Abstract
Movement of transposable elements is often accompanied by replication to ensure their proliferation. Replication is associated with both major classes of transposition mechanisms: cut-and-paste and cointegrate formation (paste-and-copy). Cut-and-paste transposition is often activated by replication of the transposon, while in cointegrate formation replication completes integration. We describe a novel transposition mechanism used by insertion sequence IS911, which we call copy-and-paste. IS911 transposes using a circular intermediate (circle), which then integrates into a target. We demonstrate that this is derived from a branched intermediate (figure-eight) in which both ends are joined by a single-strand bridge after a first-strand transfer. In vivo labelling experiments show that the process of circle formation is replicative. The results indicate that the replication pathway not only produces circles from figure-eight but also regenerates the transposon donor plasmid. To confirm the replicative mechanism, we have also used the Escherichia coli terminators (terC) which, when bound by the Tus protein, inhibit replication forks in a polarised manner. Finally, we demonstrate that the primase DnaG is essential, implicating a host-specific replication pathway.
Keywords: host factors, IS911, replication, transposase, transposition
Introduction
To be successful, transposable elements must proliferate through populations but exhibit controlled activity to avoid damaging their host genomes. The factors involved in their proliferation are not well characterised. Simply maintaining transposon copy number by passive replication would appear to be a poor strategy for promoting transposon spread within and between genomes. A more efficient strategy would be to couple transposition with replication of the element, and indeed a replication event is associated with movement of many transposons within the cell.
This can be achieved in several ways. One, adopted by the bacterial insertion sequence IS10, is to activate transposition following its passive replication by passage of a host replication fork during a normal host cycle. Activation depends on Dam methylation sites in the ends of IS10, which become transiently hemimethylated following passage of the fork. This state renders the ends more active in transposition and activates the Tpase promoter located in one end, resulting in increased Tpase synthesis (Roberts et al, 1985). Transposition is therefore effectively preceded by duplication of the element. In this case, if one of the two daughter ISs undergoes transposition, it is not difficult to imagine how the cleaved donor chromosome branch from which the IS has been excised might undergo repair by a copy-choice pathway using the intact sister copy. This would replace the excised IS, therefore effectively increasing its copy number.
A second strategy is to assure that precise replication of the transposon occurs as an integral part of the transposition reaction itself. This strategy has been adopted by bacteriophage Mu (Chaconas and Harshey, 2002), the Tn3 family of transposons (Sherratt, 1989; Grindley, 2002), certain IS elements such as members of the IS6 family (Mahillon and Chandler, 1998) and in certain circumstances with IS1 (Turlan and Chandler, 1995). For these, only one of the transposon DNA strands is cleaved at each end and transferred, leaving the transposon covalently attached to both donor and target replicons. In the case of phage Mu, a specific set of host replication enzymes is assembled at the fork created by the initial strand transfer from the transposon to the target DNA (Kruklitis and Nakai, 1994; Nakai and Kruklitis, 1995). Replication circumvents the need to resolve the second transposon strand. It generates an intermediate in which one end of each transposon is attached to the donor DNA and the other end to the target. The resulting cointegrate structure carries two copies of the transposon. Donor and target molecules are joined with a single transposon copy in a direct orientation at each donor–target junction. This intimate relationship between transposon and host is highlighted by the observation that, under conditions in which they fail to undergo their normal excisive transposition cycle due to Tpase mutation (e.g. Tn7; May and Craig, 1996) or mutation in the ends (e.g. IS903; Tavakoli et al, 1997; Tavakoli and Derbyshire, 2001), these elements are processed by the host replication machinery to generate cointegrates.
We address here the role of replication in the transposition of IS911, a member of a widespread group of transposable elements, the IS3 family of bacterial insertion sequences.
During the first step of IS911 transposition, a single DNA strand of one end is cleaved and transferred to the same strand 3 bases from the opposite end (Figure 1; see Rousseau et al, 2002) to generate a single-strand bridge between the two transposon ends. If this is located on a circular plasmid, the molecules assume a figure-eight structure, where the IS911 terminal inverted repeats (IRL and IRR) are separated by three bases derived from external flanking DNA. If supplied with high levels of Tpase (OrfAB) in vivo, figure-eight molecules are found together with free circularised transposons (circles) in which the ends are abutted. These transposon circles are intermediates in IS911 transposition and have been shown to be highly competent for integration into a DNA target in vivo (Ton-Hoang et al, 1997) and in vitro (Ton-Hoang et al, 1998). Preliminary kinetic studies, in which chloramphenicol was used to stop Tpase synthesis, suggested that the figure-eight forms were precursors of the transposon circles (Polard and Chandler, 1995). However, in vitro, enriched preparations of OrfAB generate figure-eight molecules but not transposon circles (Polard et al, 1996). This observation raised the possibility that transposon circle formation required host factors not present in the in vitro reactions. This notion was reinforced by the observation that circularised transposons could be detected following introduction of preformed figure-eight molecules into naïve cells in the absence of Tpase (Turlan et al, 2000).
Figure 1.
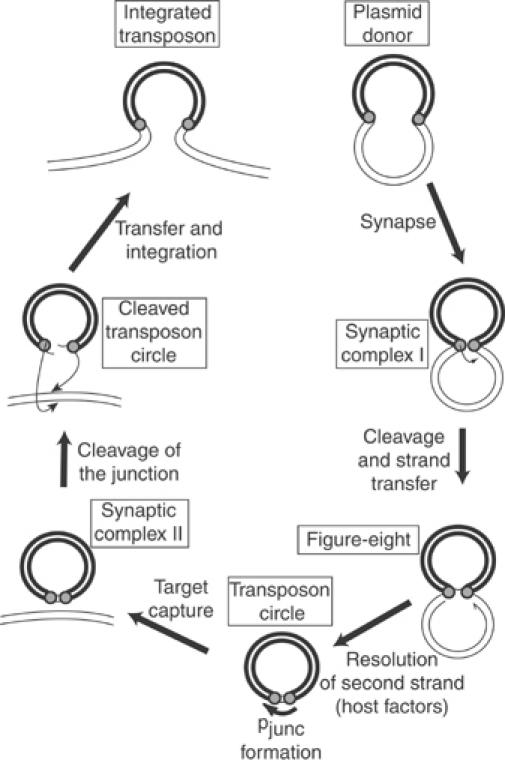
IS911 transposition pathway. The transposon (heavy line), plasmid backbone (lighter line), and transposon termini (grey circles) are indicated. The transposon ends are brought together in a synapse (synaptic complex I). A first single-strand cleavage occurs initially at one end of the IS, catalysed by the low level of Tpase expressed from a weak promoter, pIRL, present in IRL. The free 3′-OH generated is then directed to attack the opposite end on the same DNA strand (strand transfer) to generate a figure-eight form in which both ends are joined by a single-strand bridge, leaving an unjoined DNA strand with a free 3′-OH (half arrow) on the vector plasmid. The figure-eight is converted into a transposon circle where the formation of a strong promoter, pjunc, induces high-level synthesis of transposon proteins (Ton-Hoang et al, 1997; Duval-Valentin et al, 2001). A second Tpase-mediated synaptic complex (synaptic complex II) is formed upon target capture. The Tpase then undergoes the final steps of cleavage and strand transfer into the target DNA (integration). Insertion results in the disassembly of the strong promoter and returns to a low level of transposition activity.
