Abstract
The chromodomain is a conserved motif that functions in the epigenetic control of gene expression. Here, we report the functional characterization of a chromodomain protein, Chp1, in the heterochromatin assembly in fission yeast. We show that Chp1 is a structural component of three heterochromatic regions—centromeres, the mating-type region, and telomeres—and that its localization in these regions is dependent on the histone methyltransferase Clr4. Although deletion of the chp1+ gene causes centromere-specific decreases in Swi6 localization and histone H3-K9 methylation, we show that the role of Chp1 is not exclusive to the centromeres. We found that some methylation persists in native centromeric regions in the absence of Chp1, which is also true for the mating-type region and telomeres, and determined that Swi6 and Chp2 are critical to maintaining this residual methylation. We also show that Chp1 participates in the establishment of repressive chromatin in all three chromosomal regions. These results suggest that different heterochromatic regions share common structural properties, and that centromeric heterochromatin requires Chp1-mediated establishment steps differently than do other heterochromatic regions.
Keywords: chromodomain, fission yeast, heterochromatin, RNAi, silencing
Introduction
In eukaryotic cells, the organization of the chromatin into higher-order structures plays an important role in diverse chromosomal processes. Heterochromatin, one of the most distinct chromatin structures originally defined by cytological studies, is involved in various nuclear functions, including nuclear organization, chromosome segregation, dosage compensation, and gene regulation (Grewal and Elgin, 2002; Richards and Elgin, 2002). Heterochromatin is generally transcriptionally inert and its packaging state is epigenetically inherited during mitosis and meiosis, independent of the primary DNA sequence (Grewal and Klar, 1996; Nakayama et al, 2000). Euchromatic genes placed next to heterochromatin are subjected to a heritable inactivation leading to variegated patterns of gene expression, which is recognized as the spreading of the condensed higher-order structure (Grewal and Elgin, 2002).
In the fission yeast Schizosaccharomyces pombe, higher-order chromatin structure is also critical for the functional organization of heterochromatic domains, such as centromeres, the mating-type region, and telomeres (Allshire, 1996; Grewal, 2000). At centromeres, large tandem and inverted repeats (dg and dh) surrounding a unique central core are assembled into the heterochromatin structure (Takahashi et al, 1992). Similarly, at the mating-type region, mat2 and mat3 silent donor loci and an interval called the K region are packaged into a heterochromatin-like structure. Marker genes inserted within these regions are transcriptionally silenced (Allshire et al, 1994; Grewal and Klar, 1996). Several trans-acting factors are required for the complete repression of these heterochromatic regions (Allshire, 1996; Grewal, 2000). Among these factors, Swi6 and Clr4 play a crucial role in the formation of heterochromatin (Lorentz et al, 1994; Ivanova et al, 1998). Swi6, a homolog of heterochromatin protein 1 (HP1), contains two evolutionally conserved motifs, the chromo- and chromoshadow domains (Paro and Hogness, 1991), and functions as a dosage-critical component of heterochromatin (Nakayama et al, 2000). Clr4 is a fission yeast homolog of Su(var)3–9, a protein containing both chromo- and SET domains (Ivanova et al, 1998). Recently, it has been shown that Clr4 and mammalian Suv39h1 are histone H3–lysine 9 (H3-K9)-specific methyltransferases, and that Swi6 and HP1 specifically bind the methylated histone H3 through its amino-terminal chromodomain (Rea et al, 2000; Bannister et al, 2001; Lachner et al, 2001; Nakayama et al, 2001b). These findings reveal a mechanism in which the methylation of histone H3 and subsequent binding of Swi6/HP1 form a specialized higher-order chromatin structure (Nakayama et al, 2001b; Grewal and Elgin, 2002).
Several lines of evidence support a role for RNA in the formation of heterochromatin (Maison et al, 2002; Grewal and Moazed, 2003). The RNA interference (RNAi) pathway is generally thought to be responsible for post-transcriptional gene silencing (PTGS); in this process, a small (21–23 nucleotides) interfering RNA molecule (siRNA) triggers the degradation of the homologous mRNA (Hannon, 2002). Factors involved in RNAi include an RNase III-like enzyme (Dicer; Dcr1), an RNA-dependent RNA polymerase (RdRP: Rdp1), and an Argonaute (Ago1) protein. In S. pombe, deletion of any of the genes encoding the RNAi components (Ago1, Rdp1, Dcr1) causes centromere-specific defects in heterochromatin assembly, accompanied by decreases in H3-K9 methylation and Swi6 localization, and also results in the aberrant accumulation of unprocessed transcripts from centromeres (Volpe et al, 2002). Specifically, the RNAi components are involved in the establishment of heterochromatin assembly at the mating-type region but are dispensable for its maintenance (Hall et al, 2002). It has also been shown that expression of a short hairpin RNA induces the formation of repressive chromatin in a euchromatic region (Schramke and Allshire, 2003).
In S. pombe, at least nine chromodomain-containing proteins including Swi6 and Clr4 are present in the genome (Wood et al, 2002). Among these proteins, Chp1 and Chp2 are involved in transcriptional silencing at the heterochromatic regions (Figure 1A). Previous studies showed that the deletion of chp1+ causes defects in chromosome segregation and centromeric silencing, but not in the mating-type region or telomeric silencing (Doe et al, 1998; Thon and Verhein-Hansen, 2000). In addition, Chp1 associates with centromeric heterochromatin and is required for H3-K9 methylation at both endogenous centromeres and an ectopic site with cis-acting centromeric DNA (Partridge et al, 2000, 2002). Consistent with the centromere-specific phenotype, a recent report showed that Chp1 associates with Ago1, Tas3, and siRNAs to form a RITS (RNA-induced initiation of transcriptional silencing) complex (Verdel et al, 2004). Another protein, Chp2, shares extensive sequence similarity with Swi6 and contains both a chromo- and a chromoshadow domain (Figure 1A). In contrast to the obvious phenotypes of the Δswi6 mutants, deletion of the chp2+ gene causes weak silencing defects at centromeres, the mating-type region, and telomeres (Halverson et al, 2000; Thon and Verhein-Hansen, 2000). These previous reports argue that each chromodomain protein has a specialized role for different heterochromatic regions, which may be determined by differential associations. However, in terms of the domain structure characterized by H3-K9 methylation and Swi6 localization, the three heterochromatic regions, the centromeres, mating-type region, and telomeres, are fairly similar. How these chromodomain proteins cooperatively or differentially function in the formation of higher-order chromatin structure remains elusive.
Figure 1.
