Abstract
While tractography is widely used in brain imaging research, its quantitative validation is highly difficult. Many fiber systems, however, have well-known topographic organization which can even be quantitatively mapped such as the retinotopy of visual pathway. Motivated by this previously untapped anatomical knowledge, we develop a novel tractography method that preserves both topographic and geometric regularity of fiber systems. For topographic preservation, we propose a novel likelihood function that tests the match between parallel curves and fiber orientation distributions. For geometric regularity, we use Gaussian distributions of Frenet-Serret frames. Taken together, we develop a Bayesian framework for generating highly organized tracks that accurately follow neuroanatomy. Using multi-shell diffusion images of 56 subjects from Human Connectome Project, we compare our method with algorithms from MRtrix. By applying regression analysis between retinotopic eccentricity and tracks, we quantitatively demonstrate that our method achieves superior performance in preserving the retinotopic organization of optic radiation.
Keywords: probabilistic tractography, Bayesian inference, visual path-way
1 Introduction
Tractography is a widely used technique for studying brain connectomes with diffusion MRI (dMRI) and has provided many exciting results in brain imaging research [1]. The lack of rigorous validation for in vivo human brain studies, however, has long been a critical challenge to push tractography toward a quantitative tool [2, 3]. On the other hand, the regular topographic organization of many fiber systems in human brains provide a surprisingly untapped anatomical knowledge for the improvement and validation of tractography techniques. Some of the well-known examples include the retinotopic organization of the visual pathway[4], the somatotopic organization of the somatosensory pathway[5], and the tonotopic organization of the auditory pathway[6]. In this paper, we incorporate this insight on anatomical regularity to develop a novel probabilistic tractography algorithm for studying the connectome of these topographically organized systems.
Conventional streamline methods rely on step size and curvature parameters to control the regularity of fiber tracks [7]. This type of local mechanism is difficult to account for the topographic organization of fiber pathways. It becomes particularly challenging when we want to reconstruct fiber tracks that are topographically arranged but with highly bended segments such as the Meyer’s loop of the optic radiation. More recently, global tractography techniques were developed to improve the robustness of streamline techniques [8, 9], but their focus is on balancing the fitting of dMRI signals and the regularity of local stick models. These developments are followed by microstructure based optimization techniques that start with a large set of tracks and assign weights or prune them so that resultant set of tracks best fit the dMRI signal [10, 11].
In this work we propose a novel probabilistic tractography method that incorporates both topographic and geometric regularity. Each fiber track is represented with the Frenet-Serret frame in our method. Fiber orientation distributions (FODs) [7, 12] are used in our work to represent the connectivity information at each voxel. For topographic regularity, the key idea in our method is the development of a novel likelihood function that tests the match of parallel curves to the FODs in the neighborhood of each point. The geometric prior is modeled as a Gaussian distribution of the Frenet-Serret frame. Taken together, we use a Bayesian approach with rejection sampling to propagate the fiber tracks and reconstruct highly organized fiber bundles.
In our experimental results, we validate the proposed technique on the reconstruction of the optic radiation using the multi-shell diffusion imaging data of the Human Connectome Project (HCP)[13]. The retinotopy of the visual system means there is a corresponding point on the cortex for each point of the retinal space[4]. This correspondence between the topography of the retina and axonal projections provides a great opportunity for localized mapping of retina disease to visual pathway integrity. Another distinct aspect of this fiber system is the Meyer’s loop that bends sharply toward the anterior aspect before moving toward the visual cortex[14]. Given these anatomical knowledge about the topography and geometric organization of the optic radiation, we believe it is an ideal testbed for tractography algorithms. On multi-shell imaging data from 56 HCP subjects, we compare our method with three tractography algorithms of the MRtrix package[7]. We demonstrate that our method is able to generate highly organized tracks while capturing the challenging Meyer’s loop. Using regression analysis, we quantitatively demonstrate that our method performs better in preserving the retinotopy of the optic radiation.