Here we describe a series of kinetic experiments, which clearly show that figure-eight molecules are precursors of transposon circles and that the host replication machinery is involved in this step. Figure-eight molecules appear before transposon circles and disappear with a concomitant increase in transposon circles. In vivo labelling experiments using 3H-thymidine and plasmids with a temperature-sensitive replication apparatus showed that the process does not require replication initiated from the plasmid replication origin, but that it does require active Tpase presumably to generate figure-eight molecules. The results also suggest that the replication pathway not only produces circles from figure-eight molecules, but also regenerated the transposon donor molecule, implying that both strands are replicated. We have used the Escherichia coli replication terminator sequences, terC, which, if bound by the host Tus protein, inhibit replication forks in a polarised manner. When introduced within the transposon in an appropriate orientation, terC was found to inhibit figure-eight resolution to circles in the presence of Tus, but not in its absence. Finally, we have initiated studies to determine which host enzymes are involved in the replicative formation of circles from figure-eight forms. Present experiments demonstrate that the primase DnaG is essential. Transposition of IS911 can therefore be considered to occur by a copy-and-paste mechanism (Curcio and Derbyshire, 2003).
Results
Figure-eight molecules are precursors of transposon circles
In the model of the IS911 cycle (Figure 1), figure-eight intermediates were proposed to form before and to give rise to transposon circles. To confirm this, we have used an in vivo kinetic approach to determine the chronology of appearance of the two types of molecule. For this, we have exploited the observation that the activity of IS911 Tpase is temperature sensitive (Haren et al, 1997).
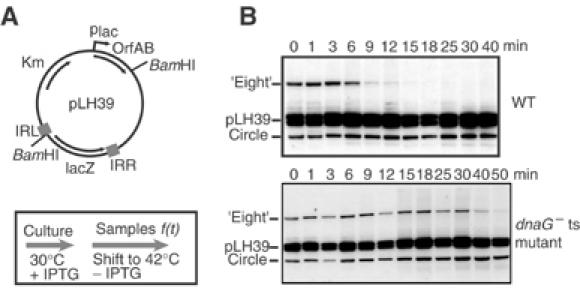
The kinetics of figure-eight and circle formation were determined using pLH39, a p15A-based plasmid carrying an artificial IS911-derived transposon and an orfAB gene controlled independently by the placuv5 promoter and located outside the element (Figure 2). Cultures were grown at 42°C to eliminate residual activity of the naturally temperature-sensitive OrfAB Tpase. This prevents accumulation of intermediates before Tpase induction. The culture was transferred into LB medium, pre-warmed at 30°C, and production of Tpase was induced. Samples were withdrawn at different times and plasmid DNA was isolated, digested with BamHI, which cleaves once within the transposon and once within the donor plasmid backbone. The enzyme therefore generates two fragments from the donor plasmid, linearises transposon circles, and converts the figure-eight structures into χ forms which migrate high in an agarose gel (Polard and Chandler, 1995). The results shown in Figure 2 revealed that figure-eight molecules arose before 5 min of induction, whereas transposon circles were visible after only 12–16 min. This suggests that figure-eight molecules are generated before transposon circles.
Figure 2.
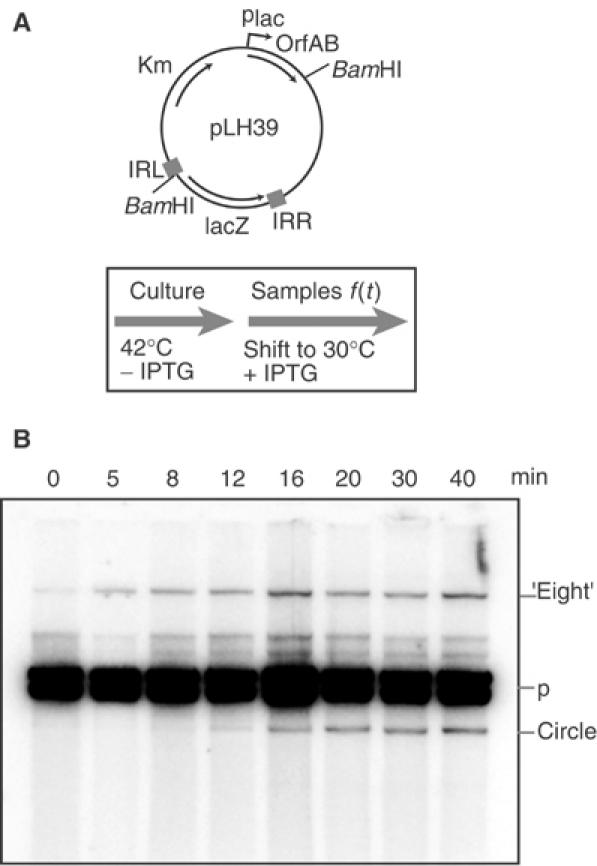
In vivo kinetics of figure-eight and circle formation. (A) Plasmid structure and experimental regime. Plasmid pLH39 is a p15A-based plasmid carrying an artificial IS911-derived transposon and an independent OrfAB gene under control of placuv5. The position of BamHI sites to separate different molecules is shown. The culture was grown at 42°C (OrfAB activity is temperature sensitive) to ensure that there was no residual Tpase activity. At t=0 (OD600=0.6), the population was transferred into LB medium prewarmed at 30°C and production of Tpase was induced with IPTG. Samples were withdrawn at different times, growth was stopped by addition of an sodium azide/ice mixture, and cleared lysates were then made. (B) Separation of plasmid species. Agarose gel of BamHI-digested DNA samples removed at times 0–40 min (shown above the lanes). The separated species were visualised by hybridisation directly in the dried gel with a transposon-specific oligonucleotide probe. The position of the χ forms generated by BamHI digestion of the figure-eight molecules is shown (‘eight'), together with the two bands obtained from the donor plasmid (p) and the BamHI-linearised transposon circles. Note that some additional minor bands are due to incomplete digestion of plasmid molecules.
To determine if the figure-eight molecules are precursors of transposon circles, the experimental regime was inversed: cultures of pLH39-carrying cells were grown at 30°C in the presence of 0.1 mM IPTG to induce Tpase expression and accumulate figure-eight and circle intermediates. The culture was centrifuged and the cells transferred into fresh LB medium pre-warmed at 42°C without IPTG (Figure 3A). This inactivates any residual Tpase activity and prevents further expression. Samples were withdrawn at different times and treated as described above. This experiment showed (Figure 3B) that figure-eight forms disappeared, while levels of transposon circle increased concomitantly. Quantitation of the amounts of each species (Materials and methods) showed that the reduction in figure-eight molecules was mirrored by a similar increase in the transposon circle species (Figure 3C). This strongly suggests that figure-eight molecules are precursors of transposon circles and, since Tpase is largely inactivated under these growth conditions, it suggests that host factors are responsible for this conversion.
Figure 3.
