Abstract
Iron is required for many biological processes but is also toxic in excess; thus, body iron balance is maintained through sophisticated regulatory mechanisms. The lack of a regulated iron excretory mechanism means that body iron balance is controlled at the level of absorption from the diet. Iron absorption is regulated by the hepatic peptide hormone hepcidin. Hepcidin also controls iron release from cells that recycle or store iron, thus regulating plasma iron concentrations. Hepcidin exerts its effects through its receptor, the cellular iron exporter ferroportin. Important regulators of hepcidin, and therefore of systemic iron homeostasis, include plasma iron concentrations, body iron stores, infection and inflammation, and erythropoiesis. Disturbances in the regulation of hepcidin contribute to the pathogenesis of many iron disorders: hepcidin deficiency causes iron overload in hereditary hemochromatosis and nontransfused β-thalassemia, whereas overproduction of hepcidin is associated with iron-restricted anemias seen in patients with chronic kidney disease, chronic inflammatory diseases, some cancers, and inherited iron-refractory iron deficiency anemia. This review summarizes our current understanding of the molecular mechanisms and signaling pathways involved in the control of hepcidin synthesis in the liver, a principal determinant of plasma hepcidin concentrations.
Keywords: iron, hepcidin, inflammation, erythropoiesis, anemia
Introduction
Iron is an essential trace metal involved in oxygen transport, cellular metabolism, DNA synthesis, innate immunity, growth, and development (1). The ability of iron to cycle between 2 stable oxidation states, ferrous iron [iron (II) or Fe2+] and ferric iron [iron (III) or Fe3+], equips iron to participate in a wide array of biochemical processes. The iron content in men and women ranges from 35 to 55 mg Fe/kg body weight (2, 3). The majority of iron in the body is found in the erythroid compartment, within heme in hemoglobin, and is used to deliver oxygen to every cell in the body. Mature erythrocytes have a life span of ∼120 d, after which point the senescent erythrocytes are phagocytosed by specialized macrophages that recycle iron (4). This recycling of iron provides most of the daily iron requirement in humans (∼20–25 mg; most of it used for hemoglobin synthesis in bone marrow) (2). Typically, adults lose between 1 and 2 mg Fe/d, mainly due to desquamation of epithelial cells or minor bleeding; the absorption of iron from the diet compensates for this basal iron loss (1–3). Absorption can increase severalfold when iron needs to be increased—for example, after hemorrhage or during pregnancy. Iron stores of ∼400–1000 mg in the adult liver (2) represent another important compartment that is mobilized in times of accelerated iron usage.
When iron losses chronically outpace iron intake, iron deficiency develops. Iron deficiency is the most common micronutrient deficiency in the world and is the most prevalent cause of anemia worldwide (5). Health consequences of insufficient iron availability include weakness and fatigue; decreased physical and cognitive performance; adverse pregnancy outcomes, including increased newborn and maternal mortality with severe iron deficiency anemia (IDA)4; delayed mental and motor development in children; and worse outcomes in patients with coexisting pathologic conditions (6). Conversely, iron in excess is toxic to cells because it catalyzes the formation of reactive oxygen species, resulting in oxidative stress (7–9). Oxidative stress has been implicated in the pathogenesis of numerous human diseases, including cancer (10), cardiac disease (11), diabetes (12), and neurodegenerative diseases including Alzheimer and Parkinson diseases (13, 14). There is no known mechanism for the physiologic clearance of excess iron from the body; therefore, body iron content is tightly regulated at the level of absorption from the diet. However, extracellular and plasma iron concentrations are determined by the net balance between cellular iron uptake, mainly in the erythropoietic marrow, and iron release from cells storing and recycling iron (15).
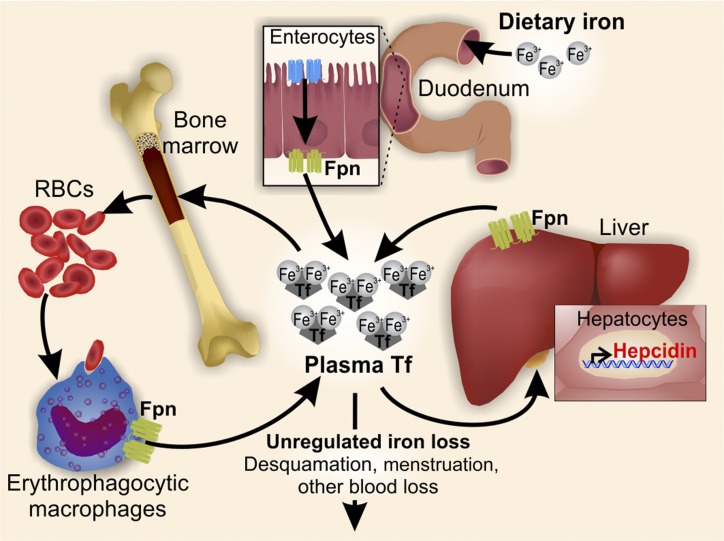
The delivery of iron to every cell in the body depends on circulating iron, bound to the plasma protein transferrin. Iron is supplied into the circulation from macrophages recycling senescent erythrocytes, from duodenal enterocytes absorbing dietary iron, and from hepatic stores. All of these cells release iron into the circulation through the only known iron exporter, ferroportin (commonly referred to as FPN1 and encoded by the gene SLC40A1) (16–18). Therefore, the rate of iron entry into the circulation is proportional to the amount of ferroportin on iron-exporting cells. The key iron-regulatory hormone, hepcidin (HAMP or HEPC), controls systemic iron homeostasis through its ability to negatively regulate ferroportin, its cognate receptor (1, 19) (Figure 1).
FIGURE 1.
Hepcidin regulates systemic iron homeostasis. The hormone hepcidin regulates plasma iron concentrations by controlling ferroportin concentrations on iron-exporting cells, including duodenal enterocytes, recycling macrophages of the spleen and liver, and hepatocytes. Hepcidin production by hepatocytes is the main source of plasma hepcidin. Hepatocyte hepcidin synthesis is regulated at the transcriptional level by multiple stimuli. Fpn, ferroportin; Tf, transferrin.
Hepcidin was first isolated from urine and plasma on the basis of its structural similarities to antimicrobial peptides and named for its origin in the liver and its antimicrobial properties in vitro (20, 21). Northern blot analyses of multiple organs showed that HAMP expression predominates in the liver and to a much lesser extent in the heart and spinal cord (20). Initial experiments showed minor antimicrobial activity of hepcidin in vitro; the role of hepcidin in the regulation of iron was suggested when Pigeon et al. (22) reported hepcidin overexpression during iron overload in mice. The bioactive form of hepcidin is the 25–amino acid peptide hormone secreted by hepatocytes, the main cell type in the liver (20–22). Hepcidin inhibits the release of iron into the circulation by postranslationally regulating its cognate receptor ferroportin, the iron transporter expressed on the basolateral membrane of duodenal enterocytes, on macrophages, placental syncytiotrophoblasts, and hepatocytes (Figure 1) (16–18). Independently of hepcidin, FPN1 production is also regulated transcriptionally by hypoxia-inducible factor 2α (HIF2α) and heme-dependent transcription factors (23), and post-transcriptionally by the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network (24) and iron-responsive micro-RNA, miR-485-3p (25). Of all of these mechanisms, post-translational regulation by hepcidin is the ultimate and decisive step in controlling iron homeostasis. At the molecular level, binding of hepcidin to FPN1 causes receptor ubiquitination (26), internalization, and subsequent lysosomal degradation in vitro (19). On the basis of the recently described structure of the ferroportin bacterial ortholog (27), it is also possible that hepcidin controls iron export from FPN1 by gating the transporter, without inducing its endocytosis. It still remains to be experimentally determined whether and under which conditions the gating may occur. Consequently, the removal of FPN1 from the cell surface (or gating of FPN1) inhibits the release of iron into the plasma and causes iron retention in any cell that expresses FPN1 (19).
Hepcidin is produced at a high rate (∼10 mg/d at baseline) (28) and is rapidly cleared from the circulation by the kidneys, with a half-life of several minutes (29). Thus, altering the hepcidin production rate can rapidly change its circulating concentrations, and consequently the flow of iron into plasma. So far, the regulation of hepcidin production has only been shown at the level of its transcription. The major stimuli regulating hepcidin transcription include blood plasma iron concentrations, liver iron stores, inflammation, and erythropoiesis (30).