2 Methods
Frenet-Serret apparatus
we use the Frenet-Serret frame and formulas to represent fiber tracks as differentiable curves. Let γ(s) represent a non-degenerate curve parameterized by its arc length s. The Frenet-Serret frame can then be defined with three orthonormal vectors: tangent (T), normal (N) and binormal(B) that are given as T= dγ/ds, N = (dT/ds)/ | dT/ds | and B = T × N. The Frenet-Serret formulas express the derivatives of T, Nand B in terms of themselves with a system of first order ODEs: dT /ds = κN, dN/ds = −κT + τ B and dB/ds = −τ N. Here κ and τ are the curvature and torsion of the curve, respectively. Given the initial conditions, this system can be uniquely solved.
Curve spaces
Curve parameters can be put in a vector form to represent the space of differentiable curves that pass through with . Here denotes the Frenet-Serret frame and is a short notation for the space of curves, , where S2 is the unit sphere. We will use the notation to denote where cp traces in space and denote (s) = {T cp (s),Ncp(s), B cp(s)}, κ cp(s) and τ cp(s) as the parameters of the curve at s.
Parallel curves
Let cp and cp′ be two curves. If p′ lies on the normal plane of cp and cp = cp′ then we call cp and cp′ parallel curves and denote them as cp ‖ cp′. These are also known as offset curves in computer graphics and computer aided design.
Fiber model
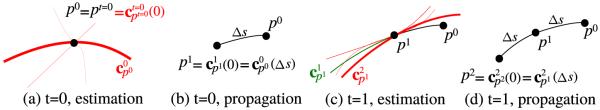
We model a fiber track as a finite length, arc length parameterized curve that starts from a seed point pt=0 ∈ ℝ3 at time step zero (t = 0). In a nutshell, our algorithm initializes by estimating for s = 0 and It solves the Frenet-Serret ODE system and moves to t = 1 by taking a step of Δs which gives s = Δs, and . We then compute the priors, likelihood and posterior using Bayesian inference. Lastly we pick the next curve, , randomly by rejection sampling and iterate until a stopping condition is met. Our track is a train of points traced by the curve . Fig.1 explains our notation and the propagation technique.
Fig. 1.
(a) At t = 0, without prior information, we select a random curve among the red candidate curves based on their likelihood. The thicker the curve, the higher posterior probability it has. (b) By solving the Frenet-Serret ODE, we propagate by Δs. (c) At t = 1, we calculate the prior probability for candidate curves for a smooth transition from the previous curve shown in green. (d) Propagate to p2.
Bayesian inference
Given a curve at time t, , and data , we estimate the posterior probability for the next curve using Bayesian inference as follows:
| (1) |
Prior estimation
In order to preserve smoothness between the transition of curves that involves the changes in κ, τ and 3D rotation of , we use Gaussian distributions to define the geometric prior. The variances for curvature and torsion are denoted with and . 3D rotation is realized by rotating around T, N and B axes in order,with variance σT, σN and σB respectively. The overall prior probability is expressed in Eq.2:
| (2) |
The functions used for computing the prior probability are given in Eq.3.
| (3) |
In Eq.3, 〈 , 〉 is the dot product, T′, N′ and B′ are the intermediary rotations, and Ψ (κ) = asin (κ) is used to linearize change in curvature.
Likelihood estimation
We estimate the data support for the next curve using the parallel curve definition and fiber orientation distribution (FOD). The field of FODs over an image volume can be expressed as a spherical function where is the position of a point in the dMRI image and . We define our likelihood expression as follows:
| (4) |
In Eq.4, r is the radius of a sphere centered at p, which is the integration domain. Constant terms in front of Eq.4 are used for normalization. The likelihood expression computes the data support for a candidate curve by integrating contributions of FODs belonging to parallel curves.