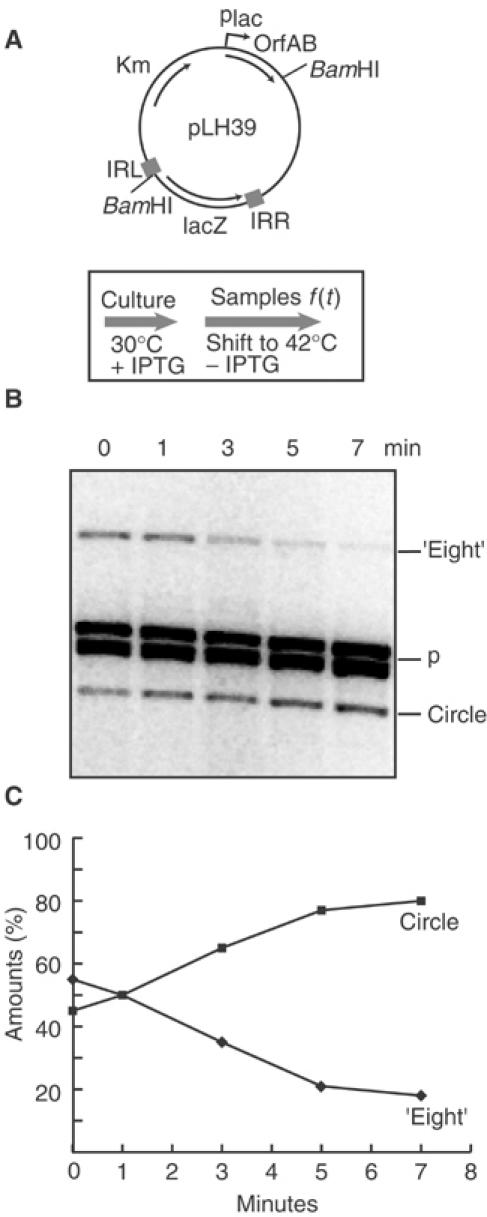
In vivo kinetics of figure-eights to circles conversion. (A) Plasmid structure and experimental regime. The plasmid used was the same as described in Figure 2 (A). The culture was grown at 30°C in the presence of IPTG to induce Tpase synthesis and accumulate figure-eight. At t=0, the culture was transferred into fresh LB medium prewarmed at 42°C without IPTG to inactivate Tpase and stop the induction. Samples were withdrawn at different times and treated as described in Figure 2. (B) Separation of plasmid species. Agarose gel showing details of samples taken between 0 and 7 min. The separated species were visualised by hybridisation as in Figure 2. The symbols are as described in Figure 2. (C) Quantitation of gel shown in (B). The values shown were calculated as follows: the intensities of figure-eight and circle bands were measured and the sum was used to define a value of 100%. The level of parental plasmid was used to normalise sampling variations from well to well.
OrfAB-dependent DNA synthesis
Previous studies had shown that transposon circles could be detected when pre-formed and deproteinised figure-eight molecules were introduced into naïve cells which do not contain IS911 proteins (Turlan et al, 2000). This implied that host factors are involved in this step of circle formation. Thus, transposon circles might be generated from figure-eight molecules either by a recombination process, which would effectively excise the transposon as a circle from the donor plasmid, or by replication of the transposon, which would lead to circle production and regenerate the parental donor plasmid (Figure 4; Polard et al, 1996). In the experiments described above, the absence of detectable levels of appropriate DNA fragments representing the donor backbone lacking the transposon suggested that transposon circles are not formed by simple excision from the parental donor plasmid (Figure 4C). Alternatively, transposon circle formation could involve replication. This could use either the plasmid origin of replication (Figure 4A) or the 3′-OH generated in the donor backbone by the initial strand transfer event, which generated the figure-eight single strand bridge (Figure 4B).
Figure 4.
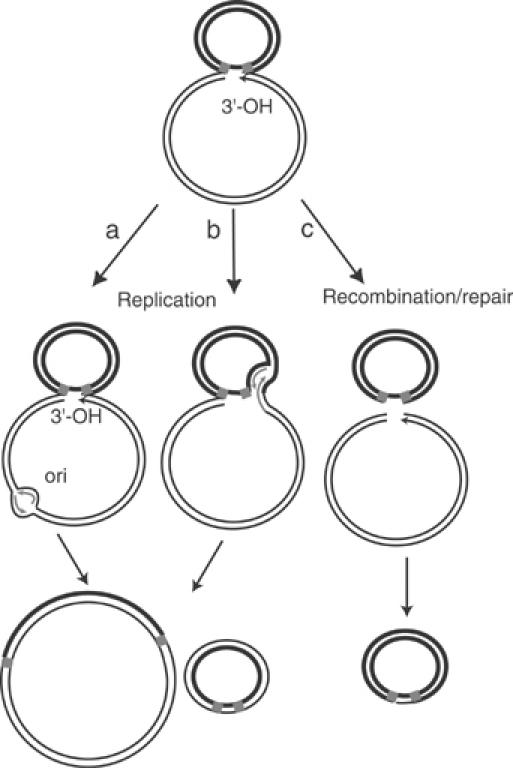
Model of three possible pathways for resolving figure-eight molecules to circles. The figure-eight molecule carrying a 3′-OH is shown above. The two left-hand drawings show circle formation driven by replication from the plasmid origin of replication (left) or from the Tpase-generated 3′-OH (centre). In both cases, replication would generate a double-stranded transposon circle and regenerate the original parental plasmid. Conversion using a recombination/repair pathway is shown on the right. Host-mediated repair would be expected to lead to either destruction or recircularisation of the donor plasmid backbone.
To test the replicative model and to distinguish between these possibilities we investigated circle formation using a temperature sensitive replication mutant of the low copy number plasmid pSC101, pPR4, carrying an artificial IS911-based transposon and a compatible p15A-derived plasmid, pAPT111, which carries the OrfAB gene under control of the plac promoter or pAPT110 without the Tpase gene (Figure 5A).
Figure 5.
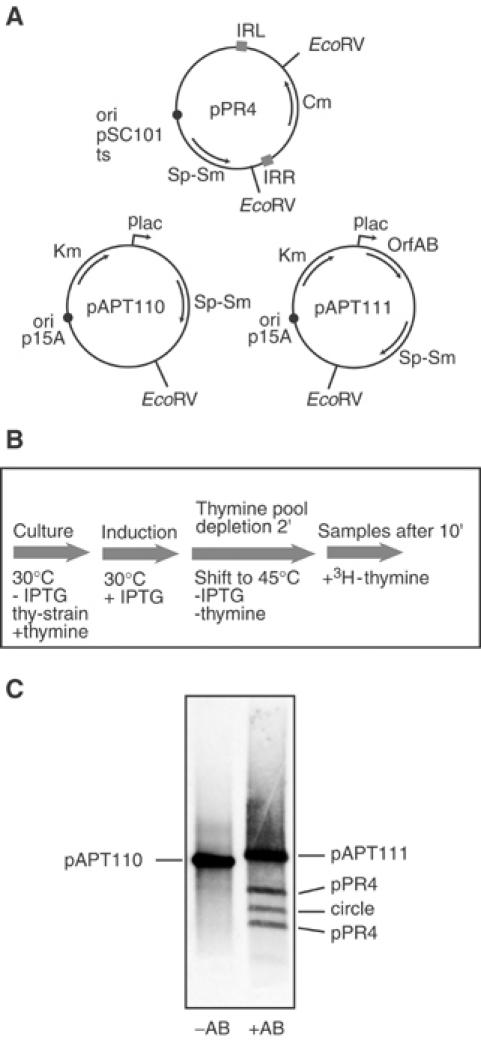
3H-thymidine incorporation in vivo during circle formation. (A) Plasmid structure. The temperature-sensitive plasmid replication mutant, pPR4, carrying the IS911-based transposon with a chloramphenicol resistance gene, Cm, located between correctly oriented IS911 ends (IRL and IRR), is shown together with both the p15A-derived Tpase vector plasmid, pAPT111, carrying the orfAB gene, and its isogenic parent, pAPT110, lacking this gene. (B) Experimental regime. A culture of a thyA− strain carrying a temperature-sensitive pSC101 derivative as a transposon vector (pPR4) and either pAPT110 or pAPT111 was grown at 30°C in minimal medium complemented with thymine, and OrfAB synthesis was induced with IPTG. After growth to OD600=0.6, the culture was transferred into fresh medium without thymine or IPTG at 45°C in order to rapidly inactivate OrfAB production and replication of the transposon-carrying plasmid. After 2 min of incubation to deplete the internal pool of thymine, labelling with 3H-thymidine was performed for 10 min, alkaline lysates were prepared, plasmid DNA was digested with EcoRV and analysed by agarose gel electrophoresis. (C) Agarose gel electrophoresis. The species were visualised directly by radio-luminescence as described in Materials and methods. Left lane: control experiment in the absence of OrfAB. Only the p15A-based vector plasmid pAPT110 is labelled, confirming that the temperature-sensitive pPR4 is not replicated at the nonpermissive temperature and that no circles were detected. Right lane: both the OrfAB-carrying p15A-based vector plasmid and the temperature-sensitive pPR4 plasmid with the transposon are labelled.
