIAS proteins. Pli1p notably shares with these two well-conserved domains: an amino-terminal SAP (SAF-A/B, Acinus and PIAS) domain and a central SP-RING (Siz/PIAS-RING) domain (Hochstrasser, 2001), essential for the SUMO E3 ligase activity associated with the Siz/PIAS proteins (Seeler and Dejean, 2003) (Figure 1). Western blot analysis of WCE of cells containing an endogenous HA (data not shown) or CFP-tagged version of the protein (Figure 2B) confirmed the existence and size of the Pli1p protein. Further immunofluorescence analysis using an anti-GFP antibody to detect the endogenous Pli1-CFP protein revealed that Pli1p is localized in the nucleus, where it forms numerous spots (Figure 2A).
Figure 1.

Sequence alignment of the SAP and SP-RING domains of Pli1p and several (putative) SP-RING SUMO ligases. Alignment of S. pombe Pli1p, S. cerevisiae Siz2p, Siz1p, D. melanogaster Zimp-B and Homo sapiens hPIASy protein sequences (GenBank accession numbers CAA22599.1, NP_014799, NP_010697, AAD29288, Q8N2W9, respectively), using CLUSTAL X algorithm. Identical residues are boxed and similar residues are shaded. Overall sequence identity of Pli1p with the other four orthologs is 26–27% (33–38% similarity), whereas the SAP domain has 22–35% identical residues (75–68% similarity) and the SP-RING domain 39–51% (70–75% similarity). Asterisks indicate the positions of the cysteiyl and histidyl residues forming the C2HC3 conserved SP-RING domain.
Figure 2.
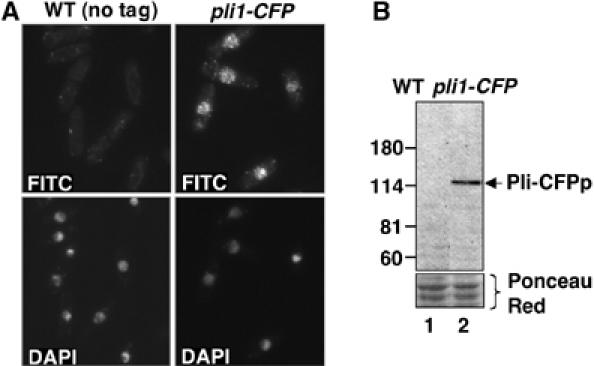
Pli1p is localized in the nucleus and forms numerous foci. (A) WT (PB46) and pli1-CFP (BX4) cells grown exponentially to 5 × 106 cells in MM at 33°C were fixed with paraformaldehyde and stained with anti-CFP rabbit polyclonal antibody, followed by FITC-conjugated anti-rabbit IgG secondary antibody and DAPI. Z-scanning of cells every 0.4 μm showed that the CFP-specific spots observed in pli1-CFP cells were present throughout the nucleus. (B) Anti-CFP Western blot of denaturing protein whole-cell extracts of WT (PB46) and pli1-CFP (BX4) cells grown in the same conditions as above.
Pli1p promotes SUMO/Pmt3p conjugation in vivo and in vitro
The homology of Pli1p to the SIZ1/2 budding yeast and mammalian PIAS proteins suggests that Pli1p might act as a SUMO E3 ligase. We therefore first analyzed the effect of pli1 deletion on the in vivo pattern of SUMO conjugates (Figure 3A) Wild-type and pli1Δ cells containing or not an N-terminal His6 tag fused in frame with the endogenous pmt3 gene were analyzed by Western blotting using a rabbit antibody raised against the S. pombe SUMO/Pmt3p protein. pli1Δ extracts showed an increase in the intensity of free Pmt3p and an overall severe decrease in the intensity of Pmt3p-specific bands corresponding to Pmt3p conjugates, indicating a significant reduction of global sumoylation in pli1-deleted cells.
Figure 3.
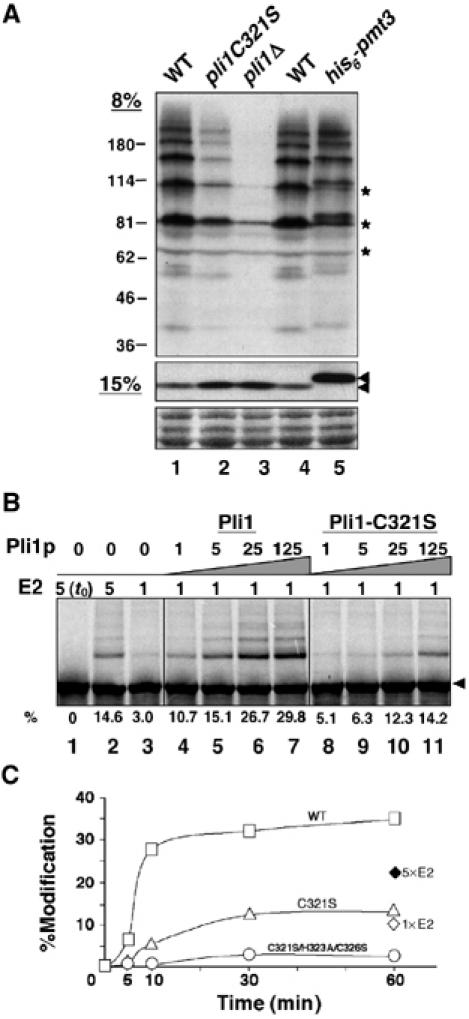
Pli1p promotes SUMO conjugation in vivo and in vitro. (A) Whole-cell extracts of WT, pli1C321S and pli1Δ cells were separated on denaturing 8% (top panel) and 15% (middle panel) SDS–polyacrylamide gels and Western blotted with anti-Pmt3p antiserum. The specificity of bands revealed by the anti-Pmt3p polyclonal antibody is shown by their slight upward shift in the His6-pmt3 strain carrying the endogenous pmt3 gene fused in frame with an amino-terminal His6 tag. The arrows indicate unconjugated Pmt3p; asterisks indicate crossreacting bands. Strains used: PB46 (WT); BX6 (pli1C321S); BX2 (Δpli1); BX5 (his6-pmt3). (B) Sumoylation reactions with in vitro-translated, 35S-labeled Rad22p substrate (4 μl per reaction in 22 μl total) and recombinant, bacterially produced Pmt3p (Pmt3-GG, 1 μg in lanes 2–7), E1 (GST–Rad31p–Fub2p, 32 ng), E2 (Hus5p, 80 ng in lanes 1–2 and 4–7; 400 ng in lane 3) and WT or Pli1C321S (4.8; 24; 120; 600 ng in lanes 4–7, respectively) proteins. The fastest migrating band of the modified species (i.e. mono-modified Rad22) was quantitated by PhosphorImager analysis and expressed as a percentage of total Rad22p. (C) Reaction time course of Rad22p in vitro modification. Standard reactions (4 μl 35S-Rad22p, 1 μg Pmt3-GG, 32 ng E1, 80 ng E2) were assembled, containing 8 ng of WT Pli1p (open squares), Pli1pC321S (open triangles) or Pli1pC321S/H323A/C326S (open circles), and incubated for the indicated times. Open and closed diamonds represent modification without added E3 at 80 and 400 ng of E2 (Hus5p), respectively. Percent total modification was quantitated by PhosphorImager analysis.