Implementation details
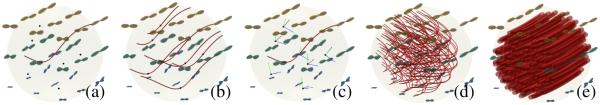
The double integral in Eq.4 is intractable. In order to estimate the likelihood numerically, we discretize the integral domain and add the contributions of FODs for a set of points as illustrated in Fig. 2. In this case, we used an actual human dMRI and picked a point in corpus callosum. For better visualization, we show here 7 random points as black dots within a radius of a few voxels, but in practice we use 27 points. The likelihood of a curve is computed by estimating the parallel curves that pass through these points. We then compute the tangents and interpolate the FODs at the points. The final likelihood is obtained by adding and averaging the data support contributed by these points. In Fig. 2(d) and (e), the estimated likelihoods of two different sets of parallel curves are visualized, where the thickness of fibers are in proportion to their likelihood.
Fig. 2.
(a) To estimate the likelihood of a curve, we randomly pick a number of points within the integration radius, r. (b) Parallel curves passing through the random points. (c) We compute the tangents of parallel curves for each point and obtain the average FOD. (d)(e) Estimated likelihoods are shown in proportion to the thickness of curves.
3 Test subjects and experimental setup
In our experiments we used multi-shell images from Q1 release of HCP that includes 74 subjects, 56 of which completed both T1 and dMRI scans. For all 56 subjects, we computed FODs following the algorithm in [12] for multi-shell data. The FODs from this method are represented by spherical harmonics (SPHARM) and fully compatible with MRtrix that we compare with. We focused on the reconstruction of optic radiation that connects the lateral geniculate nucleus (LGN) and primary visual cortex (V1). One salient feature of this bundle is the retinotopic organization. Following the method in [16], we automatically generate V1 ROI and its retinotopic map that assigns each vertex in V1 cortex two coordinates: angle and eccentricity. ROI for LGN was generated using the method proposed in [14]. Inferior bundle of the optic radiation has an unconventional trajectory which first courses anteriorly before it runs posteriorly towards the visual cortex forming the elusive Meyer’s loop. Because of the quantitative coordinates provided by the retinotopic map and the challenges in reconstructing the Meyer’s loop, we picked the optical radiation in our tests. We conducted qualitative and quantitative comparisons of our technique with algorithms in MRtrix3 [7], which includes two probabilistic tractography algorithms: iFOD2 [15], iFOD1 and a deterministic, SD STREAM, approach. Table 1 shows the parameters used for each technique. For our technique, we typically use a very small step size together with small and values since we walk along curves.In order to capture the Meyer’s loop, for MRtrix3 algorithms we used a higher angular threshold than the default parameters. Cut-off values were adjusted so that all the techniques can capture the Meyer’s loop. A lower cut-off threshold and a much higher angle threshold are used for SD STREAM to achieve this.
Table 1.
Tractography parameters used for each technique. vs is voxel size, ° is degree.
| Step(vs) | Angle | Cutoff | r(vs) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Our Method | 0.001 | 0.04 | 60° | 1.25° | 1.25° | 0.2 | 0.2 | 2 | |
| MRtrix3 iFOD2 | 0.2 | 22° | 0.04 | ||||||
| MRtrix3 iFOD1 | 0.1 | 11° | 0.04 | ||||||
| MRtrix3 SD_STREAM | 0.1 | 60° | 0.02 |
4 Results
Qualitative evaluation
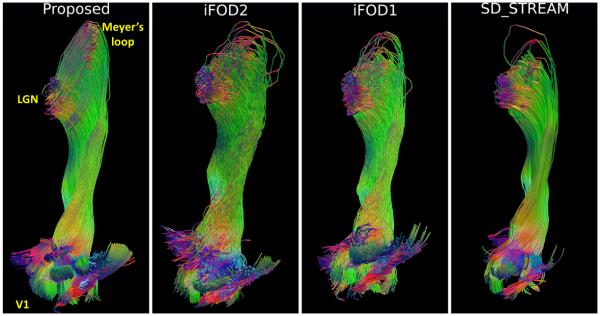
Reconstruction results of the left optic radiation of an HCP subject by our method and MRtrix algorithms are shown in Fig. 3. We can clearly see that our results are more desirable as they are able to successfully capture the Meyer’s loop while exhibiting highly organized trajectories. As the tracks approach the V1 cortex, we can see the probabilistic tractography results from iFOD2 and iFOD1 start to become topographically less organized.