Importantly, dysregulation of hepcidin production results in common iron disorders. Hepcidin deficiency causes iron overload (hereditary hemochromatosis and β-thalassemia) (30–32). Conversely, overproduction of hepcidin is associated with iron-restricted anemias (in chronic kidney disease, autoimmune inflammatory disorders, some cancers, and inherited iron-refractory IDA) (28, 30, 33). Therefore, appropriate regulation of hepcidin is essential for the maintenance of iron homeostasis and prevention of disease; and a better understanding of the pathways involved in hepcidin regulation are key to understanding the pathobiology of iron-related disorders.
Hepcidin Regulation: Current Understanding
Stimulatory pathways for hepcidin production
Hepcidin production is stimulated by iron loading and inflammation (Table 1). Hepcidin increase by iron prevents further iron loading to ensure the maintenance of body iron balance. Inflammatory increase in hepcidin causes acute hypoferremia, which likely has a role in nutritional immunity. Iron-mediated hepcidin regulation occurs via the bone morphogenetic protein–SMAD (BMP-SMAD) pathway, whereas inflammation-mediated regulation occurs via both the IL-6/Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling axis and the BMP-SMAD pathway.
TABLE 1.
Hepcidin regulators1
| Pathway | Ligands and modulators | Receptors | Signaling components |
| Hepcidin inducers | |||
| Iron status | |||
| BMP-SMAD | BMP6, TFR1, TFR2, HFE, TMPRSS6, neogenin | ALK2, ALK3, BMPR2, ACTR2A, HJV | SMAD1/5/8 |
| Inflammation | |||
| JAK-STAT | IL-6 | IL-6R, GP130 | JAK2, STAT3 |
| BMP-SMAD (noncanonical) | Activin B | ALK2, ALK3 | SMAD1/5/8 |
| Hepcidin suppressors | |||
| Erythropoiesis | ERFE | ? | ? |
| GDF15 | ? | ? | |
| TWSG1 | ? | ? | |
| Other regulators | |||
| Sex hormones | Testosterone | EGFR (?) | ? |
| 17β-Estradiol | ? | ? | |
| Progesterone | PGRMC1 | SFKs | |
| Growth factors | HGF | c-Met | MEK, PI3K, TGIF |
| EGF | EGFR | ||
| PDGF-BB | PDGFR | CREB/H |
The table summarizes inducers and suppressors of hepcidin production and the respective signaling pathways involved, including ligands and modulators, their receptors, and additional signaling components. ACTR2A, activin A receptor type 2A; ALK, activin receptor-like kinase; BMPR2, bone morphogenetic protein receptor 2; BMP-SMAD, bone morphogenetic protein-mothers against decapentaplegic homolog; BMP6, bone morphogenetic protein 6; c-Met, mesenchymal-epithelial transition factor (MET) proto-oncogene, receptor tyrosine kinase; CREB/H, cAMP-response element binding protein H, EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; ERFE, erythroferrone; GDF15, growth differentiation factor 15; GP130, glycoprotein 130; HFE, human hemochromatosis protein; HGF, hepatocyte growth factor; HJV, hemojuvelin; IL-6R, IL-6 receptor; JAK, Janus kinase; MEK, mitogen-activated protein kinase kinase; PDGF-BB, platelet-derived growth factor BB; PDGFR, platelet-derived growth factor receptor; PGRMC1, progesterone receptor membrane component 1; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; SFK, Src family kinase; STAT, signal transducer and activator of transcription; TFR, transferrin receptor; TGIF, transforming growth-interacting factor; TMPRSS6, transmembrane protease serine 6; TWSG1, twisted-gastrulation 1; ?, unknown.
Iron status and the BMP-SMAD pathway.
Iron sensing.
Hepcidin is produced in the liver by hepatocytes and its expression is feedback regulated by iron. The appropriate regulation of hepcidin expression is dependent on the ability of the liver to sense intracellular and extracellular iron and relay these signals to the hepatocyte nucleus where hepcidin expression can be appropriately modulated to maintain homeostasis. Hepatocyte transferrin receptor (TFR) 1, TFR2, and human hemochromatosis protein (HFE) are believed to function as the extracellular iron-sensing mechanism, and to specifically sense circulating concentrations of transferrin-bound iron (iron-TF). These proteins function by potentiating the signaling through the BMP pathway to stimulate hepcidin transcription in proportion to the concentrations of iron-TF. Hfe−/− or Tfr2 mutant mice failed to increase hepcidin in response to acute iron loading when iron-TF concentrations increased, although they still showed increased hepcidin in response to chronic iron loading when hepatic iron stores were increased (34). HFE is a major histocompatibility complex (MHC) class I–like protein; it was originally identified as a gene mutated in the most common form of hereditary hemochromatosis (35). Structural and functional studies identified TFR1 as a protein interacting with HFE (36). The HFE binding site on TFR1 overlaps with that of iron-transferrin; therefore, binding of HFE to TFR1 is competitively inhibited by iron-TF. When circulating iron-transferrin concentrations are high, HFE is displaced from TFR1 and likely interacts with activin receptor-like kinase (ALK) 3 (37), a BMP receptor type I known to regulate hepcidin expression. HFE stabilizes ALK3 protein by preventing its ubiquitination and proteasomal degradation, thus increasing ALK3 expression at the cell surface. HFE was also reported to associate with TFR2 in overexpressing cellular systems, and TFR2 itself is stabilized by binding iron-TF (38). However, whether an iron-TF/TFR2/HFE complex forms in vivo with endogenous proteins remains to be shown, and it is unclear whether TFR2 also interacts with ALK3 (37). Like Hfe knockout mice, Tfr2-deficient mice also show decreased hepatic SMAD signaling, and double-knockout Hfe/Tfr2 mice show even lower SMAD phosphorylation and hepcidin expression than either of the single knockouts (39). Although the individual roles of iron-TF, TFR1, HFE, TFR2, and ALK3 in iron regulation are well established, it remains to be shown how all of these components interact to modulate BMP signaling.
A distinct pathway appears to be involved in hepcidin regulation by iron stores (34), but the exact mechanism of intracellular iron sensing by hepatocytes is still unclear. One likely mediator is increased production of BMP6 by nonparenchymal cells of the liver in response to iron loading (40, 41). Thus, both extracellular and intracellular iron leads to hepcidin production via the stimulation of the BMP-SMAD pathway. Additional studies are needed to understand the detailed mechanisms of iron sensing that govern hepcidin regulation.
The BMP-SMAD pathway.
The TGF-β superfamily of signaling molecules participates in fundamental cell processes, including proliferation and differentiation (42). Members include TGF-β proteins, activins, nodals, BMPs, and growth and differentiation factors (GDFs). These signaling molecules bind to complexes of type I and type II serine and threonine kinase receptors, forming heteromeric signaling complexes (43), which phosphorylate and activate downstream SMAD transcription factors. BMPs signal through the receptor-activated SMADs 1/5/8, whereas TGF-β proteins, activins, and nodals signal through SMAD2/3. The receptor-activated SMADs then associate with a common SMAD mediator, SMAD4, forming an activated SMAD transcription factor complex, which translocates to the nucleus to modulate gene transcription (44). Studies have shown that multiple members of this family, including TGF-β, activins, and BMPs, are able to induce hepcidin expression, but their individual roles in iron regulation, if any, remain to be delineated.
The initial connection between hepcidin regulation and the TGF-β superfamily pathway was made in mice with liver-specific knockout of Smad4; the disruption of Smad4 resulted in 100-times lower hepcidin expression and iron accumulation in multiple organs (45). Another key finding was the identification of hemojuvelin [HJV, known also as repulsive guidance molecule C (RGMc) or HFE type 2 protein (HFE2)] as a BMP coreceptor (46). Mutations in HJV result in profoundly decreased hepcidin expression and juvenile (type 2) hemochromatosis (47). Experiments that used radiolabeled BMPs showed that the extracellular domain of HJV binds BMPs, and coimmunoprecipitation experiments revealed a direct interaction between HJV and BMP type I receptors, together showing that HJV is a BMP coreceptor (46). HJV was shown to interact with multiple BMP type I receptors (ALK2, 3, and 6), enhance utilization of BMP type II receptor activin A receptor type 2A (ACTR2A) (48), and bind BMP ligands 2, 4, 5, 6, and 7 (49). HJV was also reported to form a membrane complex with HFE and TFR2 in an in vitro overexpression system; however, whether this regulatory complex is formed by endogenous proteins remains to be determined (50).