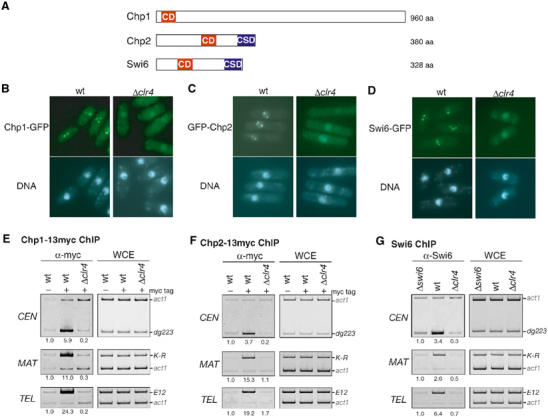
Chromodomain proteins Chp1 and Chp2 associate with centromeres, the mating-type region, and telomeres in a clr4+-dependent manner. (A) Schematic diagram showing chromodomain proteins Chp1, Chp2, and Swi6. The chromodomain (CD) and its relative, the chromoshadow domain (CSD), are shown. (B–D) GFP-tagged Chp1 (B), Chp2 (C), and Swi6 (D) formed several nuclear spots in a clr4+-dependent manner. Cells were stained with Hoechst33342. (E–G) Chp1, Chp2, and Swi6 associated with centromeres (CEN), the mating-type region (MAT), and telomeres (TEL) in a clr4+-dependent manner. (E, F) DNA isolated from an anti-myc-immunoprecipitated chromatin fraction (α-myc) or whole-cell extract (WCE) was used as a template for PCR-amplifying CEN-dg223, MAT-K-R, or TEL-E12. The samples were prepared from either wild-type or Δclr4 cells expressing Chp1-13myc or Chp2-13myc (myc tag+). As a control, wild-type cells in which Chp1 or Chp2 was not tagged (myc tag−) were used. The ratio of act1 to CEN-dg223, MAT-K-R, or TEL-E12 signals in the ChIP results was calculated, and the relative fold enrichments are shown beneath each lane. (G) Association of Swi6 with the indicated heterochromatic regions was assayed by ChIP as in (E, F), using anti-Swi6 antibodies. As a control, swi6+-deleted cells (Δswi6) were used.
In this study, we started to analyze the localization of the Chp1 and Chp2 proteins. We demonstrated that Chp1 and Chp2 are structural components of the three heterochromatic regions. Although deletion of the chp1+ gene caused centromere-specific silencing defects, Chp1 was essential for the establishment of repressive chromatin structure in all three heterochromatic regions. The establishment function has previously been described for the components of the RNAi machinery. These results suggest that different heterochromatic regions share common structural features and imply that centromeric heterochromatin has a dynamic structure and requires frequent establishment steps.
Results
Chp1 and Chp2 associate with three heterochromatic regions
Genetics experiments revealed that Chp1 and Chp2 are involved in heterochromatic silencing (Thon and Verhein-Hansen, 2000). Although both Chp1 and Chp2 associate with centromeres (Halverson et al, 2000; Partridge et al, 2000), their precise cellular or chromosomal distribution or intermolecular dependency is not well understood. To investigate the function of these chromodomain proteins, we first analyzed their cellular distribution. In fission yeast, three heterochromatic regions, centromeres, the mating-type region, and telomeres, are visualized by immunostaining of Swi6 or GFP-fused Swi6 (Ekwall et al, 1995; Sadaie et al, 2003). Two to five discrete Swi6 foci in interphase cells represent one large cluster of all centromeres, several clusters of telomeres, and the mating-type region (Ekwall et al, 1995). GFP was fused to the C-terminus of Chp1 or Chp2 by introducing a gfp+-kanMX6 cassette, and the fusion proteins were expressed from their native promoter. Chp1-GFP formed two to three discrete spots in interphase nuclei (Figure 1B). This is consistent with a previous report in which GFP-Chp1 was expressed from the nmt1 promoter (Doe et al, 1998). In the cells expressing Chp2-GFP protein, we were unable to detect any signals, presumably because of its low abundance (data not shown). However, when GFP-Chp2 was expressed from the nmt1 promoter on an episomal pREP41 plasmid, it appeared to form several spots in the nuclei (Figure 1C). These localization patterns were indistinguishable from the signals for Swi6 (Figure 1D and Supplementary Figure 1), suggesting that both Chp1 and Chp2 localize to the three heterochromatic regions in fission yeast. To test whether these localizations were dependent on histone H3-K9 methylation, clr4-deleted strains (Δclr4) were used for the analyses. In the Δclr4 strain, Chp1-GFP and GFP-Chp2 did not form nuclear spots, as was previously observed for Swi6-GFP (Figures 1B–D, Δclr4; Ekwall et al, 1996). These results suggested that the Chp1 and Chp2 proteins target heterochromatic regions through interactions with each chromodomain and methylated histone H3-K9 (Supplementary Figure 2).
To further examine whether Chp1 and Chp2 associate with the centromeres (CEN), mating-type region (MAT), and telomeres (TEL), a chromatin immunoprecipitation (ChIP) assay was performed. For this purpose, we constructed strains expressing 13myc-tagged Chp1 or Chp2 from its native promoter. These strains did not show any of the obvious phenotypes associated with the Δchp1 or Δchp2 mutation, suggesting that the myc-tagged proteins functionally replaced the wild-type proteins (data not shown). In the ChIP assay, immunoprecipitated DNA was subjected to polymerase chain reactions (PCRs) in which each primer set amplified the centromeric, mating-type region, or telomeric DNA (dg223, K-R, or E12 in Figures 1E–G). In addition, a primer set to amplify the act1+ gene was included as an internal control to verify the enrichment. Since each cell contains multiple copies of dg223 (CEN) or E12 (TEL), and a single copy of K-R (MAT), we optimized the concentration of each primer set to obtain approximately the same amount of product as that of the act1 gene in the control PCR experiments (Figures 1E–G, WCE). Consistent with previous reports (Halverson et al, 2000; Partridge et al, 2000), an enrichment of CEN was observed in the anti-myc immunoprecipitates for both the Chp1- and Chp2-13myc-tagged strains, compared with the total input chromatin (WCE) or precipitates from untagged strains (Figures 1E and F). Interestingly, the Chp1- and Chp2-immunoprecipitated complexes were also enriched for MAT and TEL (Figures 1E and F). The same results were obtained for Swi6 ChIP using anti-Swi6 antibodies (Figure 1G; Nakayama et al, 2000, 2001a; Partridge et al, 2000). In agreement with the cytological analyses, the high levels of CEN, MAT, and TEL in the ChIP assays were abolished in Δclr4 (Figures 1E–G; Partridge et al, 2000). Taken together, these results indicate that both Chp1 and Chp2 associate with the three heterochromatic domains through clr4+-mediated H3-K9 methylation.