Fig. 3.
Qualitative comparison of our method with MRTrix algorithms on the reconstruction of a left optic radiation of an HCP subject.
Quantitative evaluation
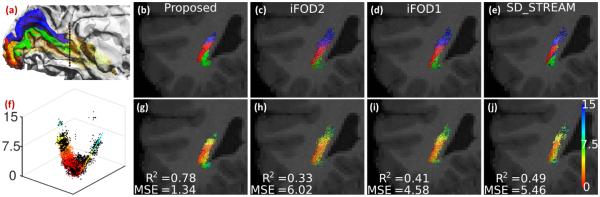
In Fig.4 we illustrate our approach for quantitative evaluation. Using the eccentricity of the V1 cortex, we can divide it into three parts: fovea (red), superior-peripheral (green) and inferior-peripheral (blue) regions as shown in Fig.4(a), which also allow us to split the fiber bundle into three sub-bundles. By cutting the bundle with a coronal plane (black dashed lines in Fig.4(a)), we can visualize the topographic organization of the fiber tracks from different methods as shown in Fig. 4 (b)-(e). Because of the topographic organization of the fovea and peripheral bundles, the eccentricity of the fiber tracks and their coordinates on the cross-section should follow a U-shape relation as plotted in Fig. 4 (f), where the black dots are the raw data and the colored points are the fitted value with quadratic regression. In Fig. 4(g)-(j), the eccentricity values of each bundle on the cross-section are visualized, where the color bar for eccentricity is shown on the right most image. To quantitatively assess this relation, we applied quadratic regression to model the relation between eccentricity and the cross-sectional coordinates of fiber bundles. We report both the mean square error (MSE) and coefficient of determination (R2) to measure how well the fiber tracks preserve the retinotopy of the fiber bundle. For each technique, the mean value of these two measures from 56 HCP subjects are listed in Table 2, where we can see that our method achieves the best performance in both measures.
Fig. 4.
Quantitative comparison of the bundles show in Fig.3. Top row shows the labels for three sub-bundles of the optic radiation. Bottom row shows the eccentricity values and the quality of quadratic fit using MSE and R2. The low MSE and high R2values obtained by the proposed technique corroborate the qualitative observation.
Table 2.
Quantitative evaluation of the retinotopic organization of the optic radiation bundle. Best results are in bold.
| Our Method | iFOD2 | iFOD1 | SD STREAM | |
|---|---|---|---|---|
| Mean R2 | 0.53 | 0.27 | 0.4 | 0.46 |
| Mean MSE | 7.44 | 14.83 | 9.03 | 12.5 |
5 Discussions and Conclusion
Our current implementation is non-optimal and written in MATLAB and C++. Typically it takes several hours to generate tracks on the orders of thousands which we are working to improve. Our technique uses more parameters as compared to the conventional algorithms, but they are geometrically intuitive and their effects are not very difficult to understand. For quantitative comparison, we took a different approach than in [2] which suggests counting valid, invalid and no connections. This is because we focus on the reconstruction of individual bundles. We also did not use synthetic phantoms or simulated tracks because of the availability of retinotopic maps for in vivo validation.
In summary, we developed a novel probabilistic tractography technique that aims to capture the topographic organization of fiber bundles. A key idea in our method is the use of parallel curves to examine the local fitting of fiber tracks to the underlying field of FODs. Using the retinotopic mapping on V1 cortex, we have conducted quantitative evaluations and demonstrated that our method is able to generate more organized fiber tracks that follows known anatomy of the visual system. For future work, we will conduct more extensive validations on the visual pathway, its connectivity maps and other bundles that also follow topographic organizations such as the auditory and somatosensory pathways.