Although many BMP ligands are able to induce hepcidin expression in vitro (51–54), a genomewide liver transcription profile of livers from animals fed varying iron diets determined that only BMP6 mRNA levels correlate with body iron stores (55), implicating it as an effector in iron homeostasis. The injection of BMP6 into mice increased Hamp expression and decreased serum iron (56). Conversely, studies that used targeted disruption of Bmp6 in mice revealed a severe iron overload phenotype, significantly decreased hepcidin expression, and diminished receptor-regulated SMAD (R-SMAD) phosphorylation and nuclear translocation (54, 56). In addition, in vivo inhibition of BMP6 with the use of soluble HJV fused to the Fc portion of human IgG (HJV.Fc) or HJV neutralizing antibody diminished hepcidin expression and increased serum iron, whereas treatment with the extracellular domain of repulsive guidance molecule B (RGMb or DRAGON) fused to the Fc portion of human IgG (DRAGON.Fc), a more potent inhibitor of BMP2 and BMP4 compared with BMP6, was unable to recapitulate the effects (56). It is now widely accepted that liver BMP6 is the main BMP regulating iron metabolism in mice. Recently, heterozygous human mutations in the BMP6 propeptide have been linked to iron loading in humans (57). Daher et al. (57) identified 3 missense mutations (p.Pro95Ser, p.Leu96Pro, and p.Gln113Glu) in patients with unexplained iron overload. The mutations were clustered in conserved regions of the BMP6 propeptide. Functional analyses showed that these mutations exert a dominant-negative effect on BMP6 function, because both the heterodimers of wild-type (WT) and mutant BMP6 and homodimers of mutant BMP6 exhibited impaired protein secretion (57). Consequently, in cells treated with preconditioned medium from cells expressing mutant BMP6 protein, SMAD1/5/8 phosphorylation and Hamp mRNA induction were lower in comparison to cells treated with preconditioned medium from WT BMP6-expressing cells (57). These recent findings support the role of BMP6 as an important regulator of hepcidin expression in humans.
BMP type I receptors that are expressed on hepatocytes and regulate hepcidin expression include ALK2 (ACVR1) and ALK3 [BMP receptor (BMPR) 1A]. Hepatocyte-specific deletion of either receptor results in iron overload, although the iron-loading phenotype in the Alk3 knockout was more severe (58). In hepatocytes isolated from Alk3-deficient animals, basal hepcidin expression was diminished, suggesting a role for ALK3 in basal Hamp expression (58). The maximal effective hepcidin response to iron, however, requires both ALK2 and ALK3. Treatment of either Alk2- or Alk3-deficient mice with exogenous iron failed to increase Hamp expression (58). The 2 BMP type II receptors expressed on hepatocytes are ACTR2A and BMPR2; genetic mouse models and primary human hepatocytes were utilized to determine that both receptors are involved in BMP6-induced Hamp expression (59). However, the 2 receptors have redundant roles in regulating hepcidin, and only deficiency of both ActR2a and liver Bmpr2, and not either single knockout, prevented iron-induced Hamp expression that resulted in severe iron overload (59). In aggregate, these studies show that hepcidin regulation by the BMP-SMAD pathway is dependent on BMP6 (and possibly other BMP ligands), ALK2, ALK3, BMPR2, ACTR2A, HJV, SMAD1/5/8, and SMAD4.
Matriptase-2 [transmembrane protease serine 6 (TMPRSS6)], a liver-expressed transmembrane serine protease, is an additional regulator of iron homeostasis; it functions as a negative regulator of the BMP-SMAD pathway (60–62). Matriptase-2 itself is regulated by iron: iron deficiency in animals or low intracellular iron in hepatic cells results in stabilization of matriptase-2 protein concentrations (61, 63). The ectodomain of matriptase-2 interacts directly with and cleaves cell surface HJV, suppressing the BMP-SMAD signaling and thus decreasing Hamp expression (64). Patients with mutations in TMPRSS6 develop iron-refractory IDA (60). Similarly, Tmprss6 deficiency in mice causes microcytic anemia and iron deficiency–related hair loss (65). Anemia in both cases is a consequence of inappropriately high hepcidin concentrations that cause decreased dietary iron absorption (65).
Not surprisingly, hepcidin expression was also shown to be affected by other known modulators of BMP signaling, including BMP binding endothelial regulator (BMPER) (66), neogenin (67, 68), endofin (69), and SMAD6 and SMAD7 (70, 71). Additional studies are necessary to develop a more comprehensive understanding of how this system integrates signals from multiple other pathways known to regulate hepcidin.
Hepcidin regulation by inflammation.
Infection causes a decrease in plasma iron concentrations, as described >80 y ago (72). The hypoferremia of inflammation likely evolved as a host defense against extracellular pathogens, which, like all organisms, require iron for replication and other biological functions. Chronically, infection and inflammation can also cause anemia of inflammation, a condition characterized by low serum iron, reduced transferrin saturation, normal to elevated ferritin concentrations, and low reticulocyte counts (73, 74). Hepcidin is thought to be a key mediator of hypoferremia and anemia of inflammation because increased hepcidin sequesters iron in splenic and hepatic macrophages and duodenal enterocytes, preventing export into the plasma. Hepcidin was also shown to mediate resistance to some infections (75, 76), although the spectrum of microbes affected by this mechanism is poorly understood. The link between hepcidin and inflammation was made early on when hepcidin was first cloned: a >100-fold increase in hepcidin concentrations was observed in a septic subject (77). Importantly, in a mouse model of turpentine-induced inflammation, a decrease in serum iron did not occur in hepcidin-deficient mice (78). Furthermore, hepcidin deficiency ameliorated anemia in mouse models of anemia of inflammation (caused by heat-killed Brucella abortus) and chronic kidney disease (79), providing strong evidence for the role for hepcidin as a mediator of hypoferremia and anemia of inflammation. However, hepcidin-independent mechanisms may also contribute to hypoferremia of inflammation via the direct suppressive effect of inflammation on FPN1 (80, 81).
IL-6 was identified as an important upstream mediator of hepcidin induction by inflammation (82). IL-6 infusion in humans induced hepcidin excretion and decreased serum iron and transferrin saturation (73). Subsequent studies showed that IL-6 is required for hepcidin induction by multiple microbial molecules (PAMPs) in primary hepatocytes and in mice, and by bacterial or viral infection in vivo; induction was ablated in the presence of IL-6–neutralizing antibodies and in interleukin-6 (Il6)–deficient mice (73, 83).
Binding of IL-6 to its receptor results in the formation of a hexameric complex of IL-6 receptors [IL-6 receptor (IL-6R) or CD126] with glycoprotein 130 (GP130), inducing a downstream signaling cascade (84–86). The receptor complex lacks intrinsic kinase activity and utilizes the tyrosine kinase, JAK2; formation of the complex results in JAK2 activation, enabling it to phosphorylate downstream targets including STATs. Phosphorylated STATs subsequently dimerize and translocate to the nucleus where they activate gene transcription (84–86). Induction of HAMP transcription by IL-6 is specifically dependent on STAT3: the disruption of the STAT3 binding region in the HAMP promoter prevented its induction by inflammatory stimuli (87). Hepatocyte-specific deletion of the Gp130 region for STAT1 and STAT3 activation (but not a Gp130 mutation that impaired RAS-MAPK activation) prevented hepcidin induction by IL-6 in mice (33). In addition, hepatocyte-specific deletion of Stat3 confirmed that hypoferremia and anemia in a chronic turpentine model were dependent on the hepatocyte STAT3-enhanced transcription of hepcidin (88). Therefore, the signal transduction pathway necessary for inflammation-mediated iron restriction involves IL-6 activation of JAK2 and STAT3 in hepatocytes to upregulate hepcidin transcription.
Signaling pathways rarely act in isolation, and some level of interaction and crosstalk exists between different pathways. Studies suggest that the BMP-SMAD and JAK/STAT pathways interact to induce hepcidin during inflammation. Hepatocyte-specific disruption of SMAD4 abrogated not only BMP but also IL-6–mediated hepcidin induction, indicating that the BMP pathway also contributes to hepcidin regulation by inflammation (45). The inhibition of SMAD1/5/8 signaling with soluble HJV or a small molecule BMP type I inhibitor blunts hepcidin induction and associated hypoferremia in animal models of anemia of inflammation (89). Genetic studies in hepatocyte-specific knockout of the BMP type I receptors showed a role for ALK3: Alk3-deficient animals did not induce hepcidin or suppress serum iron in response to IL-6 stimulation (90). How SMAD and STAT3 signaling interacts at the level of hepcidin transcription remains to be determined.