Chromodomain proteins make distinct contributions to the formation of heterochromatin
We next examined whether the heterochromatin association of Chp1, Chp2, or Swi6 was affected in each deletion mutant. It has been shown that Chp1 localization to centromeres is not affected in the swi6 mutant, but Swi6 localization to centromeres depends on Chp1 (Partridge et al, 2000). The association of Chp1-13myc, Chp2-13myc, or Swi6 with heterochromatin was analyzed in the Δchp1, Δchp2, or Δswi6 background by ChIP assay (Figure 2). We found that the association of Chp1-13myc with the three heterochromatic regions (CEN, MAT, and TEL) was not affected in the absence of swi6+ or chp2+ (Figure 2A). In contrast, the centromeric localization of Swi6 or Chp2-13myc was specifically decreased in the Δchp1 cells (Figures 2B and C, Δchp1). When compared with the Δswi6 strain, the centromeric association of Swi6 was not completely abolished in the Δchp1 strain. Some Swi6 appeared to persist at the centromeric sequence. Interestingly, Swi6 was required for the localization of Chp2 to the mating-type region or telomeres but not to the centromeres (Figure 2B, Δswi6). Swi6 may act upstream of Chp2 at the mating-type region and telomeres. From these results (summarized in Figure 2D), it appears that Chp1, Chp2, and Swi6 make distinct contributions to heterochromatin formation. The changes in their heterochromatin localization were not caused by a reduction in the amount of each protein in the mutant cells, since the expression of Chp1, Chp2, or Swi6 protein was not affected in the Δchp1, Δchp2, Δswi6, or Δclr4 cells (data not shown).
Figure 2.
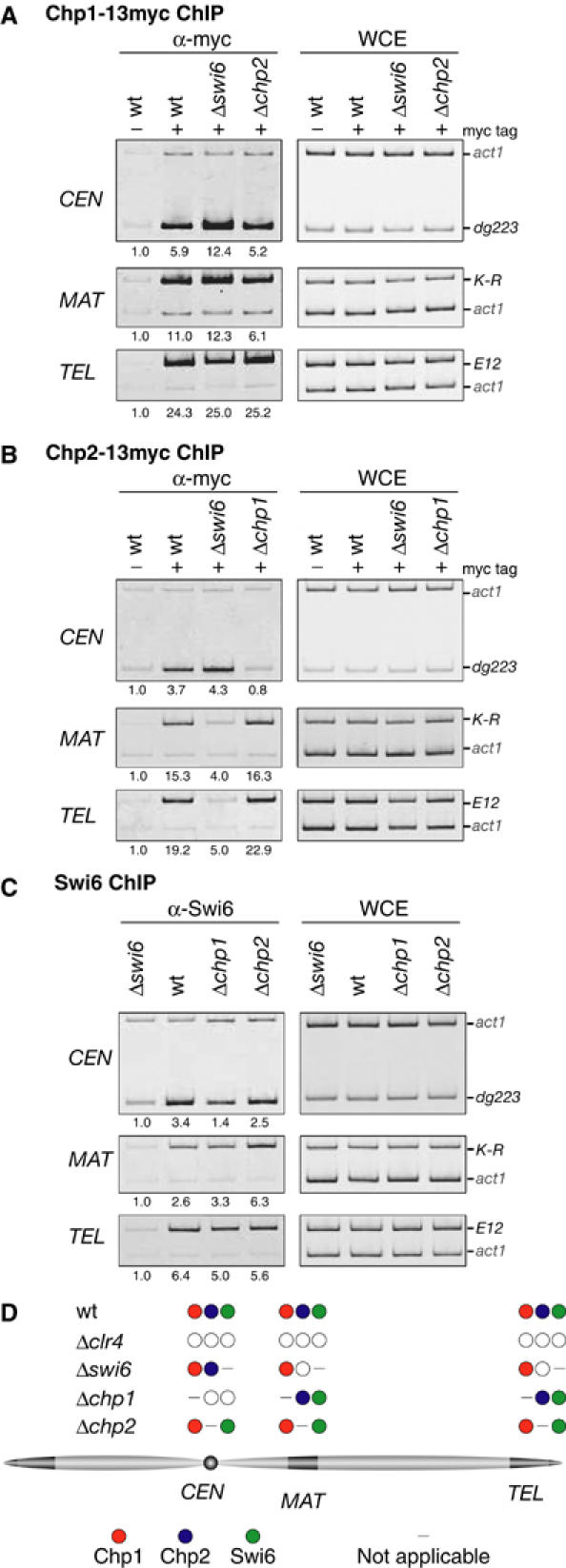
Interdependency of chromodomain proteins to associate with heterochromatin. (A) Association of Chp1 with heterochromatic regions in the absence of swi6+ or chp2+. Association of Chp1-13myc with the indicated heterochromatic regions was assayed by ChIP using anti-myc antibodies as in Figure 1. As a control, wild-type cells in which Chp1 was not tagged were used. (B) Association of Chp2 with heterochromatic regions in the absence of swi6+ or chp1+. ChIP using anti-myc antibodies was performed as in (A). (C) Association of Swi6 with heterochromatic regions in the absence of chp1+ or chp2+. ChIP was performed as in (A), using anti-Swi6 antibodies. As a control, Δswi6 cells were used. (D) Summary of the results shown in (A–C). The localization of chromodomain proteins (filled circles) to each heterochromatic region in the indicated strains is depicted. A reduction in the heterochromatin association of the protein is indicated by an open circle.
Chp1 is required for the establishment of heterochromatin
Chp1 protein plays a critical role in the localization of the Swi6 and Chp2 proteins to centromeric heterochromatin. It has been shown that a centromere-specific silencing defect is correlated with a deficiency in the establishment of heterochromatin assembly. Factors involved in the RNAi machinery (rnai) are required for the establishment of heterochromatin, and all of the Δrnai strains display defective gene silencing at centromeric heterochromatin, but not at the mating-type region or telomeres (Hall et al, 2002, 2003; Volpe et al, 2002). We therefore tested whether chp1+ is required for the establishment of gene silencing. In wild-type cells, a DNA fragment from a centromere (L5), or the mating-type region (cenH) attracts heterochromatin factors when integrated at a euchromatic locus, resulting in the ectopic silencing of a neighboring reporter gene (Ayoub et al, 2000, 2003; Partridge et al, 2002). A previous report showed that Chp1 is required for the silencing and H3-K9 methylation at an ectopic site containing centromeric L5 DNA (Partridge et al, 2002). If Chp1 plays a central role in the establishment of silent heterochromatin and its role is not specific for the centromeric sequences, the ectopic silencing mediated by cis-acting DNA from other heterochromatin regions might also be abrogated in the absence of Chp1. To test this possibility, we introduced the L5cen1 or cenHE/H fragment with a ura4+ reporter gene into a euchromatic ade6 locus in the wild-type or Δchp1 strain (Figures 3A and B). As the control, the same DNA constructs were introduced into the Δclr4, Δswi6, Δchp2, and Δrnai strains. The cells were spotted onto media containing 5-fluoroorotic acid (FOA, which is toxic to ura4+-expressing cells) to evaluate the ura4+ reporter expression. As previously reported, the wild-type ade6∷L5cen1-ura4+ cells grew on FOA medium, but the other mutant cells did not, suggesting that ectopic silencing mediated by L5cen1 was induced in the wild-type but not in the Δchp1, Δclr4, Δswi6, Δchp2, or Δrnai strains (Figure 3A; Partridge et al, 2002; Volpe et al, 2003). The same assays were performed with strains carrying ectopic cenHE/H (ade6∷cenHE/H-ura4+). We found that only the wild-type strain carrying ade6∷cenHE/H-ura4+ grew on the FOA medium (Figure 3B), indicating that ectopic silencing mediated by cenHE/H was not induced in the Δchp1 and Δrnai strains, as is the case for ectopic L5cen1 (Hall et al, 2002; Volpe et al, 2002). These results indicated that Chp1 protein is required for the silencing at the ectopic L5cen1 or cenHE/H sequence, and also demonstrated that the silencing defect by Δchp1 is not exclusive to the centromeric sequence. Although these results did not clarify Chp1's function in the establishment or maintenance step, subsequent experiments clearly showed that Chp1 participates in the establishment of the silent state (see below).