To demonstrate that Pli1p possesses SUMO E3 ligase activity, we employed a modification system consisting of recombinant Pmt3p and the E1 (Rad31p-Fub2p) and E2 (Hus5p) enzymes to modify Rad22p, a previously described S. pombe SUMO substrate (Ho et al, 2001). As shown in Figure 3B, 35S-labelled, in vitro translated Rad22p is efficiently modified in the absence of Pli1p at high concentrations of E2 enzyme (lane 2). At one-fifth the concentration of E2, Rad22p modification was limited (lane 3) but could be restored by the addition of recombinant Pli1p (lanes 4–7) in a dose-dependent manner. In vitro binding assays showed that Pli1p interacts with both the E2 enzyme (Hus5p), as well as with the Rad22p substrate (data not shown), thus demonstrating that Pli1p fulfills the criteria for SUMO E3 ligase function. Furthermore, by introducing point mutations of either C321S or C321S/H323A/C326S within the SP-RING finger (Figure 1), we obtained Pli1 proteins which partially or totally lost the capacity to enhance the modification of Rad22p in the in vitro sumoylation system (Figure 3B and C). Indeed, addition of the inactive Pli1p C321S/H323A/C326S mutant to the reaction reduced sumoylation to levels somewhat below those obtained in its absence, possibly because Pli1p itself is a SUMO substrate in vitro (data not shown) and, when inactive, behaves as a competitor for sumoylation. When introduced in S. pombe cells, the pli1C321S WCE showed intermediate levels of global sumoylation between WT and pli1Δ cells (Figure 3A), whereas the C321S/H323A/C326S mutant behaved like the pli1Δ cells (data not shown).
pli1 is involved in centromeric function
In S. pombe, the pmt3 null mutant displays severe morphological abnormalities and hypersensitivity to various stress conditions (Tanaka et al, 1999), while the hus5 (encoding Ubc9p) null allele is almost inviable (al-Khodairy et al, 1995). In contrast, pli1-deleted strain grew well and lacked hypersensisitivity to DNA-damaging agents like UV, MMS and hydroxy-urea (data not shown). However, pli1Δ cells were sensitive to the microtubule-destabilizing drug thiabendazole (TBZ) (Figure 4A). As TBZ sensitivity is frequently associated with defects in centromeric function, we then tested the chromosome stability in pli1Δ cells, which indeed showed a 10-fold increase in the frequency of the Ch16 minichromosome loss (Figure 4B). We further analyzed the effect of pli1 deletion on the heterochromatic function of S. pombe centromeres. In fission yeast, the centromeres are large DNA structures (40–100 kb) composed of outer repetitive regions (otr) corresponding to the pericentromeric heterochromatin and central regions (innermost repeat imr and central cnt), which form specialized SpCENP-A-dependent chromatin, and previous studies have shown that transcription of genes inserted into these domains is silenced (Chikashige et al, 1989; Niwa et al, 1989; Clarke and Baum, 1990; Allshire et al, 1994, 1995; Baum et al, 1994; Ekwall et al, 1999; Partridge et al, 2000). We assayed silencing of the ura4+ gene inserted at three different loci in CEN1 (Centromere of Chromosome I): the center region (cnt1∷ura4+), the innermost repeat (imr∷ura4+) and the outer repeat (otr∷ura4+) (Figure 4C) in WT and pli1Δ strains, by the ability of cells to survive in the presence of 5-fluoro-orotic acid (FOA), which generates a toxic metabolite in (ura+) strains. In WT cells, the inserted ura4+ variegates between expressed and repressed states as cells can grow both on −URA and FOA plates. However, deletion of pli1 causes a significant decrease in silencing at the imr1∷ura4+ and, in a milder way, at the somewhat more expressed cnt1∷ura4+ insertion sites, as indicated by the lower efficiency of plating on FOA plates of these cells (Figure 4D). These results were also confirmed by Northern blot analysis using an ura4 probe (data not shown). The use of the pli1C321S and pli1C321S/H323A/C326S mutants in these experiments showed a strong correlation between SUMO E3 ligase activity and the centromeric phenotypes, since the pli1C321S single mutant showed intermediate levels of TBZ sensitivity, minichromosome loss and silencing at the central region between WT and pli1Δ cells (Figure 4) and the triple mutant exactly reproduced the pli1 deletion phenotypes (data not shown).
Figure 4.