More recently, activin B has been proposed as an IL-6–independent inflammatory mediator of hepcidin expression (91). Canonical activin B signaling occurs downstream of ALK4 or ALK7 (92, 93) through SMAD2/3 (44). However, it has been shown that activin B is also able to induce phosphorylation of SMAD1/5/8 in both hepatic cell lines and in primary mouse hepatocytes (91); phosphorylation of SMAD1/5/8 was specific to activin B and was not observed after activin A stimulation (91, 94). Furthermore, in mouse models of inflammation, liver expression of the gene encoding the activin βB subunit (Inhbb) is highly upregulated (75, 91, 94), suggesting a potential role for activin B in inflammation-mediated hepcidin gene expression through noncanonical SMAD1/5/8 signaling. Activin B did not induce phosphorylation of STAT3 in vitro, and the treatment of cells with both IL-6 and activin B had a synergistic effect on Hamp expression (91). Systematic small interfering RNA (siRNA) knockdown of endogenously expressed members of the TGF-β superfamily signaling pathways followed by activin B stimulation showed that activin B induction of HAMP is facilitated by the BMP type I receptors ALK2 and ALK3, the activin type II receptors ACVR2A and ACVR2B, the BMP coreceptor HJV, and SMAD5 (94). Finally, activin inhibitor blunted the induction of Hamp expression in mice injected with LPS or B. abortus (94). However, a recent study that used activin B–deficient mice reported that activin B was not required for hepcidin induction in a mouse model of Escherichia coli sepsis (95). Additional studies are needed to clarify the role of activin B in different pathophysiologic conditions associated with inflammation.
Multiple other cytokines were reported to increase hepcidin production, including IL-1β (96, 97), IL-22 (98–100), oncostatin M (101, 102), leukemia inhibitory factor (LIF) (102), and leptin (103), but how much they contribute to the regulation of hepcidin and iron metabolism in vivo remains to be shown.
Inhibitory pathways of hepcidin production
Under homeostatic conditions, hepcidin expression prevents iron overload. However, certain physiologic conditions, including stress erythropoiesis, hypoxia, growth, and pregnancy, necessitate increased iron supply. To meet this increased demand for iron, hepcidin must be suppressed; this allows for increased iron absorption from the diet and release of iron from stores, thereby increasing iron bioavailability in the circulation (Table 1).
Erythropoiesis.
The production of RBCs is critically dependent on the bioavailability of iron. Iron is an important component of heme in hemoglobin, with each heme containing 1 Fe2+ ion necessary for oxygen binding. Hemoglobin, the oxygen transport protein in RBCs, is composed of 4 heme groups, and each RBC contains ∼300 million hemoglobin molecules, with >1 billion atoms of iron (104). Consequently, approximately two-thirds of body iron is found in the erythroid compartment (105). Normal turnover of RBCs requires ∼20–25 mg Fe/d (106), and much of this is requirement is fulfilled through recycling of senescent RBCs (2). However, after blood loss or RBC destruction (hemolysis), the requirement for iron increases to allow for the replacement of lost RBCs. To make iron available from stores and increased dietary absorption, hepcidin must be rapidly suppressed; it has long been hypothesized that this is accomplished by an “erythroid regulator” of iron absorption and release (105). Pathologic effects of the “erythroid regulator” are evident in β-thalassemia, where ineffective erythropoiesis results in high erythropoietin concentrations and an increased number of erythroid precursors that fail to mature into functional RBCs. Consequently, the augmented production of the “erythroid regulator” suppresses hepcidin, which often leads to pathologic iron overload due to hyperabsorption from the diet (107).
Erythropoietin (EPO), a cytokine produced by the kidney, is the primary driver of proliferation and differentiation of erythroid progenitor cells. It was previously thought that EPO could directly regulate hepcidin production; hepcidin and EPO concentrations are inversely correlated in β-thalassemia (108). However, ablation of the bone marrow in the presence of an erythropoietic stimulus (exogenous EPO or hemorrhage) prevented Hamp suppression (109, 110). This suggested that the hepcidin-suppressing “erythroid factor” is produced in the bone marrow. Several years ago, 2 members of the TGF-β superfamily were postulated to function as putative erythroid regulators of hepcidin: GDF15, produced by late and apoptotic erythroblasts (111), and twisted-gastrulation 1 (TWSG1), produced by early erythroblasts (112). However, unequivocal confirmation of their role is still lacking. The difficulty, at least in part, lies in important differences between murine and human erythropoiesis (113): in contrast to the very high expression of GDF15 in human erythroid cells (among the top 25 genes), Gdf15 is not expressed in murine erythroid cells. Studies in humans showed that GDF15 levels are massively elevated in patients with β-thalassemia and other diseases of dysfunctional erythropoiesis (111, 114) and that treatment of primary human hepatocytes with high concentrations of GDF15 or TWSG1 suppressed HAMP expression (111, 112). On the other hand, studies in animals showed that Gdf15−/− mice are still capable of suppressing hepcidin in response to phlebotomy (115), and similarly, neither Gdf15 nor Twsg1 expression increases in response to hemorrhage (116). Therefore, a definitive role of these proteins in hepcidin regulation by erythropoiesis in humans still remains to be determined.
More recently, erythroferrone (ERFE) was identified as a physiologic and pathologic “erythroid regulator” of hepcidin (116). ERFE is a member of the C1q/TNF-related protein (CTRP) family; it is encoded by the family with sequence similarity 132 member B (Fam132b) gene (116) and has alternatively been named CTRP15 and myonectin. Because both mouse and human erythroid precursors regulate ERFE expression in a similar manner (116), animal studies provided robust evidence for the role of ERFE as a key erythroid regulator of hepcidin. Baseline production of ERFE is very low, and the protein does not appear to have a role in steady state erythropoiesis because ERFE-deficient mice showed normal hematologic and iron variables in adulthood. However, after phlebotomy or EPO injection, Fam132b mRNA levels were greatly increased in erythroblasts and were accompanied by a large increase in serum Erfe concentrations, suggesting a role in stress erythropoiesis. EPO directly induces Fam132b expression in erythroblasts via the JAK2/STAT5 signaling pathway; thus, in stress erythropoiesis, Fam132b levels mirror EPO changes (116). The treatment of mice or isolated hepatocytes with recombinant ERFE suppressed Hamp mRNA, showing that ERFE acts directly on hepatocytes to regulate hepcidin expression. However, the receptor and associated signaling pathway are still unknown. Erfe-deficient mice are unable to suppress hepcidin after phlebotomy and have a delay in recovery from hemorrhage. Furthermore, Erfe-deficient mice showed a more severe anemia in a mouse model of inflammation, associated with inappropriately high hepcidin in the face of anemia (117).
Indicative of its possible role as a pathologic erythroid regulator, Erfe mRNA and serum concentrations are greatly elevated in a mouse model of β-thalassemia (116). Genetic deletion of Fam132b in a β-thalassemia mouse model rescued hepcidin expression and partially improved the iron overload phenotype (without any changes in anemia) (118). These data provide strong evidence that ERFE is indeed a long sought-after “erythroid regulator” of iron metabolism and suggests that Erfe may be responsible for hepcidin dysregulation in β-thalassemia. Importantly, human erythroid cells respond to EPO in the same manner as mouse erythroid cells (116), but measurements of circulating ERFE in human disease await the development of a validated ERFE assay. It remains to be determined whether pathologic or physiologic erythroid suppression of hepcidin is dependent on a single cytokine or if it is a concerted effort between multiple signaling pathways. Studies show that stimulation of the BMP pathway, either by the administration of parenteral iron (116) or by deletion of Tmprss6 (119), interferes with effective hepcidin suppression by hemorrhage or EPO. Although ERFE does not appear to be acting by directly altering the BMP pathway signaling (116), a parallel attenuation of the BMP-SMAD signaling is necessary for maximal hepcidin suppression in response to erythropoietic activity. The identification of the receptor will provide much-needed insight into the mechanism governing ERFE-mediated suppression of hepcidin.
Growth factors.