Figure 3.

chp1+ is required for L5cen1- or cenHE/H-mediated gene silencing. (A) Deletions of chp1+ or components of the RNAi machinery disrupt the silencing at ectopic L5cen1. A schematic representation of the L5cen1-ura4+ construct inserted into the ade6 locus is shown (top). Five-fold-diluted cultures of the indicated strains carrying L5cen1-ura4+ were plated onto AA medium containing FOA (FOA) and nonselective (N/S) medium. (B) Deletions of chp1+ or components of the RNAi machinery disrupted silencing at ectopic cenHE/H. ura4+ silencing at cenHE/H inserted at the ade6 locus was examined as in (A). (C) Transcripts derived from centromeric repeats were detected by Northern blotting using probes specific for centromeric dg repeats (top). Ethidium bromide-stained RNAs are shown as a loading control. The two major bands correspond to rRNAs (bottom).
It has been shown that deletion of components of the RNAi machinery results in the aberrant accumulation of long centromeric RNA transcripts (Volpe et al, 2002). To verify the relationship between Chp1 and the RNAi factors, the centromeric transcripts in mutant strains were analyzed by Northern blotting. We found that the long centromeric transcripts accumulated in Δchp1 and Δclr4 (Figure 3C). The level of accumulated transcripts in the Δchp1 cells was comparable to that in the Δrnai cells. Therefore, Chp1 protein was also involved in the production or processing of centromeric RNA transcripts, which might be linked to heterochromatin establishment.
We next investigated the effect of chp1 deletion on the establishment of H3-K9 methylation at native heterochromatic regions. For this purpose, we performed a clr4+ deletion/reintroduction experiment (Figures 4A and B, left panels), in which clr4+ was deleted from either the wild-type or Δchp1 strain (clr4 deletion, Δclr4) followed by the integration of functional clr4+ into the original locus (clr4+ reintroduction, clr4+ (Re)). At each step, the level of H3-K9 methylation at CEN (dg223), MAT (K-R), or TEL (E12) was monitored by ChIP analysis. In the wild-type (chp1+) background, clr4+ deletion (Δclr4) caused a loss of the H3-K9 methylation at the three heterochromatic regions (Figure 4A). When clr4+ was reintroduced, the methylation was restored to a level comparable to that of the original strain (Figure 4A). These results indicated that the reintroduced clr4+ was functional and that the reintroduction was sufficient to establish H3-K9 methylation at the three heterochromatic regions. Similar to the wild-type strain, a deletion of clr4+ in Δchp1 cells caused the loss of H3-K9 methylation at all three heterochromatic regions (Figure 4B, Δchp1Δclr4). However, in sharp contrast, the reintroduction of clr4+ into the Δchp1Δclr4 strain did not lead to the restoration of H3-K9 methylation in any of the three heterochromatic regions (Figure 4B, Δchp1clr4+ (Re)), except for a small amount of restored methylation in the telomeres (see Discussion). The expression of clr4+ mRNA in the Δchp1clr4+ (Re) strains was comparable to that in the original Δchp1 strain (data not shown). We confirmed these results by testing another independently isolated Δchp1clr4+ (Re) clone to rule out the possibility that defects in the establishment of H3-K9 methylation were due to the colony isolation steps (data not shown). These results further supported the notion that Chp1 is required for the establishment of repressive chromatin at the native heterochromatic regions. The present data also demonstrate that Chp1 function is required not only for the centromeres but also for the mating-type region and telomeres.
Figure 4.
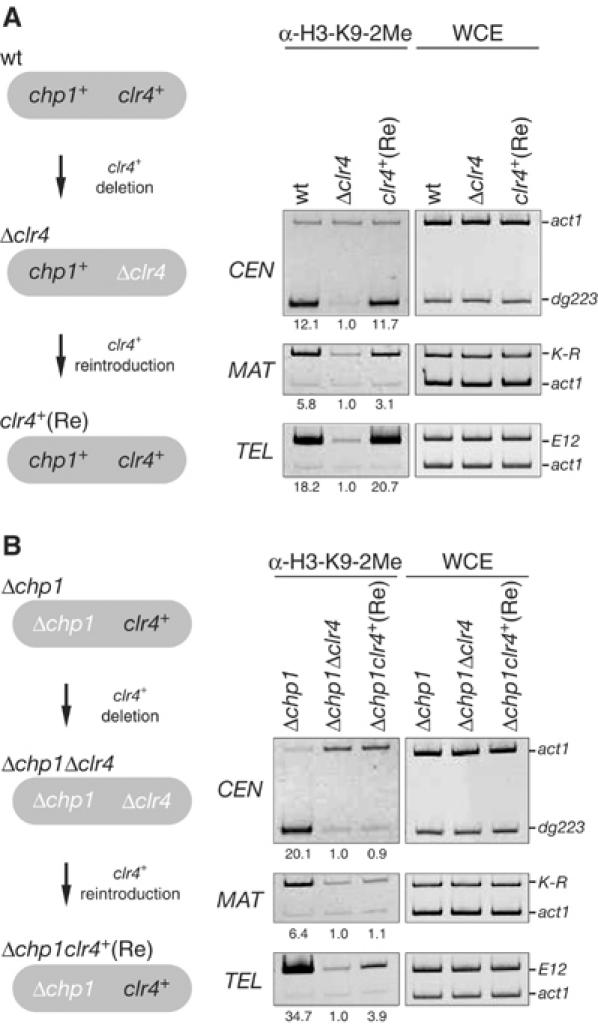
chp1+ is required for the establishment of H3-K9 methylation at the three heterochromatic regions. clr4+ was deleted from wild-type (wt) or Δchp1 cells to generate Δclr4 or Δchp1Δclr4, respectively. Functional clr4+ was then reintroduced (Re) and the level of H3-K9 methylation was assayed by ChIP using anti-H3-K9-2Me antibodies.