Effects of pli1 mutants on centromeric function. (A) pli1 cells are sensitive to the microtubule-destabilizing drug thiabendazole (TBZ): Serial five-fold dilutions of exponential cultures of WT (PB10), pli1C321S (BX7) and pli1Δ (BX1) cells were spotted to YES and YES plates containing TBZ (17.5 or 20 μg/ml) and incubated for 4 days at 33°C. (B) pli1 cells show an increased frequency of minichromosome loss. Independent colonies of WT (JS161), pli1 C321S (BX9) and pli1Δ (BX8) strains grown in adenine selective media, to select for the presence of the Ch16 minichromosome, were plated to YE plates at a concentration of 103 cells/plate and incubated for 4 days at 33°C. The values shown are the average of the percentage of red colonies calculated for 16 independent colonies in two independent experiments. (C, D) pli1 deletion affects silencing at the central core of centromeres. Expression of the ura4+ gene inserted at three different positions of centromere 1, schematically represented in (C), was assayed by plating efficiency of serial three-fold (cnt) or five-fold (imr and otr) dilutions of cell suspensions spotted on selective media shown in (D). Strains used: cnt1TM1(NcoI)∷ura4+: WT=FY336, pli1C321S=BX10, pli1Δ=PG2964; imr1R(NcoI)∷ura4+: WT=FY498, pli1C321S=BX12, pli1Δ=PG2972; otr1R(SphI)∷ura4+: WT=FY648, pli1C321S=BX14, pli1Δ=PG2976.
Implication of pli1 in the sequence stability of centromeres
While assaying silencing of the ura4+ gene inserted at cnt1 and imr1R, we noted that pli1 mutant cells recurrently gave rise to large colonies on FOA plates (Figure 4D) that turned out to be unable to grow on −URA plates. As this does not correspond to a variegation phenotype (Allshire et al, 1994), we analyzed them at the DNA level. The central domain of CEN1 is composed of two 5.6 kb inverted repeats (imr1R and imr1L) surrounding a 4.1 kb center (cnt1), which otherwise partially shares its sequence with the central domain of CEN 3 (Takahashi et al, 1992) (Figure 5A). The ura4 gene was inserted at the NcoI sites of cnt1 and imr1R (Allshire et al, 1994, 1995). Challenging independent FOA papillae from WT and pli1Δ strains carrying the ura4+ gene at cnt1 and imr1R for growth on −URA plates indicated that most, if not all, papillae from pli1 mutants were stable (ura4−), whereas papillae from WT could grow both on FOA and −URA plates as expected (Allshire et al, 1994). Genomic DNA from 20 independent (ura−) clones of the pli1Δ, CEN1-imr1R(NcoI)∷ura4+ (PG2972) strain was digested with KpnI and analyzed by a Southern blot sequentially using a cnt1 and an ura4 probe (Figure 5B). This analysis revealed that all isolated clones had deleted the ura4 gene at imr1R. Further Southern blot analysis using an NcoI digestion and an imr1 probe revealed that the deletion of the ura4 gene had restored the WT sequence, as indicated by the presence of an NcoI site at imrR1 (Figure 5B). Since imr1R and imr1L are identical over 99% of their sequence, the simplest interpretation would be that the ura4 gene inserted at imr1R was deleted following gene conversion events using the homologous sequence at the inverted repeat imr1L as template. A similar result was obtained with 19 over 20 independent (ura−) clones of the pli1Δ, CEN1-cnt1(NcoI)∷ura4+ (PG2964) strain (see Supplementary Figure S1). Since cnt1 also shares 100% identity over 3.3 kb with the central domain of CEN3, these latter clones likely arose from interchromosomal gene conversion events. The frequency of ura4 loss was evaluated at ≈10−3 for both domains in pli1Δ cells and, as stable (ura−) cells could not be isolated from 104 wild type cells, ura4 deletion is therefore increased by at least 10-fold in pli1Δ compared to WT cells. This effect seems to be specific for the centromeric loci, since an artificial intrachromosomal recombination reporter (van den Bosch et al, 2002) at a euchromatic locus displayed no difference between pli1Δ and WT cells (see Supplementary Figure S2).
Figure 5.
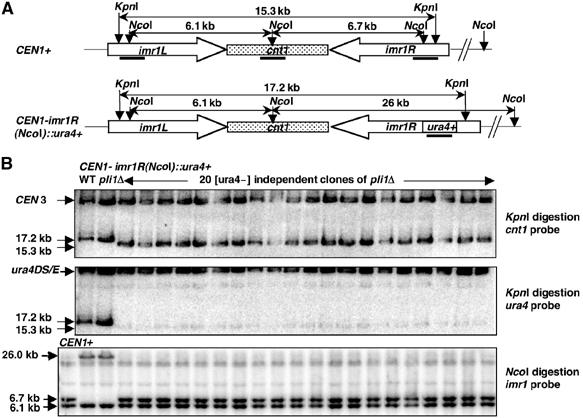
Effect of pli1 deletion on the stability of ura4+ gene inserted at imr1R. (A) Structure of centromere 1 central domain in CEN1+ and CEN1: imr1R(NcoI)∷ura4+ strains. KpnI and NcoI sites, sizes of their respective digestion fragments and positions of the probes used for the Southern blot analysis are indicated. Note that the cnt1 and ura4 probes also recognize homologous sequences, respectively, at CEN3 and ura4D/SE loci, thus serving as internal controls. (B) Southern blot analysis of stable [ura−] independent clones raising from CEN1-imr1R(NcoI)∷ura4 deleted for pli1 gene. After digestion with KpnI, genomic DNA prepared in agarose plugs was subjected to pulsed field electrophoresis on 1.2% agarose gel and analyzed by a Southern Blot using the cnt1 probe. The membrane was then stripped and reprobed with the ura4 probe. Genomic DNA was also digested with NcoI, subjected to electrophoresis on 0.75% agarose gel, transferred to a nylon membrane and hybridized to the imr1 probe. Sizes or identities of bands are indicated. Strains used: CEN1+: PB10; CEN1-imr1R(NcoI)∷ura4+: WT=FY498, pli1Δ=PG2972; and 20 [ura−] independent clones isolated from PG2972 strain=BX13_1–20.