Growth factors, including hepatocyte growth factor (HGF) and epidermal growth factor (EGF), were shown to suppress Hamp expression in primary mouse hepatocytes or in mice (120). These growth factors in parallel suppressed the BMP-regulated gene Id1, suggesting that HGF and EGF regulate hepcidin, at least in part, by interfering with the BMP-SMAD pathway. How this is achieved is not entirely clear. The activation of mitogen-activated protein kinase kinase (MEK, MAP2K, or MAPKK) and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) signaling by the growth factors seems to be required, because inhibition of either kinase pathway interfered with the suppression of Hamp and Id1 by HGF. PI3K-mediated suppression of hepcidin correlated with AKT phosphorylation, suggesting a possible role of AKT in hepcidin regulation (120). Growth factor treatment also increased nuclear levels of transforming growth-interacting factor (TGIF), a SMAD transcriptional corepressor, suggesting a likely point of the crosstalk between the growth factor and the BMP pathways. Not all growth factors suppressed hepcidin: platelet-derived growth factor (PDGF) and insulin-like growth factor (IGF) I and IGF-II had no effect on hepcidin expression in primary hepatocytes (120).
It is not yet clear in which pathophysiologic processes hepcidin suppression by HGF or EGF plays a role. Chronic liver disease may be one such situation. Chronic liver disease is characterized by relative hepcidin deficiency (121), as well as by increased concentrations of HGF or EGF, both known mediators of the hepatic regenerative response. Growth factor–dependent hepcidin suppression may be the cause of excessive accumulation of iron that is frequently observed in this disease, thus worsening the liver injury. Periods of rapid growth may be another condition in which hepcidin regulation by growth factors could be important, because growth and development are known to have increased iron requirements. Low hepcidin production was indeed shown in young growing mice (122), but it remains to be determined to which extent this is dependent on the growth factor signaling and whether the same regulation occurs in humans.
A different growth factor, platelet-derived growth factor BB (PDGF-BB) (123), was implicated in hepcidin regulation by hypoxia. A negative correlation was observed between hepcidin suppression and PDGF-BB in humans after 6 h at simulated altitude (123), and experiments in mice and cells suggested that signaling downstream of the PDGF receptor contributed to hepcidin suppression in hypoxia. However, hypoxia is also a strong stimulus of EPO production and erythropoiesis (124, 125), which would be associated with increased production of the hepcidin suppressor ERFE. The relative contribution of different factors to the hypoxic suppression of hepcidin still remains to be established experimentally.
Sex hormones.
The sex hormone testosterone has also been reported to have suppressive effects on hepcidin, whereas the effects of estrogen are less clear. Daily administration of testosterone to female mice significantly reduced hepcidin concentrations and increased serum iron and transferrin saturation (126, 127). Similarly, in a heat-killed B. abortus model of chronic inflammation, the administration of testosterone reversed anemia, possibly due to Hamp suppression (50). The inhibitory effect on Hamp expression seems to be driven by androgen receptor sequestration of SMAD1 and SMAD4, preventing association with and downstream signaling of BMP receptors (126). The suppressive effect of testosterone may also be mediated by increased hepatic EGF signaling, because testosterone administration in female mice induced hepatic Egf receptor (Egfr) expression, and selective inhibition of EGFR signaling in male mice significantly increased Hamp expression (127). Testosterone administration has been shown to induce a transient but significant increase in Epo expression; however, EPO and erythropoiesis are not responsible for testosterone-mediated suppression of Hamp (126, 127). The testosterone-dependent suppression of hepcidin helps explain the clinical observations of erythrocytosis after androgen treatment (128). It is also speculated that this mechanism may be a modifier of disease severity in conditions in which hepcidin production is already inadequate (127).
The effects of estrogen on hepcidin expression have proven to be more challenging to understand. A functional estrogen-responsive element half-site has been detected in the Hamp promoter region (129, 130). Treatment with 17β-estradiol (E2) reduced hepcidin expression in vitro and in vivo and was blocked by treatment with an E2 antagonist (130) (129). In humans, increased endogenous E2 during the in vitro fertilization protocol was associated with significantly reduced circulating hepcidin concentrations (131). However, another study reported decreased Hamp in ovariectomized animals, and in cell lines E2 actually increased Hamp expression. Estrogen effect was dependent on the activation of G protein–coupled receptor 30 (GPR30) and subsequent BMP6 expression (132). Another study found no changes in HAMP expression in vitro after E2 dose response (133). These conflicting results highlight the need for additional studies to delineate the role of estrogen in hepcidin regulation. More recently, multiple steroid molecules, including progesterone and mifepristone, were reported to increase hepcidin production. Their effect was mediated via the progesterone receptor membrane component 1 (PGRMC1), exerting its effects through Src-family tyrosine kinases (SFKs) (134). Further studies are necessary to understand the role of this pathway in iron pathophysiology or in conditions in which the steroid drugs are used as treatment.
Conclusions
The identification of hepcidin as the master regulator of systemic iron homeostasis has greatly advanced our understanding of iron biology. It is now well appreciated that hepcidin has a nonredundant role in controlling the dietary absorption of iron, its storage, and its release into the circulation. Hepcidin concentrations are tightly controlled and their pathologic dysregulation underlies a number of human diseases. Regulation of hepcidin is multifaceted and complex, with numerous positive and negative regulators converging to fine-tune its expression. The well-defined regulatory pathways of hepcidin include the BMP-SMAD pathway (mediating the effect of iron and inflammation on hepcidin), the IL-6 pathway (mediating the effect of inflammation), and the EPO-ERFE axis (mediating hepcidin suppression by erythropoietic activity). Our understanding of hepcidin regulation is increasing, but numerous questions related to hepcidin pathobiology remain to be addressed, providing opportunities for important studies in the future.
Acknowledgments
Both authors read and approved the final manuscript.
Footnotes
Abbreviations used: ACTR2A, activin A receptor type 2A; ACVR1, activin A receptor type I; AKT, protein kinase B; ALK, activin receptor-like kinase; BMP, bone morphogenetic protein; BMPER, BMP binding endothelial regulator; BMPR, bone morphogenetic protein receptor; CTRP, C1q/TNF-related protein; E2, 17β-estradiol; EGF, epidermal growth factor; Egfr, epidermal growth factor receptor; EPO, erythropoietin; ERFE, erythroferrone; Fam132b, family with sequence similarity 132 member B; FPN, ferroportin; GDF, growth and differentiation factor; GP130, glycoprotein 130; GPR30, G protein–coupled receptor 30; HAMP, hepcidin antimicrobial peptide; HFE, human hemochromatosis protein; HGF, hepatocyte growth factor; HIF2α, hypoxia-inducible factor 2α; HJV, hemojuvelin; Id1, inhibitor of DNA binding 1; IDA, iron deficiency anemia; IGF, insulin-like growth factor; Il6, interleukin 6; IL-6R, IL-6 receptor; Inhbb, inhibin βB; IRE/IRP, iron-responsive element/iron-regulatory protein; iron-TF, transferrin-bound iron; JAK, Janus kinase; LIF, leukemia inhibitory factor; MHC, major histocompatibility complex; PAMP, pathogen-associated molecular pattern; PDGF, platelet-derived growth factor; PDGF-BB, platelet-derived growth factor BB; PGRMC1, progesterone receptor membrane component 1; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; RAS-MAPK, rat sarcoma viral oncogene homolog-mitogen-activated protein kinase; RGMc, repulsive guidance molecule C; SFK, Src-family tyrosine kinase; SLC40A1, solute carrier family 40 member 1; STAT, signal transducer and activator of transcription; TFR, transferrin receptor; TGIF, transforming growth-interacting factor; TMPRSS6, transmembrane protease serine 6; TWSG1, twisted-gastrulation 1.