Histone H3-K9 methylation in heterochromatin is maintained without Chp1 or the RNAi machinery
Previous reports demonstrated that a mutation in chp1 or any of the three RNAi-related genes abolishes H3-K9 methylation at a ura4+ reporter gene inserted in centromeric outer repeat (otr) heterochromatin (Partridge et al, 2002; Volpe et al, 2002). Furthermore, the methylation level at centromeric dg repeats is reduced in each Δrnai strain (Volpe et al, 2002). Intriguingly, we found that, even in the Δchp1 cells, histone H3 in native centromeric heterochromatin (CEN-dg223 locus) remained methylated at lysine 9 (Figure 4B, Δchp1). To clarify these differences, we compared the methylation level of H3-K9 at the CEN-ura4+ reporter and CEN-dg223 loci. Chromatin prepared from a strain in which a ura4+ reporter gene was inserted into centromeric otr heterochromatin was immunoprecipitated with anti-dimethylated-H3-K9 antibodies, and then competitive PCR was used to determine whether the immunoprecipitates were enriched for the CEN-ura4+ or CEN-dg223 sequence (Figure 5). At the CEN-ura4+ locus, H3-K9 methylation in the Δchp1 strain was reduced to the same level as in the Δclr4 strain (Figure 5B, ura4+). In contrast, at the CEN-dg223 locus, the H3-K9 methylation level in the Δchp1 strain was reduced, but the level was clearly higher than in Δclr4 (Figure 5B, top panel). Several other otr loci, CEN-dg660, -dg760, and -dh383, were also subjected to the same analysis. Although the levels of the retained H3-K9 methylation were varied, we obtained similar results for all the endogenous loci (Figure 5B, bottom panel). The chp1+ deletion did not affect H3-K9 methylation at the MAT and TEL heterochromatin (Figure 5B, top MAT and TEL). These results demonstrate that H3-K9 methylation at CEN-dg-dh loci is maintained, at least in part, without chp1+, while the H3-K9 methylation at the ura4+ reporter locus apparently depends on chp1+. In Δswi6 or Δchp2 cells, the three heterochromatic regions were enriched in K9-methylated H3 at the same level as in wild-type cells (Figure 5B). We next examined whether H3-K9 methylation was maintained in the strains lacking RNAi genes. The results were similar to those observed in the Δchp1 strain (Figure 5C). H3-K9 methylation was lost at the ura4+ reporter region but was present to some extent at CEN-dg223 in the Δrnai strains. There was no obvious decrease in the methylation level at MAT and TEL. Taken together, these results demonstrated that, even in the centromeric heterochromatin, some of the H3-K9 methylation is retained in the absence of Chp1 or RNAi components. Furthermore, the differentially reduced H3-K9 methylation level at the CEN-dg223 versus CEN-ura4+ loci in the Δchp1 and Δrnai strains suggests that both Chp1 and the RNAi machinery are involved in spreading and/or enhancing the H3-K9 methylation in centromeric heterochromatin.
Figure 5.
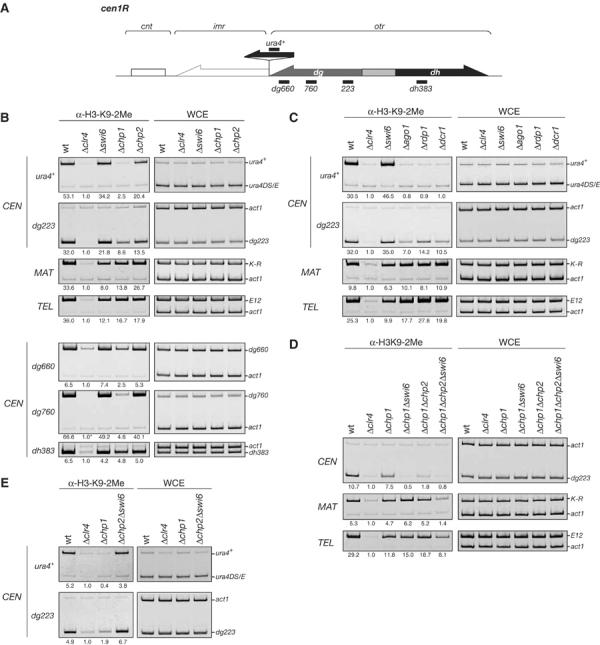
H3-K9 methylation is maintained at the three heterochromatic regions in the absence of Chp1 or components of the RNAi machinery. (A) Schematic diagram of the right half of centromere 1. Positions of the PCR products used to detect H3-K9 methylation in (B, C) are indicated by thick bars. (B, C) ChIP analysis was used to compare the levels of H3-K9 methylation in the indicated strains at centromeres (CEN), the mating-type region (MAT), or telomeres (TEL). (Asterisk) The ratio of act1 to CEN-dg760 signals present in whole-cell extract (WCE) instead of the ChIP results was used to calculate the relative fold enrichment, since the CEN-dg760 signal in the ChIP of Δclr4 was undetectable. (D) Swi6 and Chp2 are required for the maintenance of H3-K9 methylation. H3-K9 methylation at centromeres (CEN), the mating-type region (MAT), and telomeres (TEL) in wild-type, Δclr4, Δchp1, Δchp1Δswi6, Δchp1Δchp2, or Δchp1Δchp2Δswi6 cells is shown. ChIP analysis was used to compare the level of H3-K9 methylation in the indicated strains. (E) The levels of H3-K9 methylation at centromeres in the indicated cells.
Swi6 and Chp2 are required for the maintenance of H3-K9 methylation
We have shown that H3-K9 methylation is maintained at all three heterochromatic regions even in the absence of Chp1 or RNAi components. We next sought to determine the factors responsible for maintaining the H3-K9 methylation. In the mating-type region, Swi6 has been shown to be required for the maintenance of H3-K9 methylation (Hall et al, 2002). Thus, we compared the methylation level of H3-K9 at the heterochromatic regions in the Δchp1 and Δchp1Δswi6 strains (Figure 5D). Interestingly, we found that swi6+ deletion caused a loss of the H3-K9 methylation at CEN (dg223) in the Δchp1 background (Figure 5D, Δchp1Δswi6), suggesting that Swi6 is required for the maintenance of centromeric H3-K9 methylation in the Δchp1 strain. Unexpectedly, the centromeric H3-K9 methylation was also severely decreased in the Δchp1Δchp2 strain (Figure 5D, Δchp1Δchp2). These results showed that both Swi6 and Chp2 are critical for maintaining the residual methylation at the centromeres. We also examined a triple-mutant strain (Δchp1Δchp2Δswi6) for the maintenance of H3-K9 methylation. Interestingly, we found that, although deletion of either swi6+ or chp2+ did not affect the H3-K9 methylation at MAT (K-R) and TEL (E12) in the Δchp1 background, the methylation level in these regions was severely decreased in the triple-mutant strain (Figure 5D, Δchp1Δchp2Δswi6). Again, these results demonstrate that Swi6 and Chp2 are required for the maintenance of H3-K9 methylation at the three heterochromatic regions, and also indicate that Swi6 and Chp2 have overlapping functions in the maintenance of H3-K9 methylation. The synergistic effects of Δswi6 and Δchp2 were observed only in the Δchp1 background (Figures 5D and E). Deletion of chp2+ did not enhance the accumulation of centromeric transcript in the Δchp1, Δswi6, or Δchp1Δswi6 strains (Supplementary Figure 3). It should also be noted that, even in the triple-mutant strain, a reduced but detectable level of H3-K9 methylation persisted at the telomere, suggesting that another mechanism might function at the telomere (see Discussion).