Pli1p is involved in the control of telomere length
While examining silencing at three other heterochromatic loci: the mat2/3 inactive sexual locus, the rDNA and the telomeres, we noticed that pli1 mutants slightly increased the silencing of the ura4+ gene next to the telomeric repeat of the Ch16m23∷ura4+ TEL[72] minichromosome without affecting silencing at the two other loci (data not shown). As the frequency of transcriptional repression at the telomere can be increased by longer telomeres (Kyrion et al, 1993) and, further, since deletion of pmt3 was previously reported to lead to telomere elongation (Tanaka et al, 1999), we assessed telomere length in pli1 cells. Genomic DNA digested with EcoRI was analyzed by a Southern blot using a 32P end-labeled telomeric oligonucleotide (Cooper et al, 1997) (Figure 6B). The EcoRI site is located ≈0.8 kb away from the telomeric repeats sequences, giving rise to a broad telomere hybridization signal centered at 950 bp in the WT strain (Figure 6A, lane 1). The pli1Δ strain showed a clearly upshifted, more intense signal that ranges from 1 to 1.4 kb (lane 3), corresponding to a telomere length intermediate between that of WT and pmt3Δ cells (Tanaka et al, 1999). The pli1C321S single mutant again showed a milder effect on the increase of telomere length (lane 2), while the pli1C321S/H323A/C326S triple mutant showed the same effect as the pli1 deletion (data not shown).
Figure 6.
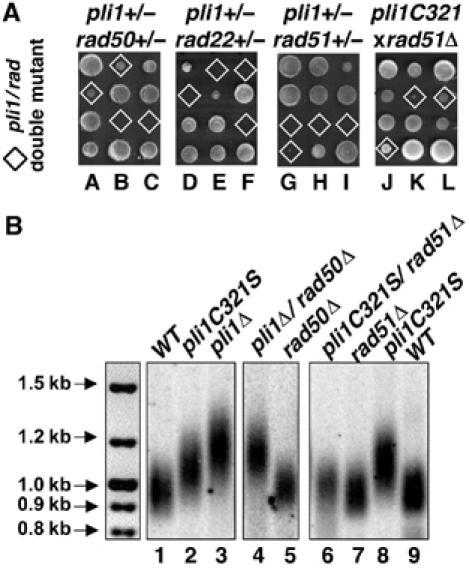
Genetic interactions of pli1 and recombination genes: effects on telomere length. (A) Genetic interaction of pli1 with homologous recombination mutants. Three representative tetrads from diploids heterozygous for disruption of pli1 and either rad50 (BX16), rad22 (BX18) or rad51 (BX19), and three representative tetrads from the rad51(JAC1/51Δ) × pli1C321S (BX7) cross, as indicated. The genotype of each segregant was determined (or inferred) by replica plating on appropriate selective media and/or by PCR; double-mutant clones are indicated by diamonds. Given the close physical linkage of the rad22 and pli1 loci (0.05 Morgans), tetrad analysis of the rad22+/− pli1+/− diploïd (250 tetrads) was complemented by extensive random spore analysis (data not shown). (B) Southern blot analysis of telomere length in (1) WT (PB10); (2) pli1C321S (BX7); (3) pli1Δ (BX1); (4) pli1Δ/rad50Δ (BX20); (5) rad50Δ (BX21); (6) pli1C321S/rad51Δ (BX22_1); (7) rad51Δ (BX22_2); (8) pli1C321SΔ (BX22_3); (9) WT (BX22_4). BX22_1–4 correspond to the four clones of a tetra-type tetrad from the rad51(JAC1/51Δ) × pli1C321S (BX7) cross. After digestion with EcoRI, genomic DNA was subjected to electrophoresis on 1% agarose gel, transferred to a nylon membrane, and hybridized to a telomeric DNA probe (Cooper et al, 1997).
To test whether the increase in telomere length in pli1 mutants could have resulted from normally repressed gene conversion events, we performed a genetic analysis involving pli1 and three major genes involved in homologous recombination: rad22 (fission yeast RAD52 homolog), rad50 and rad51/rhp51. We therefore produced diploids doubly heterozygous for pli1 and each of the three rad genes by sequential disruption. At least 100 tetrads from microdissected asci were genotyped for each experiment and representative examples are shown in Figure 6A. The pli1-rad50 double mutants showed a reduced viability (50%) and a significant increase in generation time independent of mating type (data not shown). They also exhibited a significant increase in telomere length compared to the rad50Δ single mutant (Figure 6B, compare lanes 4 and 5), but very similar to that observed in pli1Δ cells (lane 3). In contrast, disruptants of pli1 and either rad22 or rad51 displayed synthetic lethality, in that double-mutant clones gave rise to microcolonies of 8–150 cells that contained numerous abnormally elongated cells (data not shown). In order to possibly circumvent the lethality of pli1–rad51 double mutants, we crossed the pli1C321S and rad51 mutants and found that the obtained pli1C321S–rad51Δ double mutants were viable, although they grew slower than the rad51 single mutants of the same mating type (Figure 6A). When we next analyzed clones from tetra-type tetrads of the pli1C321S × rad51Δ cross at the telomere level (Figure 6B, lanes 6–9), we found that pli1C321S–rad51Δ clones (lane 6) showed a telomere length similar to the rad51Δ single mutant (lane 7) and not the pli1C321S single mutant (lane 8). This shows that the increase in telomere length characteristic of the pli1C321S single mutant (lane 2 and 8) requires the wild-type rad51 background, consistent with the hypothesis that telomere elongation of pli1 mutants is, at least partially, mediated by gene conversion (homologous recombination) type events.
Discussion
In this work, we show that the S. pombe homolog of the Siz/PIAS proteins, Pli1p, functions as a SUMO E3 ligase in vivo and in vitro. pli1 mutant cells display phenotypes intermediate between SUMO-deficient and WT cells with respect to telomere length and centromeric function, as witnessed by TBZ sensitivity and enhanced minichromosome loss. We furthermore provide evidence that Pli1p and hence SUMO modification might play an important structural role at the centromere domain required for kinetochore assembly (central core), and in the protection of heterochromatic repeated sequences from illegitimate recombination.