Cultures of a thyA strain carrying both plasmids were grown at 30°C in minimal medium complemented with thymine (Materials and Methods). OrfAB synthesis was induced and growth was continued to accumulate figure-eight molecules. The culture was transferred into fresh prewarmed medium without thymine or IPTG at 45°C to inactivate residual OrfAB activity, to prevent further production and to block replication of the temperature sensitive pPR4 donor plasmid. The culture was pulse-labelled with 3H-thymidine. Alkaline lysates were prepared and plasmid DNA was digested with EcoRV (which cleaves twice in pPR4, once within the transposon circle and once in pAPT111) and analysed by agarose gel electrophoresis. Note that this lysis procedure removes a majority of the figure-eight molecules since they contain a single strand break and are irreversibly denatured. In the absence of OrfAB (pAPT110, Figure 5, lane 1) only the p15A derivative was labelled. No labelling of the pSC101 derivative was observed demonstrating that no detectable vegetative replication of pPR4 occurred under these conditions. However, in the presence of OrfAB (lane 2), the Tpase donor plasmid, pAPT111, the two transposon donor plasmid DNA fragments and the linearised transposon circle were labelled. Thus under conditions in which replication from the plasmid origin is inhibited, DNA synthesis requires the presence of OrfAB. This presumably occurs using the free 3′-OH generated in the donor backbone (Figure 1) as a primer for replication.
Moreover, since both transposon circles and the transposon donor plasmid were labelled, this suggests that parental transposon bearing plasmids can be regenerated from figure-eight forms concomitantly with circle formation.
terC affects figure-eight resolution in a polar manner
To further examine the replicative nature of transposon circle formation we made use of the host ter/Tus system. This is composed of short polarised DNA sequences, ter, which bind the E. coli Tus protein in an oriented manner and delay passage of a replication fork (Hill et al, 1988; Kuempel et al, 1989). It seemed possible that placing a ter site within the transposon may affect replicative formation of transposon circles in the presence of Tus but not in its absence whereas placing the site in the donor plasmid backbone outside the transposon should have no effect on transposon circles (Figure 6).
Figure 6.
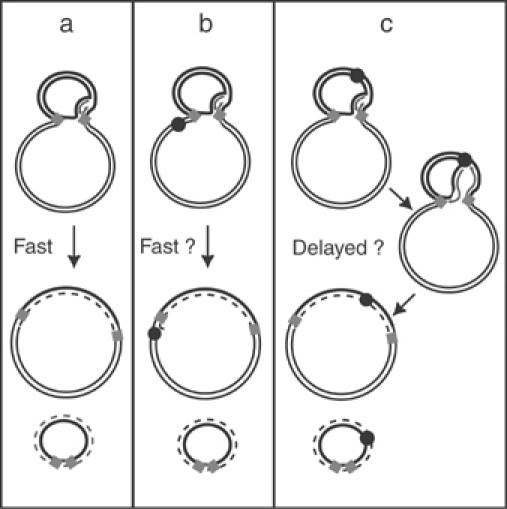
Scheme showing experiments to confirm the replicative pathway of figure-eight resolution in the presence of different locations of terC sites. ter sites and the Tus protein inhibit replication fork movement in a polar manner. terC sequences were therefore introduced either outside (scheme b) or inside (scheme c) an IS911-derived transposon in both orientations. The kinetics of circle formation would be expected to be affected differentially, depending on the orientation of ter. The predicted effects on the resolution rate are proposed. The newly replicated strand is shown as a dotted line.
One difficulty with this approach is that the initiating strand transfer event which generates a figure-eight molecule is not polarised. It can occur using either the right or left end and therefore gives rise to a mixed population of figure-eight molecules. Thus replication, to generate transposon circles, would be expected to occur from either end in the mixed figure-eight population. In order to orient the first strand transfer, we used IS911 derivatives which carried a mutation of the first two base pairs of IRR or of IRL. These mutations inhibit the ability of the mutated end to undergo strand transfer but do not affect its ability to act as a target end (Polard and Chandler, 1995). The ter sites were introduced into plasmids carrying these IS911 variants (IRL* with IRR, or IRL with IRR*). Plasmids (Table I) were constructed with terC sites inside the IS911-based transposon in either orientation (pDV41, 42, 43 with IRL*, or pDV48, 49, 50 with IRR*) or in the donor plasmid backbone (pDV45 and 46). Plasmids pDV43 and pDV50 harbour two terC sites in tandem (Figure 7 and Table I).
Table 1.
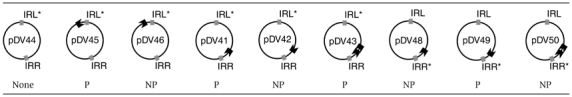 | ||||||||
| None | P | NP | P | NP | P | NP | P | NP |
Constructions showing IRs mutated for donor activity and position and orientation of terC sites  Permissive (P) or Non-Permissive (NP) orientations of terC terminators. Permissive (P) or Non-Permissive (NP) orientations of terC terminators. | ||||||||
Figure 7.