Rik1 and Clr4 function cooperatively to promote heterochromatic H3-K9 methylation
Methylation of H3-K9 is maintained without the Chp1-mediated establishment step. Although Swi6 and Chp2 cooperatively function to maintain the methyl modification, we observed residual methylation in the triple-mutant strain (Figure 5D). This suggested that other factors might help to promote methylation to the heterochromatin. Rik1, a WD-40 repeat-containing protein, is a candidate for such a factor, since it has been shown to be required for heterochromatic silencing (Allshire et al, 1995), and was later demonstrated to be critical for H3-K9 methylation at a ura4+ reporter gene inserted in the centromeres or mating-type region (Nakayama et al, 2001b). Thus, to gain further insight into the mechanisms for maintaining H3-K9 methylation at the endogenous regions, we analyzed Δrik1 strains. We found that H3-K9 methylation at the three heterochromatic regions (CEN-dg223, MAT-K-R, or TEL-E12) was reduced to a level comparable to that in Δclr4 (Figure 6A), suggesting that Rik1 has a critical role in H3-K9 methylation at the native heterochromatic regions. Since the phenotypes of Δrik1 strains are similar to those observed for the Δclr4 strain, we next determined whether Rik1 contributes to the heterochromatin association of Clr4. For this experiment, strains expressing 5FLAG-tagged Clr4 from its native promoter were generated. In wild-type cells, 5FLAG-Clr4 associated with the centromeric CEN locus (Figure 6B, left panel, wt, FLAG tag+). In contrast, the association of 5FLAG-Clr4 with CEN was disrupted in Δrik1 cells (Figure 6B, left panel, Δrik1, FLAG tag+), suggesting that functional Rik1 is required for Clr4's association with centromeric heterochromatin. We further investigated Rik1's association with centromeres using strains expressing 13myc-tagged Rik1 from its endogenous promoter. Interestingly, the association between Rik1-13myc and CEN was detected in wild-type cells but was disrupted in Δclr4 cells (Figure 6C). These results showed that Rik1 and Clr4 mutually affect each other's centromeric association. Finally, we examined whether Rik1 and Clr4 physically interact with each other. Whole-cell extracts prepared from a strain expressing both Rik1-13myc and 5FLAG-Clr4 were used for co-immunoprecipitation (co-IP) experiments. Strains expressing either Rik1-13myc or 5FLAG-Clr4 were used as controls. Rik1-13myc was efficiently co-precipitated with 5FLAG-Clr4, and the association was confirmed in the reciprocal IP experiments (Figure 6D). Collectively, these results suggested that Rik1 and Clr4 interact with each other and function cooperatively to promote H3-K9 methylation in vivo.
Figure 6.
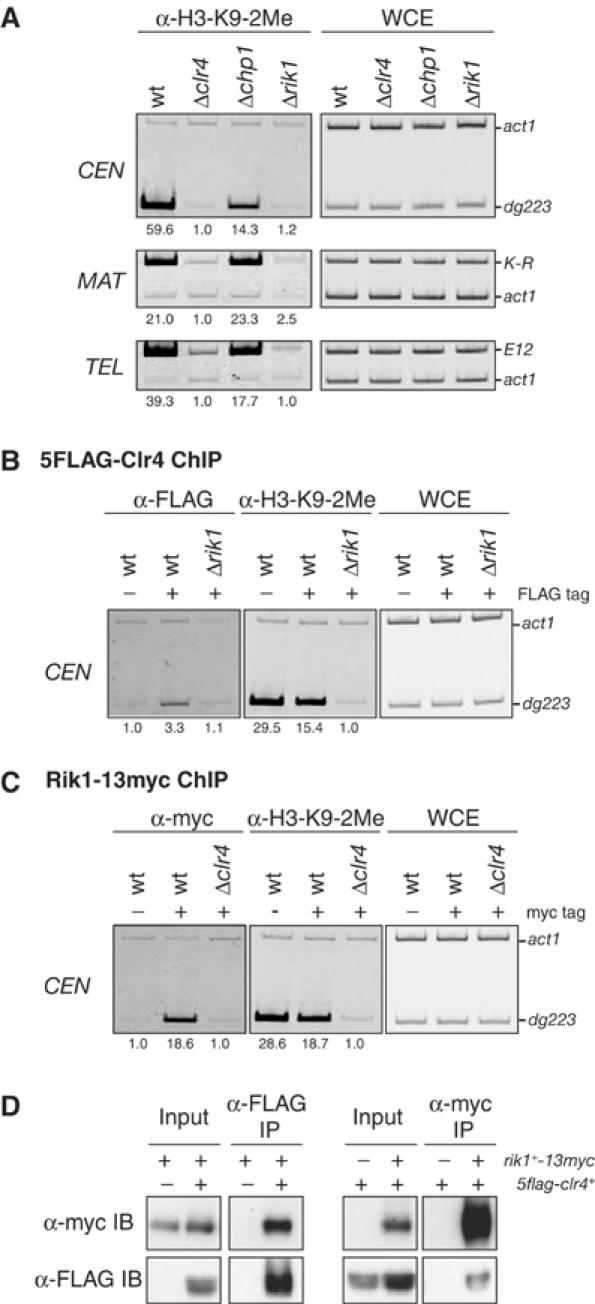
Clr4 and Rik1 interact with each other and act in concert to methylate H3-K9 at the three heterochromatic regions in vivo. (A) H3-K9 methylation at CEN, MAT, and TEL is abolished in Δclr4 or Δrik1. (B) Clr4 associates with centromeres in a rik1+-dependent manner. DNA isolated from the anti-FLAG- (α-FLAG) or anti-K9-dimethylated H3- (α-H3-K9-2Me) immunoprecipitated chromatin fraction or whole-cell extract (WCE) was used as a template for PCR. The DNA samples were prepared from either wild-type or Δrik1 cells expressing 5FLAG-tagged Clr4. As a control, wild-type cells in which Clr4 was not tagged were used. (C) The association of Rik1 with centromeres depends on clr4+. ChIP analysis with anti-myc antibodies was used to determine the presence of Rik1-13myc at centromeres. Wild-type, Δclr4 cells expressing Rik1-13myc, or wild-type cells expressing untagged Rik1 were used. (D) Clr4 is co-immunoprecipitated with Rik1 in vivo. Anti-FLAG antibodies (left panels) or anti-myc antibodies (right panels) were used to immunoprecipitate 5FLAG-Clr4 or Rik1-13myc from a strain expressing both 5FLAG-Clr4 and Rik1-13myc (IP). 5FLAG-Clr4 or Rik1-13myc was detected by immunoblotting with anti-FLAG or anti-myc antibodies, respectively (IB).
Discussion
Chromodomain proteins play an important role in the formation of repressive chromatin domains. In the present study, we report the functional characterization of Chp1 protein and show that it is required for the establishment of heterochromatin and is functionally correlated with RNAi factors in S. pombe. Our analyses also reveal that two chromodomain proteins, Swi6 and Chp2, are involved in the maintenance of histone H3-K9 methylation.