Pli1p and the centromere
The centromere is the chromosomal site of kinetochore assembly which allows spindle microtubule attachment and chromosome segregation to daughter cells. Centromeres of fission yeast and higher eucaryotes are composed of large repetitive DNA domains packaged into a heterochromatic structure (Clarke, 1990; Pluta et al, 1995; Bjerling and Ekwall, 2002). S. pombe centromeres (40–100 kb) exhibit a symmetrical organisation with a central core (cnt+imrs) flanked by outer repeats (otrs). Although the whole centromeric region is refractory to transcription, the molecular mechanisms underlying silencing seem to be quite distinct between the two domains. Indeed, the clr4 (Su(var)3–9 homolog) and swi6 (HP1 homolog) mutants affect silencing over the otr repeats as well as the mat2/3 inactive sexual locus, rDNA and the telomeres, but not over the central core. Moreover, in contrast to the otrs, the central core has an unusual chromatin structure lacking regularly spaced nucleosomes, likely due to the presence of the histone H3-variant Cnp1p (CENP-A), believed to form the primary kinetochore-building scaffold. Consistent with this, the central core domain is strictly required for minimal centromeric function (Chikashige et al, 1989; Niwa et al, 1989; Clarke and Baum, 1990; Allshire et al, 1994, 1995; Baum et al, 1994; Ekwall et al, 1999; Partridge et al, 2000). Our results indicate a specific role of Pli1p for silencing at the central domain of centromeres, since, of all known heterochromatic loci tested (cnt1, imr1R, otr1R, mat2-3, rdNA, telomeres), only the imr1R and cnt1 exhibited reduced silencing of the inserted ura4 gene in pli1 deficient cells. Furthermore, pli1Δ cells also exhibit TBZ sensitivity and high rate of minichromosome loss, which might result from a defect in establishment or maintenance of proper central core structure for centromere and/or kinetochore function. Interestingly, the double pli1Δswi6Δ mutant showed a striking increase in TBZ sensitivity compared to the single mutants alone (data not shown). This synergistic attenuation of centromere function suggests that Swi6p and Pli1p operate in separable pathways and thus correlates well with the differential effects of Pli1p at the central core versus the outer repeats.
Pli1p and recombination
Like in many organisms, S. pombe telomeres are composed of short tracts of repeated sequences organized into a non-nucleosomal chromatin structure by the mediation of the TRF-like protein Taz1p (Cooper et al, 1997). Telomere length is maintained by highly regulated mechanisms as mutations affecting telomerase, telomere-specific binding proteins, silencing, replication, DNA repair and checkpoint proteins lead to either lengthening or shortening of telomeres and, furthermore, to senescence (reviewed in Blackburn, 2001; Henson et al, 2002). We have shown here that, while pli1 cells exhibit only a slight increase in reporter gene (ura4) silencing, they do manifest a consistent increase of telomere length, for which Rad51p, but not Rad50p, seems to be required. These genetic interactions strongly suggest that the increased telomere length in pli1 cells might result from normally repressed gene conversion events in which the 3′ end of a telomere illegitimately invades a homologous telomeric repeat array (Henson et al, 2002). Moreover, we found that the ura4 variegation reporter gene at the central core of centromeres was lost in the pli1 genetic background by restoring the wild type imrR or cnt1 sequences. This strongly suggests that ura4 deletion had occurred upon gene conversion events using the respective homologous imr1L and cnt3 sequences as donors of information. While recombination events at the central core show an increase by at least 10-fold, recombination rates at a euchromatic context seem to be unaffected by the absence of pli1. Thus, the effect of Pli1p on recombination may be region specific and could be explained by two, albeit nonmutually exclusive mechanisms. First, Pli1p may exert an indirect effect on replication and/or chromatin structure that leads to an increased number of recombinogenic substrates at the central core and at the telomere in pli1 cells. Second, Pli1p might directly regulate the recombination machinery by actively inhibiting the activity of recombination proteins at these repeated sequences. The finding of enhanced apparent gene conversion at centromeres and telomeres is consistent with either hypothesis, while the strong genetic interaction of pli1 mutants with rad50, rad51 and rad22 (RAD52 fission yeast homolog), that range from strongly impaired mitotic growth (rad50) to colethality (rad22, rad51), mostly favors the second hypothesis and suggests that homologous recombination is necessary for survival of pli1Δ cells. Similarly, a negative genetic interaction between ulp1-I615N and all the RAD51-dependent recombination pathway mutants in S. cerevisiae has been described recently (Soustelle et al, 2004). Given that both Rad22p and Rad51p also represent possible SUMO substrates in vivo (Ho et al, 2001), it will be interesting to study the centromere and telomere phenotypes of nonmodifiable versions of these proteins.
Pli1p and sumoylation
The Pli1p protein fulfills the criteria for SUMO E3 ligase activity in that it interacts with both the E2 enzyme (Hus5p/Ubc9p) and the substrate and increases the rate of SUMO modification in an in vitro reconstituted S. pombe sumoylation assay. The in vivo significance of Pli1p is witnessed by the drastic reduction of global sumoylation in pli1-deficient cells. Despite this striking decrease of SUMO conjugates, pli1-deficient cells failed to show severe mitotic growth defects, in contrast to pmt3- (SUMO) and hus5- (Ubc9) deleted cells (al-Khodairy et al, 1995; Tanaka et al, 1999), which are almost inviable. This is reminiscent of S. cerevisiae pli1 homologs, siz1 and siz2, in that both single- and double-deletion mutants grow and have few obvious phenotypes (Johnson and Gupta, 2001). Nonetheless, the pli1 deletion phenotypes recapitulate some of the pmt3 phenotypes and characteristics, albeit in a milder way. First, pli1Δ cells have telomeres of intermediate length between wild-type and pmt3Δ cells. Second, pli1Δ cells exhibit intermediate levels of minichromosome loss and sensitivity to TBZ. Therefore, the milder pleiotropic effects of pli1Δ cells may be due to attenuated sumoylation of target proteins involved in centromeric and telomeric functions and possibly in the recombination process. It remains possible that the phenotypes observed in pli1Δ cells stem from a physiological activity of Pli1p that is independent from its role in sumoylation. We consider this unlikely since we observe a very strong correlation between the levels of SUMO E3 ligase activity of Pli1 proteins mutated at the RING domain and the described phenotypes: TBZ sensitivity, minichromosome loss, silencing at the central core, centromere sequence instability and telomere length regulation. In this regard, some of the first evidence implicating SUMO in recombination and/or centromere function was obtained by yeast two-hybrid screens with human Rad51 or Rad52 as baits and from screens for high-copy suppressors of temperature-sensitive alleles of the budding yeast MIF2 or of its human homolog, the centromeric CENP-C protein (Meluh and Koshland, 1995; Shen et al, 1996).