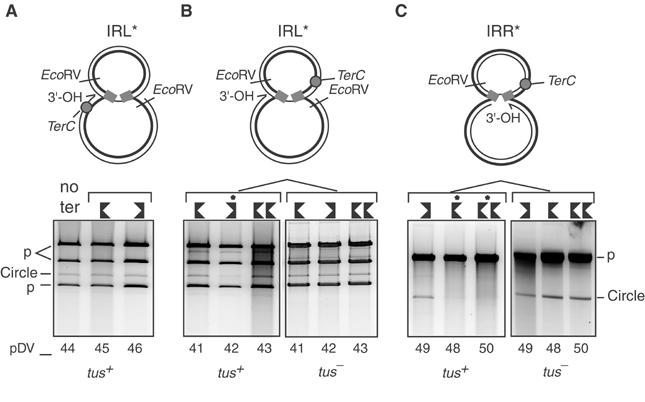
terC affects figure-eight resolution in a polar manner. IR donor mutants were used to obtain unique figure-eight populations and to orient the potential replication fork (A–C, top). The 1% agarose gels were stained with Sybr green. IRL* or IRR* donor mutants (A–C, bottom) were tested separately with different positions (shown as grey circles, top) and orientations of terC sites (shown by oriented black symbols, bottom; Table I). These were placed outside the transposon (pDV45 and 46 with IRL*; A) or inside the transposon with IRL* (pDV41, 42, 43; B) or with IRR* (pDV48, 49, 50; C). pDV43 and 50 harbour two terC sites in tandem. The right-hand panels (B, C) show the results obtained in a tus− strain. The major upper band marked p in (A, B) represents linearised pAPT111. The lower two bands marked p (A, B) represent two fragments generated from the transposon-carrying plasmids. Introduction of IRR* (C) resulting in the elimination of one of the two EcoRV sites from the parental plasmid pRP4 (Figure 5). These plasmids are therefore linearised by EcoRV and migrate at the same position as the linearised Tpase donor plasmid. The band corresponding to plasmid circles is also indicated. Additional minor bands migrate at positions consistent with partially digested products. Lanes in which transposon circle formation was delayed are indicated (*).
The appearance of transposon circles was analysed for all plasmids in both tus+ or tus− strains. Strains were grown at 42°C and then shifted to 30°C in the presence of IPTG. Since the effect of ter/Tus is to delay a replication fork rather than to completely block it, several induction periods were tested. Plasmid DNA was extracted, digested with EcoRV and analysed as for the experiments shown in Figure 5. The clearest results were obtained after 30 min of induction and are shown in Figure 7.
TerC located in either orientation in the donor backbone (pDV45 and pDV46) showed no effect compared to the parental plasmid (pDV44) in the tus+ strain (Figure 7A). However, clear effects were observed when the site was located within the transposon. In the case of the IRL*-carrying plasmids, transposon circles appeared to be reduced in pDV42 but not when the terC site occurred in the opposite orientation (pDV41 and pDV43) (Figure 7B). However, in the IRR* plasmids this pattern was inversed: plasmid pDV49 (equivalent to pDV42) continued to produce transposon circles while plasmid pDV48 and pDV50 did not (Figure 7C). Note that the EcoRV restriction site within the donor backbone was removed during the construction of these plasmids and the transposon and Tpase donor plasmids migrate as linear molecules at the same position in this gel. No inhibition was detected with any of the plasmids in the tus− control strain (Figure 7B and C, right panels).
Thus in the presence of Tus, terC exhibited permissive and nonpermissive orientations depending on the polarity of strand transfer. Furthermore, the nonpermissive orientation was that expected for replication from the free 3′-OH on the donor backbone. This result confirms that circle formation uses a host-specific replicative pathway.
Disappearance of figure-eight forms is delayed in a dnaGts mutant
In a preliminary attempt to define the host factors necessary for replicative circle formation, we tested the effect of a mutation in the dnaG primase. A temperature sensitive dnaGts mutant gave a clear effect. Figure-eight molecules were generated from pLH39 in cultures grown at 30°C as described above (Figure 3). Following a shift to 42°C and elimination of IPTG, samples were removed, cleared lysates were prepared and digested with BamHI and the resulting plasmid DNA species were separated by agarose gel electrophoresis. The results (Figure 8B) showed that the disappearance of the figure eight form was greatly delayed in the dnaGts strain compared to the wild-type strain.
Figure 8.
In vivo kinetics of figure-eight to circle conversion in a dnaGts strain. (A) Plasmid structure and experimental regime. After production of figure-eight molecules at 30°C (Tpase OrfAB and DnaG active (A)), the conversion of figure-eights to circles of a wild-type strain was compared with the dnaGts strain at 42°C. Samples were withdrawn at different times and treated as described in Figure 2. (B) Separation of plasmid species by agarose gel electrophoresis. The top panel shows results obtained with the wild-type strain, while the lower panel presents the results obtained with the dnaG mutant. Numbers above the gel indicate the time in minutes after the temperature shift.
Figure-eight molecules had disappeared by between 6–9 min in the wildtype strain whereas they persisted for 30–40 min in the dnaG mutant. In the latter case, figure-eight molecules disappeared abruptly after 30 min. This may be the result of degradation or of rescue by another mechanism (see Discussion).
These results therefore suggest a fundamental role for the DnaG primase in conversion of figure-eights to circles, and thus strongly support a replicative model for circle formation.
Discussion
The strategy adopted by IS911 and probably other IS3 family members differs notably from other known transposition mechanisms. A distinguishing feature lies in the first step of strand cleavage and transfer in which only one strand at one IS end is cleaved to generate a 3′-OH. This then attacks the same strand 3 bp into the DNA flanking the opposite end to generate a single-strand bridge between the abutted ends. Although asymmetric, either end can serve at the attacking end. This is to be contrasted with other transposition processes in which both ends are cleaved simultaneously and are either transferred to the target without cleavage of the second transposon strand (e.g. Tn3 (Grindley, 2002), Mu (Chaconas and Harshey, 2002), and members of IS6 family (Chandler and Mahillon, 2002)) or following second strand cleavage and excision of the element (e.g. Tn7 (Craig, 1996), IS10 (Kleckner et al, 1996), Tn5, (Reznikoff, 2002)).
Another particularity of IS911 transposition is that covalently closed transposon circles are generated as transposition intermediates (see (Rousseau et al, 2002)). This is also the case for IS2, IS3 and IS150 (Lewis and Grindley, 1997; Sekine et al, 1999; Haas and Rak, 2002). The experiments presented here provide evidence that circular IS911 transposition intermediates are derived from the bridged figure-eight molecules by a replicative mechanism. Kinetic experiments demonstrated that the figure-eight forms appear first, while transposon circles are produced only after a lag of about 10 min (Figure 2). Moreover, in chase experiments, preformed figure-eight molecules disappeared and transposon circles accumulated with identical kinetics and in similar quantities (Figure 3). No species with the migration properties expected of the replicative donor backbone alone (deleted for the transposon) were observed either here or in previous experiments (see for example Polard and Chandler, 1995). This suggests that simple excision of the transposon does not occur. These results extend those obtained previously (Polard and Chandler, 1995) which used a more aggressive treatment, addition of chloramphenicol, to inhibit total protein synthesis.
Experiments using plasmids in which replication initiation is temperature sensitive (pSC101ts) and which carried an artificial IS911-based transposon indicated that both transposon circles and the donor plasmid with the transposon were rapidly labelled by 3H-thymidine in vivo in the absence of plasmid replication but after prior expression of the OrfAB Tpase. No labelling was observed in the absence of OrfAB (Figure 5). Concomitant appearance of 3H-labelled circles and transposon-bearing plasmids in conditions where the initiation of replication of the transposon donor plasmid was blocked is consistent with replication of both DNA strands. The absolute dependence on the prior presence of the OrfAB and therefore presumably of figure-eight intermediates is consistent with initiation of replication from the figure eight junction. The results therefore support the replicative model shown in Figure 4B.