Chp1 and Chp2 are structural components of fission yeast heterochromatin
Consistent with the dot-like signals of the GFP fusion proteins, our ChIP analyses showed that both Chp1 and Chp2 stably associate with three known heterochromatic regions. As is the case for Swi6, this association is dependent on Clr4 histone methyltransferase. These results indicate that three chromodomain proteins, Swi6, Chp1, and Chp2, all participate in the formation of repressive chromatin at different chromosomal regions. In contrast, previous reports showed that Chp1 is crucial to centromeric silencing but not transcriptional silencing in the mating-type region or near telomeres (Thon and Verhein-Hansen, 2000). In support of this previous observation, we also found that Δchp1 causes the delocalization of Swi6 and Chp2 specifically at centromeres. In this study, we showed that Chp1 is required for the establishment of H3-K9 methylation not only at centromeres but also at the mating-type region and telomeres (see Figure 4B). Given that Chp1 plays the same role in the different chromosomal regions, it is likely that centromeric heterochromatin needs to undergo the establishment steps more routinely than do the other regions, resulting in the centromere-specific defects of the Δchp1 mutant. One possible explanation for the region-dependent effect might reside in the cis-acting DNA elements that enhance the stability of the higher-order structure. Interestingly, it has been reported that a 580-bp element REII located adjacent to mat2 functions cooperatively with the cenH fragment to enhance silencing stability (Ayoub et al, 2000). A recent report also showed that ATF/CREB family proteins directly bind another cis element, REIII, and help to nucleate heterochromatin (Jia et al, 2004). The presence of such additional cis-acting repression element(s) may cause Δchp1 to have different effects on each chromosomal region. Another possibility is that the centromeric region contains transcriptionally competent elements at a high frequency, compared with the mating-type region or telomeres. Indeed, small RNAs corresponding to centromeric repeats have been cloned from S. pombe (Reinhart and Bartel, 2002). In addition, centromeric transcripts are produced from centromeric repeats, and these transcripts are markedly increased in RNAi mutants (this study; Volpe et al, 2002). Although transcriptional competency at the mating-type region or telomeres has not been extensively characterized, it is possible that the centromeric heterochromatin competes against transcriptional activities and therefore continuously requires the establishment event. Further experiments will be needed to address the importance of heterochromatin structure in the centromere functions.
Chp1 protein is required for the establishment of heterochromatin
Previous reports showed that Chp1 associates with centromeres and is required for Swi6 localization and H3-K9 methylation at both centromeres and an ectopic site introduced by a centromeric cis-acting element (Partridge et al, 2000, 2002). These previous results indicated an important role of Chp1 in centromeric silencing. However, the general role of Chp1 in the assembly of silent chromatin has not been elucidated. From our results, it appears that the role of Chp1 is not exclusive to the centromeres. We showed that Chp1 is localized to three heterochromatic regions and is required for ectopic silencing mediated not only by the centromeric cis element L5cen1 but also by the cenHE/H sequence from the mating-type region. Moreover, we demonstrated that Chp1 is required to re-establish H3-K9 methylation at all three heterochromatic regions (Figure 4B). These results suggest that Chp1 has a general role in establishing heterochromatin and also imply that the centromere-specific defect in chp1-deleted cells might be caused by the structural features of centromeres. Interestingly, a similar centromere-specific silencing defect is observed in RNAi mutant cells (Hall et al, 2002, 2003; Volpe et al, 2002). Elements of the RNAi machinery have been demonstrated to be required for the establishment of repressive chromatin (Hall et al, 2002). We also showed that aberrant centromeric transcripts accumulate in chp1-deleted cells, as previously observed in RNAi-deleted cells (Figure 3B). Considering these results, it appears that Chp1 is functionally correlated with the RNAi machinery. This idea is further strengthened by a recent report that Chp1, Ago1, a newly identified factor Tas3, and siRNAs form a RITS complex and are required for the H3-K9 methylation at centromeric heterochromatin (Verdel et al, 2004). It is thought that the RITS complex may associate with heterochromatin through sequence-specific base pairing, that is, of an siRNA–DNA or siRNA–nascent transcript, followed by the H3-K9 methylation, which may further stabilize the association (Verdel et al, 2004). Since aberrant centromeric transcripts accumulate in Δclr4 or Δchp1 cells (Figure 3C), it is possible that a stable RITS–heterochromatin association through a Chp1-methylated H3-K9 interaction is required for the efficient processing of centromeric transcripts by the RNAi machinery.
Methylation in heterochromatin is maintained without Chp1 or the RNAi machinery
As described above, deletion of chp1+ or the RNAi factor genes causes a loss of silencing and H3-K9 methylation at centromeres (Partridge et al, 2002; Volpe et al, 2002). Interestingly, we found that, by using clr4-deleted strains as a basal-level control, H3-K9 methylation was completely abolished at an inserted marker region, but persisted at the endogenous centromeric regions in both Δchp1 and Δrnai cells. This result was confirmed using several sets of primers. These findings suggest that methyl modifications at native centromeric DNA can be maintained without the establishment step, which may partly explain why the mutations have little effect on the heterochromatin structure at the mating-type region and telomeres. For the maintenance of H3-K9 methylation, we showed that Swi6 and Chp2 play a critical role at the three heterochromatic regions. Although we did not clearly detect the centromeric localization of Swi6 or Chp2 in Δchp1 cells, it appears that they are targeted to the residual H3-K9 methylation at centromeric sequences independent of Chp1 and function in the maintenance of H3-K9 methylation. In the centromeres, both Swi6 and Chp2 are required for the maintenance of the methylation in the Δchp1 strain. It is likely that the reduced level or density of H3-K9 methylation in the Δchp1 strain changes the centromeric associations of Swi6 and Chp2 and affects their overlapping functions. The Δchp1Δchp2Δswi6 triple mutant displayed profound effects on all three heterochromatic regions. Notably, we found that H3-K9 methylation at the telomere (E12) was maintained at a clearly higher level in the triple-mutant than in the Δclr4 strain. This observation indicated that telomeric heterochromatin might use another mechanism to recruit H3-K9 methylation. This possibility is supported by another finding that, in the clr4+ deletion/reintroduction experiments, a small but detectable level of restored H3-K9 methylation was observed in the telomere sequence (Figure 4B). In addition to the heterochromatic configuration, telomeres consist of specialized DNA–protein complexes. Such telomere-specific protein(s) may have a function in recruiting H3-K9 methylation.