The discovery that Pli1p and, by extension, sumoylation, plays a role in regulating recombination, opens the way to identifying the SUMO targets involved, as well as the mechanisms by which sumoylation controls their activity. The study of such targets and of their nonmodifiable mutant counterparts should provide further valuable insight into the structure and function of centromeres and telomeres.
Materials and methods
Fission yeast strains, plasmids, media and methods
The S. pombe strains used in this study are listed in Table I. Growth, maintenance and standard genetic methods for fission yeast strains were as described by Alfa et al (1993). Details regarding strain constructions are available upon request.
Table 1.
S. pombe strains
| Strain | Genotype | Reference |
|---|---|---|
| PB10 | h− (Msmt0) leu1-32 ade6-210 | Lab stock |
| PB46 | h90 ura4-D18 leu1-32 ade6-216 | Lab stock |
| BX1 | h− (MsmtO) pli1Δ∷kanMX6 leu1-32 ade6-210 | This work |
| BX2 | h90 pli1Δ∷kanMX6 ura4-D18 leu1-32 ade6-210 | This work |
| BX3 | h90 pli1-HA∷kanMX6 ura4-D18 leu1-32 ade6-210 | This work |
| BX4 | h90 pli1-CFP∷kanMX6 ura4-D18 leu1-32 ade6-216 | This work |
| BX5 | h90 His6-pmt3 ura4-D18 leu1-32 ade6-210 | This work |
| BX6 | h90 pli1C321S leu1-32 ade6-216 | This work |
| BX7 | h− (MsmtO) pliC321S ura4-D18 leu1-32 ade6-210 | This work |
| JS161 | h− ura4-D18 leu1-32 ade6-210 [Ch16 ade6-216] | Matsumoto et al (1987) |
| BX8 | h? pli1Δ∷kanMX6 ura4-D18 leu1-32 ade6-210 [Ch16 ade6-216] | This work |
| BX9 | h? pli1C321S ura4-D18 leu1-32 ade6-210 [Ch16 ade6-216] | This work |
| FY336 | h− ura4-DS/E leu1-32 ade6-210 cnt1TM1(Nco1)∷ura4 | Allshire et al (1994) |
| PG2964 | h? pli1Δ∷kanMX6 ura4-DS/E leu1-32 ade6-216 cnt1TM1(Nco1)∷ura4 | This work |
| BX10 | h? pli1C321S ura4-DS/E leu1-32 ade6-216 cnt1TM1(Nco1)∷ura4 | This work |
| BX11_1-20 | h? pli1Δ∷kanMX6 ura4-DS/E leu1-32 ade6-216 isolated from PG2964 | This work |
| FY498 | h+ura4-DS/E leu1-32 ade6-210 imr1(Nco1)∷ura4ori1 | Allshire et al (1995) |
| PG2972 | h? pli1Δ∷kanMX6 ura4-DS/E leu1-32 ade6-210 imr1(Nco1)∷ura4ori1 | This work |
| BX12 | h? pli1C321S ura4-DS/E leu1-32 ade6-210 imr1(Nco1)∷ura4ori1 | This work |
| BX13_1-20 | h? pli1Δ∷kanMX6 ura4-DS/E leu1-32 ade6-210 isolated from PG2972 | This work |
| FY648 | h+ura4-DS/E leu1-32 ade6-210 otr1R(Sph1)∷ura4 | Allshire et al (1995) |
| PG2976 | h? pli1Δ∷kanMX6 ura4-DS/E leu1-32 ade6-210 otr1R(Sph1)∷ura4 | This work |
| BX14 | h? pli1C321S ura4-DS/E leu1-32 ade6-210 otr1R(Sph1)∷ura4 | This work |
| FY521 | h− ura4-DS/E leu1-32 ade6-210 [Ch16 ade6-216, m23∷ura4-Tel[72]] | Nimmo et al (1994) |
| PG2994 | h+ pli1Δ∷kanMX6 ura4-DS/E leu1-32 ade6-210 [Ch16 ade6-216, m23∷ura4-Tel[72]] | This work |
| BX15 | h? pli1Δ∷kanMX6 ura4-DS/E leu1-32 ade6-210 [Ch16 ade6-216, m23∷ura4-Tel[72]] | This work |
| BX16 | h+/h−(MsmtO) ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-210/ade6-216 pli1+/−∷kanMX6 rad50+/−∷LEU2 | This work |
| BX17 | h+/h−(MsmtO) ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-210/ade6-216 pli1+/−∷kanMX6 rad22+/−∷LEU2 | This work |
| BX18 | h+/h−(MsmtO) ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-210/ade6-216 pli1+/−∷kanMX6 rad22+/−∷LEU2 | This work |
| BX19 | h+/h−(MsmtO) ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-210/ade6-216 pli1+/−∷kanMX6 rad51+/−∷ura4 | This work |
| BX20 | h+ ura4-D18 leu1-32 ade210 pli1Δ∷kanMX6 rad50Δ∷LEU2 | This work |
| BX21 | h+ ura4-D18 leu1-32 ade210 rad50Δ∷LEU2 | This work |
| JAC1/51Δ | h+ ura4-D18 leu1-32 ade6-704 rad51Δ∷ura4+ | Kim et al (2002) |
| BX22_1 | h+ ura4-D18 leu1-32 ade6-? pli1C321S rad51Δ∷ura4+ | This work |
| BX22_2 | h−(Msmt0) ura4-D18 leu1-32 ade6-? rad51Δ∷ura4+ | This work |
| BX22_3 | h−(Msmt0) ura4-D18 leu1-32 ade6-? pli1C321S | This work |
| BX22_4 | h+ ura4-D18 leu1-32 ade6-? | This work |
| SL1 | h+ura4-D18 leu1-32 ade6-M26 int∷pUC8/ura4/MATa/ade6-L469 | Osman et al (1996) |
| BX23 | h+ura4-D18 leu1-32 ade6-M26 int∷pUC8/ura4/MATa/ade6-L469 pli1Δ∷kanMX6 | This work |
In vitro sumoylation assays