References
- 1.Ganz T. Systemic iron homeostasis. Physiol Rev 2013;93:1721–41. [DOI] [PubMed] [Google Scholar]
- 2.Andrews NC. Disorders of iron metabolism. N Engl J Med 1999;341:1986–95. [DOI] [PubMed] [Google Scholar]
- 3.Ponka P, Lok CN. The transferrin receptor: role in health and disease. Int J Biochem Cell Biol 1999;31:1111–37. [DOI] [PubMed] [Google Scholar]
- 4.Ganz T. Macrophages and systemic iron homeostasis. J Innate Immun 2012;4:446–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kassebaum NJ, Jasrasaria R, Naghavi M, Wulf SK, Johns N, Lozano R, Regan M, Weatherall D, Chou DP, Eisele TP, et al. A systematic analysis of global anemia burden from 1990 to 2010. Blood 2014;123:615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camaschella C. Iron-deficiency anemia. N Engl J Med 2015;372:1832–43. [DOI] [PubMed] [Google Scholar]
- 7.Galaris D, Pantopoulos K. Oxidative stress and iron homeostasis: mechanistic and health aspects. Crit Rev Clin Lab Sci 2008;45:1–23. [DOI] [PubMed] [Google Scholar]
- 8.Puntarulo S. Iron, oxidative stress and human health. Mol Aspects Med 2005;26:299–312. [DOI] [PubMed] [Google Scholar]
- 9.Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol 2001;33:940–59. [DOI] [PubMed] [Google Scholar]
- 10.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 2010;49:1603–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heitzer T, Schlinzig T, Krohn K, Meinertz T, Munzel T. Endothelial dysfunction, oxidative stress, and risk of cardiovascular events in patients with coronary artery disease. Circulation 2001;104:2673–8. [DOI] [PubMed] [Google Scholar]
- 12.Maritim AC, Sanders RA, Watkins JB III. Diabetes, oxidative stress, and antioxidants: a review. J Biochem Mol Toxicol 2003;17:24–38. [DOI] [PubMed] [Google Scholar]
- 13.Coyle JT, Puttfarcken P. Oxidative stress, glutamate, and neurodegenerative disorders. Science 1993;262:689–95. [DOI] [PubMed] [Google Scholar]
- 14.Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol 2009;7:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donovan A, Roy CN, and Andrews NC. The ins and outs of iron homeostasis. Physiology 2006;21:115–23. [DOI] [PubMed] [Google Scholar]
- 16.Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 2000;403:776–81. [DOI] [PubMed] [Google Scholar]
- 17.Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem 2000;275:19906–12. [DOI] [PubMed] [Google Scholar]
- 18.McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell 2000;5:299–309. [DOI] [PubMed] [Google Scholar]
- 19.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004;306:2090–3. [DOI] [PubMed] [Google Scholar]
- 20.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 2001;276:7806–10. [DOI] [PubMed] [Google Scholar]
- 21.Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 2000;480:147–50. [DOI] [PubMed] [Google Scholar]
- 22.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 2001;276:7811–9. [DOI] [PubMed] [Google Scholar]
- 23.Drakesmith H, Nemeth E, Ganz T. Ironing out ferroportin. Cell Metab 2015;22:777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Ann Rev Nutr 2008;28:197–213. [DOI] [PubMed] [Google Scholar]
- 25.Sangokoya C, Doss JF, Chi JT. Iron-responsive miR-485-3p regulates cellular iron homeostasis by targeting ferroportin. PLoS Genet 2013;9:e1003408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao B, Sugianto P, Fung E, Del-Castillo-Rueda A, Moran-Jimenez MJ, Ganz T, Nemeth E. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metab 2012;15:918–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi R, Kato HE, Font J, Deshpande CN, Wada M, Ito K, Ishitani R, Jormakka M, Nureki O. Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin. Nat Commun 2015;6:8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fung E, Nemeth E. Manipulation of the hepcidin pathway for therapeutic purposes. Haematologica 2013;98:1667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao JJ, Krzyzanski W, Wang YM, Li H, Rose MJ, Ma M, Wu Y, Hinkle B, Perez-Ruixo JJ. Pharmacokinetics of anti-hepcidin monoclonal antibody Ab 12B9m and hepcidin in cynomolgus monkeys. AAPS J 2010;12:646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganz T, Nemeth E. Hepcidin and disorders of iron metabolism. Ann Rev Med 2011;62:347–60. [DOI] [PubMed] [Google Scholar]
- 31.Lesbordes-Brion JC, Viatte L, Bennoun M, Lou DQ, Ramey G, Houbron C, Hamard G, Kahn A, Vaulont S. Targeted disruption of the hepcidin 1 gene results in severe hemochromatosis. Blood 2006;108:1402–5. [DOI] [PubMed] [Google Scholar]
- 32.Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet 2003;33:21–2. [DOI] [PubMed] [Google Scholar]
- 33.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, Ernst M, Klein C, Trautwein C. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology 2007;132:294–300. [DOI] [PubMed] [Google Scholar]
- 34.Ramos E, Kautz L, Rodriguez R, Hansen M, Gabayan V, Ginzburg Y, Roth MP, Nemeth E, Ganz T. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology 2011;53:1333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R Jr, Ellis MC, Fullan A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 1996;13:399–408. [DOI] [PubMed] [Google Scholar]
- 36.Feder JN. The hereditary hemochromatosis gene (HFE): a MHC class I-like gene that functions in the regulation of iron homeostasis. Immunol Res 1999;20:175–85. [DOI] [PubMed] [Google Scholar]
- 37.Wu XG, Wang Y, Wu Q, Cheng WH, Liu W, Zhao Y, Mayeur C, Schmidt PJ, Yu PB, Wang F, et al. HFE interacts with the BMP type I receptor ALK3 to regulate hepcidin expression. Blood 2014;124:1335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, Chen J, Kramer M, Tsukamoto H, Zhang AS, Enns CA. Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell Metab 2009;9:217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corradini E, Rozier M, Meynard D, Odhiambo A, Lin HY, Feng Q, Migas MC, Britton RS, Babitt JL, Fleming RE. Iron regulation of hepcidin despite attenuated Smad1,5,8 signaling in mice without transferrin receptor 2 or Hfe. Gastroenterology 2011;141:1907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enns CA, Ahmed R, Wang J, Ueno A, Worthen C, Tsukamoto H, Zhang AS. Increased iron loading induces Bmp6 expression in the non-parenchymal cells of the liver independent of the BMP-signaling pathway. PLoS One 2013;8:e60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rausa M, Pagani A, Nai A, Campanella A, Gilberti ME, Apostoli P, Camaschella C, Silvestri L. Bmp6 expression in murine liver non parenchymal cells: a mechanism to control their high iron exporter activity and protect hepatocytes from iron overload? PLoS One 2015;10:e0122696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003;113:685–700. [DOI] [PubMed] [Google Scholar]
- 43.Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol 2013;2:47–63. [DOI] [PubMed] [Google Scholar]
- 44.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev 2005;19:2783–810. [DOI] [PubMed] [Google Scholar]
- 45.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab 2005;2:399–409. [DOI] [PubMed] [Google Scholar]
- 46.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet 2006;38:531–9. [DOI] [PubMed] [Google Scholar]
- 47.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet 2004;36:77–82. [DOI] [PubMed] [Google Scholar]
- 48.Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood 2008;111:5195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Q, Sun CC, Lin HY, Babitt JL. Repulsive guidance molecule (RGM) family proteins exhibit differential binding kinetics for bone morphogenetic proteins (BMPs). PLoS One 2012;7:e46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo W, Schmidt PJ, Fleming MD, Bhasin S. Effects of testosterone on erythropoiesis in a female mouse model of anemia of inflammation. Endocrinology 2016;157:2937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest 2007;117:1933–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA 2006;103:10289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maes K, Nemeth E, Roodman GD, Huston A, Esteve F, Freytes C, Callander N, Katodritou E, Tussing-Humphreys L, Rivera S, et al. In anemia of multiple myeloma, hepcidin is induced by increased bone morphogenetic protein 2. Blood 2010;116:3635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet 2009;41:478–81. [DOI] [PubMed] [Google Scholar]
- 55.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, Deng C, Vaulont S, Mosser J, Coppin H, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood 2008;112:1503–9. [DOI] [PubMed] [Google Scholar]