This type of model was further supported by the observation that programmed IS-replication is specifically affected by the presence of the replication terminator terC. These sites are normally located around the terminus of chromosome replication in a region diametrically opposite to the replication origin (Hill et al, 1988; Mohanty et al, 1996; Bussiere and Bastia, 1999). Interaction between ter sites and the Tus protein inhibits replication fork progression in an orientation specific manner (Khatri et al, 1989; Lee et al, 1989; Mulugu et al, 2001). This property was used to test the effect of ter sites introduced within the transposon or within the donor plasmid backbone in both permissive and nonpermissive orientations for oncoming replication forks. However, IS911 can use either IRL or IRR to perform the first strand cleavage and transfer. This produces two different types of figure-eight molecules in the population with the resulting free 3′-OH on one or other strand (see Figure 1) and would result in a mixed population in which a given figure-eight molecule formed using either the left or right end (Figure 7, top). This situation complicates the use of polarised ter sites. Potential forks were therefore oriented by mutation of the terminal 5′-CA-3′dinucleotide of IRL (Figure 7A and B) or of IRR (Figure 7C). The dinucleotide is essential for the first cleavage and strand transfer (Polard and Chandler, 1995). Such mutants generate figure-eight molecules of unique polarity. The results clearly showed that circle formation was transiently paused only if ter sites were located within the transposon and in a specific orientation. In each case, the orientation was that expected to counteract replication forks initiated from the 3′-OH on the donor plasmid backbone generated by strand transfer. The observed delay in appearance of transposon circles was due to the specific action of the ter sites since it did not occur in a tus− strain (Figure 7).
While these results demonstrate that formation of transposon circles is replicative, they do not reveal the nature of the host enzymes involved. We examined the effects of the DnaG primase in figure-eight to circle conversion. Using a dnaGts strain the disappearance of figure-eight molecules was observed to be significantly delayed at the nonpermissive temperature compared to the wild-type strain (Figure 8). Since DnaG is the host primase necessary for synthesis of Okazaki fragment primer RNA involved in lagging strand replication, this confirms that circle formation involves replication.
The results obtained with DnaG and with terC raise the possibility that DnaB may be involved in transposon circle formation. Both DnaG and DnaB appear necessary for optimal primer RNA synthesis on the lagging strand (Lu et al, 1996) and Tus shows specificity for DnaB. However, in similar experiments a dnaBts strain failed to show such delays in figure eight conversion (data not shown). This behaviour might reflect the existence of alternative pathways which operate in the absence of DnaB (e.g. use of an alternative helicase or, given the relatively short segment to be replicated, involvement of a helicase independent replication mechanism—see next paragraph). Indeed, it has recently been shown, for example, that non conventional branched molecules can be formed by transfer of a single IS911 end (SET) between one end in an IR-IR junction and a second, isolated target IR (Loot et al, 2004). These partial transposition products carry a four-way DNA branched region similar to a Holliday junction but with a nick at the branch site. The structure can be resolved to generate forms which resemble the full transposition product. This involves branch migration and occurs by resolution of the second transposon strand in a process that requires the RecG helicase (Turlan et al, 2004). Although this reaction is probably much less efficient than the ‘normal' complete transposition event and would not be expected to involve extensive DNA synthesis, it shows that several parallel and nonexclusive pathways may operate within the cell.
In the replicative model shown in Figure 4A, it is assumed that leading and lagging strand synthesis are coupled to give rise directly to a double stranded transposon circle. An alternative model would be that leading and lagging strand synthesis are not coupled and that transposon circles are first produced as single strand circles by simple extension from the 3′-OH of the donor backbone and ‘extrusion' of the (unreplicated) single strand circle. The circles could then be processed to double strand circles in a second subsequent replication step similar to that which generates double strand RFII forms from circular single strand phage DNA following infection (Kornberg and Baker, 1992). Several levels of complexity for priming phage second strand synthesis are known. All involve initiation at a short hairpin structure in the phage genome. In the case of IS911, this could be provided by the 36bp abutted terminal IRs. In phage G4, for example, the DnaG primase alone accomplishes the priming step which necessitates a hairpin structure (Fiddes et al, 1978; Godson et al, 1978) close to the triplet 5′-CTG where RNA initiates (Hiasa et al, 1990). Such sequences can be found in IS911 for both polarities of IR attack. Note that single strand circles have been observed for IS91 in vivo (Del Pilar Garcillan-Barcia et al, 2001) and are thought to be intermediates in the rolling circle transposition mechanism used by this IS. This type of alternative mechanism involving a single strand circular intermediate is at present under investigation.
Another question raised by these results is whether the presence of bound Tpase is required for mobilisation of host proteins and the formation of a replisome. For bacteriophage Mu, the presence of Tpase, MuA, in the synaptic complex is necessary to prepare the template for the initiation of DNA synthesis (Nakai and Kruklitis, 1995). The MuA complex is implicated in protecting the strand transfer product and the forked junctions until host factors are recruited which then can serve as an initiator of a ‘cascade' of events leading to DNA replication. For IS911, the Tpase does not appear to be essential for initiating the replication step since purified deproteinised figure-eight intermediates were resolved following transformation into strains devoid of IS911 proteins (Turlan et al, 2000). Nevertheless, we cannot exclude that Tpase bound at the junction created after the first strand transfer affects or facilitates the following steps in a quantitative manner by blocking the action of inappropriate enzymes as has been suggested for phage Mu (Nakai and Kruklitis, 1995).
While the results presented here clearly demonstrate that IS911 circular transposition intermediates are produced from the figure-eight molecules by a replicative mechanism, many questions remain to be answered. Most importantly, the enzymology of the process has yet to be clarified. The DNA polymerase(s), clamp loading protein(s) and perhaps helicase(s) are yet to be identified and regulatory processes involved in the recombination-replication switch require analysis. Moreover, although circle formation involves replication, we have yet to determine whether this involves concomitant leading and lagging strand synthesis. In spite of the uncertainties inherent to the complexity of these events, the results presented here demonstrate that IS911 and by extension, other members of the IS3 family, have adopted a transposition mechanism different from other well known transposons such as IS10, IS50, Tn7 and bacteriophage Mu. This strategy can be viewed as a ‘copy-paste' mechanism compared to the ‘cut and paste' mechanism involved in IS10, IS50, Tn7 transposition or the replicative cointegrate formation pathway involved in Mu transposition and represents an alternative transposition paradigm. It should also be noted that this mechanism is likely to occur for many other IS families which also transpose using intermediates involving abutted IR-junctions.
Materials and methods
Bacterial strains and media
The strains for kinetic experiments were MC4100 (araD139Δ(argF-lac) u169 rpsL150 relA1 rlbB5301doC1 ptsF25 rbsR), or MC4100 dnaGts. The strain used for in vivo 3H-thymidine incorporation was LN2667 (thi thy leu deoB supE SmR (srl::Tn10) recA1). The tus strain used in Tus/ter experiment was LN4082 (HfrC del(tus:Ap) thi thy leu deoB supE). Cultures were grown in Luria broth supplemented with appropriate antibiotics. For thymidine incorporation experiments, cultures were grown in VB medium (for 1 l: 3.5 g NaNH4HPO4, 10 g K2HPO4, 2 g citric acid, 0.2 g MgSO4) supplemented with thymine (50 μg/ml), thiamine (100 μg/ml), glucose (4 mg/ml), leucine (20 μg/ml), casamino acids (1 mg/ml), and appropriate antibiotics.
Plasmids
Plasmids supplying transposition functions in trans. Plasmids pAPT110 (which carries no IS911 genes) and pAPT111 (which expresses OrfAB Tpase under the control of a placUV5 promoter) have been described previously (Polard and Chandler, 1995).