DNA sequences encoding Pmt3p, the sumoylation E1 (Rad31p/Fub2p heterodimer), E2 (Hus5p) and E3 (Pli1p) enzymes and Rad22p (the sumoylation substrate used here) were obtained by reverse transcription–polymerase chain reaction (RT–PCR) from S. pombe total RNA using specific oligonucleotide primers for first-strand synthesis and amplification. A Pmt3GG-encoding cDNA was cloned into pQE-30 (Qiagen) for the bacterial expression of a His6-Pmt3-GG fusion protein. cDNAs encoding Rad31p and Fub2p were fused in-frame with a short linker and cloned into vector pGEX2T (Pharmacia) to produce a functional GST–Rad31p–Fub2p fusion protein. Hus5p- and Pli1p-encoding cDNAs were cloned into vector pGEXcs that contains a tobacco etch virus (TEV) protease cleavage site. Site-directed mutagenesis (Stratagene QuikChange) was performed to produce Pli1 RING finger point mutations C321S and C321S/H323A/C326S. A Rad22p cDNA was cloned into PING14A (Hagemeier et al, 1993) for in vitro transcription/translation using Sp6 RNA polymerase and the TNT-coupled reticulocyte lysate system (Promega). All constructions were verified by sequencing and detailed maps are available upon request. His6-Pmt3GG, the mature form of S. pombe SUMO, was expressed in Escherichia coli strain M15 (Qiagen) and purified on Ni-NTA agarose (Qiagen) using the manufacturer's protocol. Glutathione-S-transferase (GST) fusion proteins were expressed in strain BL21(DE3)/pLysS and purified using standard methods. Hus5p and Pli1p were cleaved from GST with GST-TEV protease and separated from free GST and GST-TEV by passage over glutathione (GSH-) agarose beads. All bacterially produced proteins were concentrated, dialyzed extensively against 50 mM Tris (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, and protein concentrations were estimated colorimetrically against a BSA standard (BioRad). In vitro sumoylation reactions were carried out using 4 μl of a standard 35S-methionine-labeled, in vitro-translated Rad22p preparation, 1 μg His6-Pmt3GG and the indicated amounts of E1, E2 and E3 proteins in a 22 μl reaction volume in a buffer containing 50 mM Tris (pH 7.5), 5 mM MgCl2, 5 mM ATP and incubated for 1 h at 37°C. Reactions terminated by boiling in Laemmli buffer were subjected to SDS–PAGE and autoradiography. Percent modification was determined by PhosphorImager analysis using ImageQuant software.
Fluorescence microscopy
Cells grown in exponential conditions to 5 × 106 cells/ml in MM were fixed with 3.6% paraformaldehyde at room temperature for 45 min. Cells processed as described (Alfa et al, 1993) were sequentially stained with an anti-GFP rabbit primary antibody (Molecular Probes), a FITC conjugated anti-rabbit IgG secondary antibody (Jackson Laboratory) and DAPI. Images were acquired with a Hamamatsu ORCAII-ER cooled CCD camera controlled by the Openlab® software (version 3.2, Improvision) and processed using Adobe Photoshop® software (version 6, Adobe).
Preparation of S. pombe whole-cell extracts and Western blot analysis
In all, 108 exponentially growing cells were harvested, washed with water and TNET+ (200 mM NaCl, 0.1% Triton, 0.01% SDS, 50 mM Tris–HCl (pH 8) supplemented with protease inhibitor Cocktail (Dove et al, 1998) and 10 mM NEM), resuspended in 200 μl of TNET+, broken with 500 μl of glass beads in a Fastprep® Apparatus and clarified by 10 min centrifugation at 13 000 r.p.m. In all, 20 μl of the WCE in 1 × Laemmli buffer and heated for 5 min at 90°C was separated by SDS–PAGE and analyzed by Western blotting with anti-GFP (Molecular Probes) or anti-Pmt3p rabbit primary antibody and a horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (Amersham).
Determination of minichromosome loss rates
Ch16 is a linear minichromosome containing the ade6-216 mutation that suppresses the ade6-210 mutation by intra-allellic complementation (Matsumoto et al, 1987). The rate of Ch16 loss was estimated by a colony color assay described by Tanaka et al (1999) with slight modifications. Strains were grown in MM lacking adenine to select for Ch16-harboring cells. Single colonies were dispersed in 1 ml of MM without adenine and cell number was measured in a Coulter® cell counter. The cells were then diluted into YE medium and immediately plated onto YE plates at a concentration of ∼103 cells/plate. After 4 days growth at 33°C, the proportion of [ade−] (completely red) colonies was determined.
Silencing assays
Series of three- or five-fold dilutions of S. pombe strains grown exponentially in YES at 33°C were spotted onto AA, AA lacking uracil or 5′-FOA plates, which were then incubated at 33°C until full growth was achieved.
Isolation of stable independent [ura−] cells
S. pombe strains containing the ura4 reporter gene inserted at cnt1 or imrR1 were grown to saturation in YES at 33°C and plated onto YES plates at a concentration of ∼103 cells/plate. After 4 days growth at 33°C, colonies were replica plated onto 5′-FOA. FOAR papillae arising from independent colonies were consecutively streaked on 5′-FOA and YES plates and replica plated on AA lacking uracil.