Another interesting observation in this study was that Rik1 plays a crucial role as a cofactor of Clr4. We showed that Rik1 associates with Clr4 and is required for its association with centromeric heterochromatin. Although it remains to be tested how Clr4 or Rik1 is recruited to heterochromatic regions, it is interesting to speculate that the Rik1–Clr4 association increases the heterochromatin binding affinity of the complex and that Swi6 and/or Chp2 promote this heterochromatin binding. Finally, we should point out that the methylation levels at an endogenous element and an inserted marker region were different in the Δchp and Δrnai strains. These differences in H3-K9 methylation state might be attributable to histone exchange. It is likely that nucleosomal histone H3 in the ura4+ marker region is subjected to active replacement throughout the cell cycle, as is the case for Drosophila H3.3, which is thought to be a plausible way to remove methyl modifications (Ahmad and Henikoff, 2002). This result also supports the above idea that heterochromatin always competes against transcriptional activities, and suggests that Chp1 and RNAi factors are required for the spreading and/or enhancement of H3-K9 methylation from the cis-acting region to the surrounding region. Further studies are necessary to elucidate the molecular mechanism of how Chp1 and RNAi factors spread the H3-K9 methylation.
Materials and methods
Strains and plasmids
Deletion and tagging to produce GFP, 13myc, or 5FLAG fusion proteins of endogenous chp1+, chp2+, clr4+, and rik1+ were performed by a PCR-based gene targeting protocol (Bahler et al, 1998; Krawchuk and Wahls, 1999). To express GFP-Chp2 fusion protein from a multicopy plasmid, an EGFP coding sequence was amplified by PCR and cloned into the pREP41 plasmid (Basi et al, 1993) to create pYB228. cDNA of the chp2+ coding sequence was amplified by PCR and inserted downstream of the EGFP gene of pYB228 to construct pYB232. For integration of an L5cen1-ura4+ or cenHE/H-ura4+ fragment into the ade6 locus, PCR-amplified ura4+ (−187 to +1.24 kb) was cloned into the PstI–SalI site of the pBluescript SK(−) (pYB265). A PCR-amplified ClaI–KpnI fragment from cen1 (L5cen1) (Partridge et al, 2002) or EcoRV–HaeIII fragment (cenHE/H) (Ayoub et al, 2000) was cloned into the SmaI site of pYB265 to create pYB277 or pYB285, respectively. The BamHI–SpeI fragment from ade6+ was cloned into pBluescript SK(−) (pYB286) and BamHI–SalI fragments from pYB277 or pYB285 were inserted into the NdeI locus in ade6+ of pYB286 to generate pYB289 or pYB288, respectively. pYB289 and pYB288 were digested with XhoI and used for the transformation of wild-type and mutant cells. For the clr4+ reintroduction experiments, the clr4+ genomic sequence was PCR-amplified and cloned into the pCRII-TOPO plasmid (clr4-pTP). A hygromycin resistance gene (hygr) was inserted into clr4-pTP using the EcoRV and NotI sites (clr4-hyg-pTP). The clr4-hyg-pTP plasmid was linearized by digesting it with the HpaI site in the clr4+ genomic region and introduced into the Δclr4 strains. All other strains were constructed by standard genetic crosses. The strains used in this study are listed in Supplementary Table 1.
Chromatin immunoprecipitation
ChIP was performed as described previously (Nakayama et al, 2000). To immunoprecipitate 13myc- or 5FLAG-tagged proteins, anti-c-myc (9E10, Roche) or anti-FLAG (M2, Sigma) monoclonal antibodies were used. To immunoprecipitate Swi6 or histone H3 dimethylated at K9, anti-Swi6 or anti-H3-K9-2Me monoclonal antibodies were used. PCR products were separated on a 5% nondenaturing polyacrylamide gel, stained with ethidium bromide, and photographed by Printgraph equipped with Image Saver HR (ATTO). The primer sets used are listed in Supplementary Table 2.
RNA isolation and Northern hybridization
Total RNA was extracted from cells as described by Schmitt et al (1990). Purified total RNA samples were separated on a 1.25% agarose gel containing 6.7% formaldehyde, blotted to nylon membranes, crosslinked, and hybridized with PCR-amplified centromeric dg sequence (Volpe et al, 2002).
Co-immunoprecipitation
Cells expressing both FLAG-tagged Clr4 and myc-tagged Rik1 were collected, and extracts were prepared as described (Iida and Araki, 2004). FLAG-Clr4 or Rik1-myc was immunoprecipitated with and detected by an anti-FLAG antibody or anti-c-myc antibody, respectively.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Tables
Acknowledgments
We thank SI Grewal and F Ishikawa for strains and plasmids, Y Murakami for strains, and T Matsumoto for plasmids. We also thank C Nishimoto and J Toga for antibody preparation and strain construction, our laboratory members in the CDB, RIKEN for helpful discussion and support, and M Okano and D Sipp for critical reading of the manuscript. This research was supported by PRESTO of the JST.
References
- Ahmad K, Henikoff S (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Allshire RC (1996) Transcriptional silencing in the fission yeast: a manifestation of higher order chromosome structure and functions. In Epigenetic Mechanisms of Gene Regulation, Russo VEA, Martienssen RA, Riggs AD (eds) pp 443–466. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G (1994) Position effect variegation at fission yeast centromeres. Cell 76: 157–169 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Ayoub N, Goldshmidt I, Lyakhovetsky R, Cohen A (2000) A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics 156: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Noma K, Isaac S, Kahan T, Grewal SI, Cohen A (2003) A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol 23: 4356–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K (1993) TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123: 131–136 [DOI] [PubMed] [Google Scholar]
- Doe CL, Wang G, Chow C, Fricker MD, Singh PB, Mellor EJ (1998) The fission yeast chromo domain encoding gene chp1+ is required for chromosome segregation and shows a genetic interaction with alpha-tubulin. Nucleic Acids Res 26: 4222–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269: 1429–1431 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109: 2637–2648 [DOI] [PubMed] [Google Scholar]
- Grewal SI (2000) Transcriptional silencing in fission yeast. J Cell Physiol 184: 311–318 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC (2002) Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev 12: 178–187 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ (1996) Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86: 95–101 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI (2003) RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci USA 100: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI (2002) Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- Halverson D, Gutkin G, Clarke L (2000) A novel member of the Swi6p family of fission yeast chromo domain-containing proteins associates with the centromere in vivo and affects chromosome segregation. Mol Gen Genet 264: 492–505 [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Iida T, Araki H (2004) Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol 24: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJ (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet 19: 192–195 [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Krawchuk MD, Wahls WP (1999) High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15: 1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lorentz A, Ostermann K, Fleck O, Schmidt H (1994) Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 143: 139–143 [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30: 329–334 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Allshire RC, Klar AJ, Grewal SI (2001a) A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J 20: 2857–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI (2000) A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101: 307–317 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001b) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Paro R, Hogness DS (1991) The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA 88: 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14: 783–791 [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500 [DOI] [PubMed] [Google Scholar]
- Sadaie M, Naito T, Ishikawa F (2003) Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev 17: 2271–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V, Allshire R (2003) Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069–1074 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Verhein-Hansen J (2000) Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155: 551–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC (2003) RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11: 137–146 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schaèfer M, Muèller-Auer S, Gabel C, Fuchs M, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreâano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Saânchez M, del Rey F, Benito J, Domínguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerrutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Tables