- 56.Andriopoulos B Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet 2009;41:482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daher R, Kannengiesser C, Houamel D, Lefebvre T, Bardou-Jacquet E, Ducrot N, de Kerguenec C, Jouanolle AM, Robreau AM, Oudin C, et al. Heterozygous mutations in BMP6 pro-peptide lead to inappropriate hepcidin synthesis and moderate iron overload in humans. Gastroenterology 2016;150(3):672–83, e4. [DOI] [PubMed] [Google Scholar]
- 58.Steinbicker AU, Bartnikas TB, Lohmeyer LK, Leyton P, Mayeur C, Kao SM, Pappas AE, Peterson RT, Bloch DB, Yu PB, et al. Perturbation of hepcidin expression by BMP type I receptor deletion induces iron overload in mice. Blood 2011;118:4224–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayeur C, Leyton PA, Kolodziej SA, Yu B, Bloch KD. BMP type II receptors have redundant roles in the regulation of hepatic hepcidin gene expression and iron metabolism. Blood 2014;124:2116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet 2008;40:569–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao N, Nizzi CP, Anderson SA, Wang J, Ueno A, Tsukamoto H, Eisenstein RS, Enns CA, Zhang AS. Low intracellular iron increases the stability of matriptase-2. J Biol Chem 2015;290:4432–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frýdlová J, Prikryl P, Truksa J, Falke LL, Du X, Gurieva I, Vokurka M, Krijt J. Effect of erythropoietin, iron deficiency and iron overload on liver matriptase-2 (TMPRSS6) protein content in mice and rats. PLoS One 2016;11:e0148540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang AS, Anderson SA, Wang J, Yang F, DeMaster K, Ahmed R, Nizzi CP, Eisenstein RS, Tsukamoto H, Enns CA. Suppression of hepatic hepcidin expression in response to acute iron deprivation is associated with an increase of matriptase-2 protein. Blood 2011;117:1687–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab 2008;8:502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science 2008;320:1088–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patel N, Masaratana P, Diaz-Castro J, Latunde-Dada GO, Qureshi A, Lockyer P, Jacob M, Arno M, Matak P, Mitry RR, et al. BMPER protein is a negative regulator of hepcidin and is up-regulated in hypotransferrinemic mice. J Biol Chem 2012;287:4099–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Enns CA, Ahmed R, Zhang AS. Neogenin interacts with matriptase-2 to facilitate hemojuvelin cleavage. J Biol Chem 2012;287:35104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao N, Maxson JE, Zhang RH, Wahedi M, Enns CA, Zhang AS. Neogenin facilitates the induction of hepcidin expression by hemojuvelin in the liver. J Biol Chem 2016;291:12322–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goh JB, Wallace DF, Hong W, Subramaniam VN. Endofin, a novel BMP-SMAD regulator of the iron-regulatory hormone, hepcidin. Sci Rep 2015;5:13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryan JD, Ryan E, Fabre A, Lawless MW, Crowe J. Defective bone morphogenic protein signaling underlies hepcidin deficiency in HFE hereditary hemochromatosis. Hepatology 2010;52:1266–73. [DOI] [PubMed] [Google Scholar]
- 71.Vujić Spasić M, Sparla R, Mleczko-Sanecka K, Migas MC, Breitkopf-Heinlein K, Dooley S, Vaulont S, Fleming RE, Muckenthaler MU. Smad6 and Smad7 are co-regulated with hepcidin in mouse models of iron overload. Biochim Biophys Acta 2013;1832:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Locke A, Main ER, Rosbash DO. The copper and non-hemoglobinous iron contents of the blood serum in disease. J Clin Invest 1932;11:527–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest 2004;113:1271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weiss G, Goodnough LT. Anemia of chronic disease. N Engl J Med 2005;352:1011–23. [DOI] [PubMed] [Google Scholar]
- 75.Arezes J, Jung G, Gabayan V, Valore E, Ruchala P, Gulig PA, Ganz T, Nemeth E, Bulut Y. Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe 2015;17:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michels K, Nemeth E, Ganz T, Mehrad B. Hepcidin and host defense against infectious diseases. PLoS Pathog 2015;11:e1004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ganz T. Hepcidin and iron regulation, 10 years later. Blood 2011;117:4425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest 2002;110:1037–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akchurin O, Sureshbabu A, Doty SB, Zhu YS, Patino E, Cunningham-Rundles S, Choi ME, Boskey AL, Rivella S. Lack of hepcidin ameliorates anemia and improves growth in an adenine-induced mouse model of chronic kidney disease. Am J Physiol Renal Physiol 2016 Jul 20 (Epub ahead of print; ajprenal.00089.2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guida C, Altamura S, Klein FA, Galy B, Boutros M, Ulmer AJ, Hentze MW, Muckenthaler MU. A novel inflammatory pathway mediating rapid hepcidin-independent hypoferremia. Blood 2015;125:2265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Philip M, Chiu EY, Hajjar AM, Abkowitz JL. TLR stimulation dynamically regulates heme and iron export gene expression in macrophages. J Immunol Res 2016:4039038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood 2003;101:2461–3. [DOI] [PubMed] [Google Scholar]
- 83.Rodriguez R, Jung CL, Gabayan V, Deng JC, Ganz T, Nemeth E, Bulut Y. Hepcidin induction by pathogens and pathogen-derived molecules is strongly dependent on interleukin-6. Infect Immun 2014;82:745–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene 2007;26:6724–37. [DOI] [PubMed] [Google Scholar]
- 85.Haan C, Kreis S, Margue C, Behrmann I. Jaks and cytokine receptors–an intimate relationship. Biochem Pharmacol 2006;72:1538–46. [DOI] [PubMed] [Google Scholar]
- 86.Pellegrini S, Dusanter-Fourt I. The structure, regulation and function of the Janus kinases (JAKs) and the signal transducers and activators of transcription (STATs). Eur J Biochem 1997;248(3):615–33. [DOI] [PubMed] [Google Scholar]
- 87.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood 2007;109:353–8. [DOI] [PubMed] [Google Scholar]
- 88.Sakamori R, Takehara T, Tatsumi T, Shigekawa M, Hikita H, Hiramatsu N, Kanto T, Hayashi N. STAT3 signaling within hepatocytes is required for anemia of inflammation in vivo. J Gastroenterol 2010;45:244–8. [DOI] [PubMed] [Google Scholar]
- 89.Theurl I, Schroll A, Sonnweber T, Nairz M, Theurl M, Willenbacher W, Eller K, Wolf D, Seifert M, Sun CC, et al. Pharmacologic inhibition of hepcidin expression reverses anemia of chronic inflammation in rats. Blood 2011;118:4977–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mayeur C, Lohmeyer LK, Leyton P, Kao SM, Pappas AE, Kolodziej SA, Spagnolli E, Yu B, Galdos RL, Yu PB, et al. The type I BMP receptor Alk3 is required for the induction of hepatic hepcidin gene expression by interleukin-6. Blood 2014;123:2261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Besson-Fournier C, Latour C, Kautz L, Bertrand J, Ganz T, Roth MP, Coppin H. Induction of activin B by inflammatory stimuli up-regulates expression of the iron-regulatory peptide hepcidin through Smad1/5/8 signaling. Blood 2012;120:431–9. [DOI] [PubMed] [Google Scholar]
- 92.Bernard DJ, Lee KB, and Santos MM. Activin B can signal through both ALK4 and ALK7 in gonadotrope cells. Reprod Biol Endocrinol 2006;4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bertolino P, Holmberg R, Reissmann E, Andersson O, Berggren PO, Ibanez CF. Activin B receptor ALK7 is a negative regulator of pancreatic beta-cell function. Proc Natl Acad Sci USA 2008;105:7246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Canali S, Core AB, Zumbrennen-Bullough KB, Merkulova M, Wang CY, Schneyer AL, Pietrangelo A, Babitt JL. Activin B induces noncanonical SMAD1/5/8 signaling via BMP type I receptors in hepatocytes: evidence for a role in hepcidin induction by inflammation in male mice. Endocrinology 2016;157:1146–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Besson-Fournier C. European iron club. Innsbruck (Austria); 2016. [Google Scholar]
- 96.Inamura J, Ikuta K, Jimbo J, Shindo M, Sato K, Torimoto Y, Kohgo Y. Upregulation of hepcidin by interleukin-1beta in human hepatoma cell lines. Hepatol Res 2005;33(3):198–205. [DOI] [PubMed] [Google Scholar]
- 97.Lee P, Peng H, Gelbart T, Wang L, Beutler E. Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc Natl Acad Sci USA 2005;102:1906–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Armitage AE, Eddowes LA, Gileadi U, Cole S, Spottiswoode N, Selvakumar TA, Ho LP, Townsend AR, Drakesmith H. Hepcidin regulation by innate immune and infectious stimuli. Blood 2011;118:4129–39. [DOI] [PubMed] [Google Scholar]