Southern blot analyses
S. pombe strains were exponentially grown in MM or YES at 33°C and DNA was prepared either in agarose plugs (Young et al, 2002) or by standard methods (Moreno et al, 1991). For pulsed field electrophoresis, DNA prepared and digested in agarose plugs was resolved in 1.2% agarose gels with a Pulsafor™ system (Pharmacia) in 0.15 × TBE at 360 V with a pulse time of 0.5 s for 6 h at 9°C. After electrophoresis, the DNA was transferred to Hybond-N+ filters (Amersham) and crosslinked with UV Stratalinker (Stratagene). Following DNA–DNA hybridization, the membranes were exposed on a phosphor screen and the signal detected by PhosphorImager 445SI (Molecular Dynamics) and displayed using ImagequantNT. The sequences of all the probes used in this study are available upon request.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S1 Legend
Supplementary Figure S2
Supplementary Figure S2 Legend
Acknowledgments
We are grateful to K Tanaka for the pmt3+/− strain, A Pastink for the intrachromosomal recombination assay, SD Park for the rad51Δ strain, O Gadal, F Feuerbach-Fournier, O Danot, N Joly and M Gilbert for technical advice and useful comments on the manuscript. This work was supported by a fellowship from the Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT) to BX; the Novo Nordisk Foundation and Danish Research Council to GT; the Pasteur-Negri-Weizmann Council, the EEC 5th Framework Programme to AD, the Human Frontiers in Science Program (HFSP) and the Association pour la Recherche sur le Cancer (ARC) to BA.
References
- al-Khodairy F, Enoch T, Hagan IM, Carr AM (1995) The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J Cell Sci 108: 475–486 [DOI] [PubMed] [Google Scholar]
- Alfa CE, Gallagher IM, Hyams JS (1993) Antigen localization in fission yeast. Methods Cell Biol 37: 201–222 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G (1994) Position effect variegation at fission yeast centromeres. Cell 76: 157–169 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ (2002) The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell 9: 1169–1182 [DOI] [PubMed] [Google Scholar]
- Baum M, Ngan VK, Clarke L (1994) The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell 5: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerling P, Ekwall K (2002) Centromere domain organization and histone modifications. Braz J Med Biol Res 35: 499–507 [DOI] [PubMed] [Google Scholar]
- Blackburn EH (2001) Switching and signaling at the telomere. Cell 106: 661–673 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M (1989) Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell 57: 739–751 [DOI] [PubMed] [Google Scholar]
- Clarke L (1990) Centromeres of budding and fission yeasts. Trends Genet 6: 150–154 [DOI] [PubMed] [Google Scholar]
- Clarke L, Baum MP (1990) Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol Cell Biol 10: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- De Lahondes R, Ribes V, Arcangioli B (2003) Fission yeast sap1 protein is essential for chromosome stability. Eukaryot Cell 2: 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove JE, Brockenbrough JS, Aris JP (1998) Isolation of nuclei and nucleoli from the yeast Saccharomyces cerevisiae. Methods Cell Biol 53: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC (1999) Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153: 1153–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G (2003) Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr Opin Genet Dev 13: 108–113 [DOI] [PubMed] [Google Scholar]
- Hagemeier C, Cook A, Kouzarides T (1993) The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res 21: 4998–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari KL, Cook KR, Karpen GH (2001) The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev 15: 1334–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Neumann AA, Yeager TR, Reddel RR (2002) Alternative lengthening of telomeres in mammalian cells. Oncogene 21: 598–610 [DOI] [PubMed] [Google Scholar]
- Ho JC, Warr NJ, Shimizu H, Watts FZ (2001) SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res 29: 4179–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M (2001) SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107: 5–8 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Jackson PK (2001) A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev 15: 3053–3058 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113: 127–137 [DOI] [PubMed] [Google Scholar]
- Kim WJ, Park EJ, Lee H, Seong RH, Park SD (2002) Physical interaction between recombinational proteins Rhp51 and Rad22 in Schizosaccharomyces pombe. J Biol Chem 277: 30264–30270. Epub 32002 Jun 30265 [DOI] [PubMed] [Google Scholar]
- Kirsh O, Seeler JS, Pichler A, Gast A, Müller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, Dejean A (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 21: 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ (2002) PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol 22: 5222–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ (1993) RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev 7: 1146–1159 [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Fukui K, Niwa O, Sugawara N, Szostak JW, Yanagida M (1987) Identification of healed terminal DNA fragments in linear minichromosomes of Schizosaccharomyces pombe. Mol Cell Biol 7: 4424–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F (2000) SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol 16: 591–626 [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D (1995) Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell 6: 793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Müller S, Hoege C, Pyrowolakis G, Jentsch S (2001) SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol 2: 202–210 [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Cranston G, Allshire RC (1994) Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J 13: 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M (1989) Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J 8: 3045–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Fortunato EA, Subramani S (1996) Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics 142: 341–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14: 783–791 [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC (1995) The centromere: hub of chromosomal activities. Science 270: 1591–1594 [DOI] [PubMed] [Google Scholar]
- Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G (2002) Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J 21: 5206–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A (2003) Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4: 690–699 [DOI] [PubMed] [Google Scholar]
- Shayeghi M, Doe CL, Tavassoli M, Watts FZ (1997) Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res 25: 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ (1996) UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics 36: 271–279 [DOI] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN (2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100: 13225–13230. Epub 12003 October 13224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustelle C, Vernis L, Freon K, Reynaud-Angelin A, Chanet R, Fabre F, Heude M (2004) A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol Cell Biol 24: 5130–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V (2003) Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol 163: 729–741. Epub 2003 November 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Aravind L, Koonin EV (2001) Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Toh-e A, Kikuchi Y (2001) A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275: 223–231 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y (1999) Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol 19: 8660–8672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch M, Zonneveld JB, Vreeken K, de Vries FA, Lohman PH, Pastink A (2002) Differential expression and requirements for Schizosaccharomyces pombe RAD52 homologs in DNA repair and recombination. Nucleic Acids Res 30: 1316–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A, Perdomo J, Crossley M (2003) Modification with SUMO. EMBO Rep 4: 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR (2002) Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell 9: 253–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S1 Legend
Supplementary Figure S2
Supplementary Figure S2 Legend