Transposase substrates. Kinetic experiments: Plasmid pLH39 used in these experiments, a p15A-based plasmid carrying an independent OrfAB gene under control of placuv5, was constructed by cloning an IS911-derived transposon bearing a lacZ gene into pAPT111 (Polard and Chandler, 1995). For in vivo DNA-labelling experiments, plasmid pPR4, a derivative of a temperature-sensitive replication mutant of pSC101, was constructed as follows: the EcoRI–BamHI fragment from the temperature-sensitive pSC101- (Hashimoto-Gotoh and Sekiguchi, 1977) derived plasmid pGB2 (Churchward et al, 1984) was inserted between EcoRI and BamHI sites of plasmid pAPT99 containing chloramphenicol gene between IRL and IRR (Polard and Chandler, 1995).
Experiments with terminator terC: The parental plasmid pDV44 was obtained by BalI–BalI fragment exchange of wild-type IRL of pPR4 described above, by a 5′-CA to 5′-TC terminal dinucleotide mutant of IRL (IRL*) from plasmid pAPT177 (Polard and Chandler, 1995).
terC sequences were obtained by annealing two 36 bp complementary oligonucleotides and introduced either outside (EcoRI site) or inside (XhoI site) an IS911-derived transposon in both orientations. The sequence of the terC sites used here was: 5′-AATATAGGATGTTGTAACTAATAT-3′. Additional restriction site sequences were added at both ends to facilitate cloning into the EcoRI or XhoI sites of pDV44.
Both orientations of terC (permissive or non permissive for leading strand replication) were obtained in the EcoRI site to generate pDV45 and 46 (Table I). Insertions into the XhoI site generated pDV42, 41, and 43.
To generate IRR* terminal dinucleotide mutations, a BalI–BalI fragment exchange of an IRR* from plasmid pAPT178 (Polard and Chandler, 1995) into pDV41, 42, and 43 was used to generate pDV48, 49, and 50, respectively. These plasmids are schematised in Table I. Note that the EcoRV restriction site within the donor backbone was removed during the construction of the IRR* variant plasmids. In these cases, the transposon and Tpase donor plasmids migrate as linear molecules at the same position in this gel (Figure 7C).
Alkaline lysates were prepared as described (Birnboim and Doly, 1979). Cleared lysates were prepared as described (Clewell and Helinski, 1969), with the following modifications: the cleared supernatants collected were concentrated by isopropanol precipitation and resuspended in 100 μl of TE buffer and purified by using a Qiaquick kit as described by the supplier (Qiagen).
Kinetics of figure-eight and circle formation in vivo
Plasmid pLH39 was used in these experiments. The culture was grown at 42°C to inactivate any residual Tpase activity (OrfAB is temperature sensitive). At t=0 (OD600=0.6), the population was transferred into LB medium pre-warmed at 30°C and production of Tpase was induced with IPTG. Samples were withdrawn at different times, growth was stopped by transfer in a sodium azide mixture (50 mM, final concentration) in ice, and cleared lysates were then made. The samples were cleaved with BamHI and separated by agarose gel electrophoresis (1% agarose, TAE buffer); the gel was dried at 60°C and used directly for hybridisation. A 32P 5′-labelled oligonucleotide complementary to the 5′ end of the orfAB gene also present within the IS911-derived transposon was hybridised for 12–15 h and analysed using a Fuji BAS1000 phosphorimaging system.
Kinetics of figure-eight to circle conversion in vivo
Cells carrying pLH39 were grown at 30°C in the presence of IPTG to induce Tpase synthesis and figure-eight formation. At t=0 (OD600=0.6), the culture was centrifuged and resuspended in fresh LB medium pre-warmed at 42°C to inactivate Tpase. Samples were withdrawn at different times, growth was stopped by transfer in a sodium azide mixture (50 mM) in ice, and cleared lysates were then made. The samples were cleaved with BamHI and separated by agarose gel electrophoresis (1% agarose, TAE buffer). The values were calculated as follows: the intensities of figure-eight and circle bands were measured with a Fuji BAS1000 phosphorimaging system coupled to the PCBas software and the sum was used to define a value of 100%. The level of parental plasmid was used to normalise sampling variations from well to well. Since the parental plasmid continues to replicate during the experiment whereas transposition intermediates are no longer produced, parental plasmid DNA increases over time with the growth rate of the culture. This increase was also taken into account in the normalisation.
For experiments using mutants defective for the DnaG primase, after production of figure-eight molecules at 30°C (Tpase OrfAB and DnaG active), the conversion of figure-eights to circles of a wild-type strain was compared with the dnaGts strain at 42°C. The samples were separated by agarose gel electrophoresis (1% agarose, TAE buffer) and treated as above.
3H-thymidine DNA labelling in vivo
A culture of a thyA− strain carrying the temperature-sensitive plasmid pPR4 as a transposon vector was grown at 30°C in 100 ml of minimal VB medium complemented with thymine, thiamine, and casamino acids. OrfAB synthesis was induced with 1 mM IPTG and the culture was grown to an OD600 of 0.6. The culture was concentrated into 10 ml of the same fresh medium but depleted in thymine and IPTG, pre-warmed at 45°C in order to inactivate OrfAB production and replication of the transposon-carrying plasmid. After 2 min of incubation to deplete the internal pool of cold thymine, 500 μl of [methyl, 1′, 2′-3H]thymidine (113 Ci/mmol) was added for an additional 10 min. Alkaline lysates were then prepared (Qiagen miniprep kit), plasmid DNA was then digested with EcoRV and separated by agarose gel electrophoresis. After migration, the gel was equilibrated in 20 volumes of 95% EtOH for 30 min. This step was repeated twice to eliminate a maximum amount of water present in the gel. The gel was then soaked in a solution of autoradiography enhancer (NEN) for 3 h, and rinsed in water for 1 h. The gel was dried at 60°C under vacuum and exposed to a Kodak Biomax film for about a week.
Circle formation in the presence of terC sites in tus+ or tus− strains
IR donor mutants were used to obtain a unique figure-eight population to orient the replication fork movement in a unique polarity during the resolution. IRL* or IRR* donor mutants were tested separately with different positions and orientations of terC sites inside (pDV41, 42, 43 with IRL*, or pDV48, 49, 50 with IRR*) or outside IS911 (pDV45 and 46). pDV43 and 50 harbour two terC sites in tandem. The appearance of circles was analysed for all plasmids in both tus+ or tus− strains. Plasmid DNA was digested with EcoRV which cleaves once within the transposon, once within the donor backbone, and once in the transposon donor plasmid, except for IRR* variants which lack the EcoRV site near IRR within the donor plasmid backbone. In this case, the transposon and Tpase donor plasmids migrate as linear molecules at the same position in gel (Figure 7C).
Acknowledgments
We owe special thanks to P Rousseau for the gift of the plasmid pPR4, C Lesterlin for the tus− strain, J-M Louarn, and F Cornet for useful discussions during the progress of this work. We thank present (E Gueguen, P Rousseau, B Ton-Hoang) or ex-members (C Loot, C Normand, C Turlan) of the Mobile Genetic Elements group for discussions and comments on the manuscript. We are also grateful to CL Dalton for stimulating advice during this work and P Polard for contribution in the initial stages of this work. This work was supported by grants from the Centre National pour la Recherche Scientifique (CNRS) and Région Midi-Pyrénées, and l'Association pour la Recherche sur le Cancer (ARC).