- 99.Wallace DF, Subramaniam VN. Analysis of IL-22 contribution to hepcidin induction and hypoferremia during the response to LPS in vivo. Int Immunol 2015;27:281–7. [DOI] [PubMed] [Google Scholar]
- 100.Smith CL, Arvedson TL, Cooke KS, Dickmann LJ, Forte C, Li H, Merriam KL, Perry VK, Tran L, Rottman JB, et al. IL-22 regulates iron availability in vivo through the induction of hepcidin. J Immunol 2013;191:1845–55. [DOI] [PubMed] [Google Scholar]
- 101.Chung B, Verdier F, Matak P, Deschemin JC, Mayeux P, Vaulont S. Oncostatin M is a potent inducer of hepcidin, the iron regulatory hormone. FASEB J 2010;24:2093–103. [DOI] [PubMed] [Google Scholar]
- 102.Kanda J, Uchiyama T, Tomosugi N, Higuchi M, Uchiyama T, Kawabata H. Oncostatin M and leukemia inhibitory factor increase hepcidin expression in hepatoma cell lines. Int J Hematol 2009;90:545–52. [DOI] [PubMed] [Google Scholar]
- 103.Chung B, Matak P, McKie AT, Sharp P. Leptin increases the expression of the iron regulatory hormone hepcidin in HuH7 human hepatoma cells. J Nutr 2007;137:2366–70. [DOI] [PubMed] [Google Scholar]
- 104.Smith DW. The molecular biology of mammalian hemoglobin synthesis. Ann Clin Lab Sci 1980;10:116–22. [PubMed] [Google Scholar]
- 105.Finch C. Regulators of iron balance in humans. Blood 1994;84:1697–702. [PubMed] [Google Scholar]
- 106.Ganz T, Nemeth E. Iron metabolism: interactions with normal and disordered erythropoiesis. Cold Spring Harb Perspect Med 2012;2:a011668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pasricha SR, McHugh K, Drakesmith H. Regulation of hepcidin by erythropoiesis: the story so far. Ann Rev Nutr 2016;36:417–34. [DOI] [PubMed] [Google Scholar]
- 108.Origa R, Galanello R, Ganz T, Giagu N, Maccioni L, Faa G, Nemeth E. Liver iron concentrations and urinary hepcidin in beta-thalassemia. Haematologica 2007;92:583–8. [DOI] [PubMed] [Google Scholar]
- 109.Pak M, Lopez MA, Gabayan V, Ganz T, Rivera S. Suppression of hepcidin during anemia requires erythropoietic activity. Blood 2006;108:3730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sasaki Y, Noguchi-Sasaki M, Yasuno H, Yorozu K, Shimonaka Y. Erythropoietin stimulation decreases hepcidin expression through hematopoietic activity on bone marrow cells in mice. Int J Hematol 2012;96:692–700. [DOI] [PubMed] [Google Scholar]
- 111.Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, Moroney JW, Reed CH, Luban NL, Wang RH, et al. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med 2007;13:1096–101. [DOI] [PubMed] [Google Scholar]
- 112.Tanno T, Porayette P, Sripichai O, Noh SJ, Byrnes C, Bhupatiraju A, Lee YT, Goodnough JB, Harandi O, Ganz T, et al. Identification of TWSG1 as a second novel erythroid regulator of hepcidin expression in murine and human cells. Blood 2009;114:181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, Gallagher PG. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014;123:3466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Casanovas G, Swinkels DW, Altamura S, Schwarz K, Laarakkers CM, Gross HJ, Wiesneth M, Heimpel H, Muckenthaler MU. Growth differentiation factor 15 in patients with congenital dyserythropoietic anaemia (CDA) type II. J Mol Med 2011;89:811–6. [DOI] [PubMed] [Google Scholar]
- 115.Casanovas G, Vujic Spasic M, Casu C, Rivella S, Strelau J, Unsicker K, Muckenthaler MU. The murine growth differentiation factor 15 is not essential for systemic iron homeostasis in phlebotomized mice. Haematologica 2013;98:444–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kautz L, Jung G, Valore EV, Rivella S, Nemeth E, Ganz T. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet 2014;46:678–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kautz L, Jung G, Nemeth E, Ganz T. Erythroferrone contributes to recovery from anemia of inflammation. Blood 2014;124:2569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kautz L, Jung G, Du X, Gabayan V, Chapman J, Nasoff M, Nemeth E, Ganz T. Erythroferrone contributes to hepcidin suppression and iron overload in a mouse model of beta-thalassemia. Blood 2015;126:2031–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nai A, Rubio A, Campanella A, Gourbeyre O, Artuso I, Bordini J, Gineste A, Latour C, Besson-Fournier C, Lin HY, et al. Limiting hepatic Bmp-Smad signaling by matriptase-2 is required for erythropoietin-mediated hepcidin suppression in mice. Blood 2016;127:2327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goodnough JB, Ramos E, Nemeth E, Ganz T. Inhibition of hepcidin transcription by growth factors. Hepatology 2012;56:291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Girelli D, Pasino M, Goodnough JB, Nemeth E, Guido M, Castagna A, Busti F, Campostrini N, Martinelli N, Vantini I, et al. Reduced serum hepcidin levels in patients with chronic hepatitis C. J Hepatol 2009;51:845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nicolas G, Bennoun M, Porteu A, Mativet S, Beaumont C, Grandchamp B, Sirito M, Sawadogo M, Kahn A, Vaulont S. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci USA 2002;99:4596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sonnweber T, Nachbaur D, Schroll A, Nairz M, Seifert M, Demetz E, Haschka D, Mitterstiller AM, Kleinsasser A, Burtscher M, et al. Hypoxia induced downregulation of hepcidin is mediated by platelet derived growth factor BB. Gut 2014;63:1951–9. [DOI] [PubMed] [Google Scholar]
- 124.Liu Q, Davidoff O, Niss K, Haase VH. Hypoxia-inducible factor regulates hepcidin via erythropoietin-induced erythropoiesis. J Clin Invest 2012;122:4635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mastrogiannaki M, Matak P, Mathieu JR, Delga S, Mayeux P, Vaulont S, Peyssonnaux C. Hepatic hypoxia-inducible factor-2 down-regulates hepcidin expression in mice through an erythropoietin-mediated increase in erythropoiesis. Haematologica 2012;97:827–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guo W, Bachman E, Li M, Roy CN, Blusztajn J, Wong S, Chan SY, Serra C, Jasuja R, Travison TG, et al. Testosterone administration inhibits hepcidin transcription and is associated with increased iron incorporation into red blood cells. Aging Cell 2013;12:280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Latour C, Kautz L, Besson-Fournier C, Island ML, Canonne-Hergaux F, Loreal O, Ganz T, Coppin H, Roth MP. Testosterone perturbs systemic iron balance through activation of epidermal growth factor receptor signaling in the liver and repression of hepcidin. Hepatology 2014;59:683–94. [DOI] [PubMed] [Google Scholar]
- 128.Bachman E, Travison TG, Basaria S, Davda MN, Guo W, Li M, Connor Westfall J, Bae H, Gordeuk V, Bhasin S. Testosterone induces erythrocytosis via increased erythropoietin and suppressed hepcidin: evidence for a new erythropoietin/hemoglobin set point. J Gerontol A Biol Sci Med Sci 2014;69:725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang Q, Jian J, Katz S, Abramson SB, Huang X. 17beta-Estradiol inhibits iron hormone hepcidin through an estrogen responsive element half-site. Endocrinology 2012;153:3170–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hou Y, Zhang S, Wang L, Li J, Qu G, He J, Rong H, Ji H, Liu S. Estrogen regulates iron homeostasis through governing hepatic hepcidin expression via an estrogen response element. Gene 2012;511:398–403. [DOI] [PubMed] [Google Scholar]
- 131.Lehtihet M, Bonde Y, Beckman L, Berinder K, Hoybye C, Rudling M, Sloan JH, Konrad RJ, Angelin B. Circulating hepcidin-25 is reduced by endogenous estrogen in humans. PLoS One 2016;11:e0148802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ikeda Y, Tajima S, Izawa-Ishizawa Y, Kihira Y, Ishizawa K, Tomita S, Tsuchiya K, Tamaki T. Estrogen regulates hepcidin expression via GPR30–BMP6-dependent signaling in hepatocytes. PLoS One 2012;7:e40465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhen AW, Nguyen NH, Gibert Y, Motola S, Buckett P, Wessling-Resnick M, Fraenkel E, Fraenkel PG. The small molecule, genistein, increases hepcidin expression in human hepatocytes. Hepatology 2013;58:1315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li X, Rhee DK, Malhotra R, Mayeur C, Hurst LA, Ager E, Shelton G, Kramer Y, McCulloh D, Keefe D, et al. Progesterone receptor membrane component-1 regulates hepcidin biosynthesis. J Clin Invest 2016;126:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]