References
- Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7: 1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussiere DE, Bastia D (1999) Termination of DNA replication of bacterial and plasmid chromosomes. Mol Microbiol 31: 1611–1618 [DOI] [PubMed] [Google Scholar]
- Chaconas G, Harshey RM (2002) Transposition of phage Mu DNA. In: Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds), Washington, DC: ASM Press [Google Scholar]
- Chandler M, Mahillon J (2002) Insertion sequences revisited. In Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds), pp 305–366. Washington, DC: ASM Press [Google Scholar]
- Churchward G, Belin D, Nagamine Y (1984) A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31: 165–171 [DOI] [PubMed] [Google Scholar]
- Clewell DB, Helinski DR (1969) Supercoiled circular DNA–protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc Natl Acad Sci USA 62: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL (1996) Transposon Tn7. Curr Top Microbiol Immunol 204: 27–48 [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM (2003) The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol 4: 865–877 [DOI] [PubMed] [Google Scholar]
- Del Pilar Garcillan-Barcia M, Bernales I, Mendiola MV, De La Cruz F (2001) Single-stranded DNA intermediates in IS91 rolling-circle transposition. J Clin Microbiol 39: 8–13 [DOI] [PubMed] [Google Scholar]
- Duval-Valentin G, Normand C, Khemici V, Marty B, Chandler M (2001) Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J 20: 5802–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes JC, Barrell BG, Godson GN (1978) Nucleotide sequences of the separate origins of synthesis of bacteriophage G4 viral and complementary DNA strands. Proc Natl Acad Sci USA 75: 1081–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godson GN, Barrell BG, Staden R, Fiddes JC (1978) Nucleotide sequence of bacteriophage G4 DNA. Nature 276: 236–247 [DOI] [PubMed] [Google Scholar]
- Grindley ND (2002) The movement of Tn3-like elements: transposition and cointegrate resolution. In Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds), Washington, DC: ASM Press [Google Scholar]
- Haas M, Rak B (2002) Escherichia coli insertion sequence IS150: transposition via circular and linear intermediates. J Bacteriol 184: 5833–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Betermier M, Polard P, Chandler M (1997) IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol Microbiol 25: 531–540 [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T, Sekiguchi M (1977) Mutations of temperature sensitivity in R plasmid pSC101. J Bacteriol 131: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiasa H, Sakai H, Komano T, Godson GN (1990) Structural features of the priming signal recognized by primase: mutational analysis of the phage G4 origin of complementary DNA strand synthesis. Nucleic Acids Res 18: 4825–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TM, Pelletier AJ, Tecklenburg ML, Kuempel PL (1988) Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell 55: 459–466 [DOI] [PubMed] [Google Scholar]
- Khatri GS, MacAllister T, Sista PR, Bastia D (1989) The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell 59: 667–674 [DOI] [PubMed] [Google Scholar]
- Kleckner N, Chalmers RM, Kwon D, Sakai J, Bolland S (1996) Tn10 and IS10 transposition and chromosome rearrangements: mechanisms and regulation in vivo and in vitro. In Transposable Elements, Saedler H, Gierl A (eds), Vol. 204, pp 49–82. Heidelberg: Springer [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker T (1992) DNA Replication, 2nd edn. New York: Freeman W.H. and Company [Google Scholar]
- Kruklitis R, Nakai H (1994) Participation of the bacteriophage Mu A protein and host factors in the initiation of Mu DNA synthesis in vitro. J Biol Chem 269: 16469–16477 [PubMed] [Google Scholar]
- Kuempel PL, Pelletier AJ, Hill TM (1989) Tus and the terminators: the arrest of replication in prokaryotes. Cell 59: 581–583 [DOI] [PubMed] [Google Scholar]
- Lee EH, Kornberg A, Hidaka M, Kobayashi T, Horiuchi T (1989) Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci USA 86: 9104–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, Grindley ND (1997) Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol Microbiol 25: 517–529 [DOI] [PubMed] [Google Scholar]
- Loot C, Turlan C, Chandler M (2004) Host processing of branched DNA intermediates is involved in targeted transposition of IS911. Mol Microbiol 51: 385–393 [DOI] [PubMed] [Google Scholar]
- Lu YB, Ratnakar PV, Mohanty BK, Bastia D (1996) Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc Natl Acad Sci USA 93: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Chandler M (1998) Insertion sequences. J Bacteriol 180: 5306–53129765560 [Google Scholar]
- May EW, Craig NL (1996) Switching from cut-and-paste to replicative Tn7 transposition. 1996 272: 401–404 [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Sahoo T, Bastia D (1996) The relationship between sequence-specific termination of DNA replication and transcription. EMBO J 15: 2530–2539 [PMC free article] [PubMed] [Google Scholar]
- Mulugu S, Potnis A, Shamsuzzaman Taylor J, Alexander K, Bastia D (2001) Mechanism of termination of DNA replication of Escherichia coli involves helicase–contrahelicase interaction. Proc Natl Acad Sci USA 98: 9569–9574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H, Kruklitis R (1995) Disassembly of the bacteriophage Mu transposase for the initiation of Mu DNA replication. J Biol Chem 270: 19591–19598 [DOI] [PubMed] [Google Scholar]
- Polard P, Chandler M (1995) An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev 9: 2846–2858 [DOI] [PubMed] [Google Scholar]
- Polard P, Ton-Hoang B, Haren L, Betermier M, Walczak R, Chandler M (1996) IS911-mediated transpositional recombination in vitro. J Mol Biol 264: 68–81 [DOI] [PubMed] [Google Scholar]
- Reznikoff WS (2002) Transposition of Tn5. In Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds). Washington, DC: ASM Press [Google Scholar]
- Roberts D, Hoopes BC, McClure WR, Kleckner N (1985) IS10 transposition is regulated by DNA adenine methylation. Cell 43: 117–130 [DOI] [PubMed] [Google Scholar]
- Rousseau P, Normand C, Loot C, Turlan C, Alazard R, Duval-Valentin G, Chandler M (2002) Transposition of IS911. In Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds). Washington, DC: ASM Press [Google Scholar]
- Sekine Y, Aihara K, Ohtsubo E (1999) Linearization and transposition of circular molecules of insertion sequence IS3. J Mol Biol 294: 21–34 [DOI] [PubMed] [Google Scholar]
- Sherratt D (1989) Tn3 and related transposable elements: site-specific recombination and transposition. In Mobile DNA, Berg D, Howe M (eds), pp 163–184. Washington DC: American Society of Microbiology [Google Scholar]
- Tavakoli NP, Derbyshire KM (2001) Tipping the balance between replicative and simple transposition. EMBO J 20: 2923–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli NP, DeVost J, Derbyshire KM (1997) Defining functional regions of the IS903 transposase. J Mol Biol 274: 491–504 [DOI] [PubMed] [Google Scholar]
- Ton-Hoang B, Betermier M, Polard P, Chandler M (1997) Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J 16: 3357–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-Hoang B, Polard P, Chandler M (1998) Efficient transposition of IS911 circles in vitro. EMBO J 17: 1169–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlan C, Chandler M (1995) IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J 14: 5410–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlan C, Loot C, Chandler M (2004) Mol Microbiol 53: 1021–1033 [DOI] [PubMed] [Google Scholar]
- Turlan C, Ton-Hoang B, Chandler M (2000) The role of tandem IS dimers in IS911 transposition. Mol Microbiol 35: 1312–1325 [DOI] [PubMed] [Google Scholar]