Acknowledgements
This work was in part supported by the National Institute of Health (NIH) under Grant K01EB013633, P41EB015922, P50AG005142, U01EY025864, U01AG051218.
References
- 1.Fillard P, Descoteaux M, Goh A, Gouttard S, Jeurissen B, Malcolm J, Ramirez-Manzanares A, Reisert M, Sakaie K, Tensaouti F, Yo T, Mangin JF, Poupon C. Quantitative evaluation of 10 tractography algorithms on a realistic diffusion MR phantom. NeuroImage. 2011;56(1):220–234. doi: 10.1016/j.neuroimage.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Côté MA, Girard G, Bor A, Garyfallidis E, Houde JC, Descoteaux M. Tractometer: Towards validation of tractography pipelines. Medical Image Analysis. 2013;17(7):844–857. doi: 10.1016/j.media.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. PNAS. 2014;111(46):16574–16579. doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel SA, Glover GH, Wandell BA. Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cerebral Cortex. 1997;7(2):181–192. doi: 10.1093/cercor/7.2.181. [DOI] [PubMed] [Google Scholar]
- 5.Ruben J, Schwiemann J, Deuchert M, Meyer R, Krause T, Curio G, Vill-ringer K, Kurth R, Villringer A. Somatotopic organization of human secondary somatosensory cortex. Cerebral Cortex. 2001;11(5):463–473. doi: 10.1093/cercor/11.5.463. [DOI] [PubMed] [Google Scholar]
- 6.Morosan P, Rademacher J, Schleicher A, Amunts K, Schormann T, Zilles K. Human primary auditory cortex: Cytoarchitectonic subdivisions and mapping into a spatial reference system. NeuroImage. 2001;13(4):684–701. doi: 10.1006/nimg.2000.0715. [DOI] [PubMed] [Google Scholar]
- 7.Tournier JD, Calamante F, Connelly A. MRtrix: Diffusion tractography incrossing fiber regions. Int. J. of Imag. Syst. Tech. 2012;22(1):53–66. [Google Scholar]
- 8.Reisert M, Mader I, Anastasopoulos C, Weigel M, Schnell S, Kiselev V. Global fiber reconstruction becomes practical. NeuroImage. 2011;54(2):955–962. doi: 10.1016/j.neuroimage.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Mangin JF, Fillard P, Cointepas Y, Le Bihan D, Frouin V, Poupon C. Toward global tractography. NeuroImage. 2013;80:290–296. doi: 10.1016/j.neuroimage.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Daducci A, Dal Palu A, Lemkaddem A, Thiran JP. COMMIT: Convex Opti-mization Modeling for Microstructure Informed Tractography. IEEE Transactions on Medical Imaging. 2015;34(1):246–257. doi: 10.1109/TMI.2014.2352414. [DOI] [PubMed] [Google Scholar]
- 11.Smith RE, Tournier JD, Calamante F, Connelly A. SIFT2: Enabling dense quantitative assessment of brain white matter connectivity using streamlines trac-tography. NeuroImage. 2015;119:338–351. doi: 10.1016/j.neuroimage.2015.06.092. [DOI] [PubMed] [Google Scholar]
- 12.Tran G, Shi Y. Fiber orientation and compartment parameter estimation from multi-shell diffusion imaging. IEEE Transactions on Medical Imaging. 2015;34(11):2320–2332. doi: 10.1109/TMI.2015.2430850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Essen DV, Ugurbil K, Auerbach E, Barch D, Behrens T, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss S, Penna SD, Feinberg D, Glasser M, Harel N, Heath A, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen S, Prior F, Schlaggar B, Smith S, Snyder A, Xu J, Yacoub E. The human connectome project: A data acquisition perspective. NeuroImage. 2012;62(4):2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kammen A, Law M, Tjan BS, Toga AW, Shi Y. Automated retinofugal visual pathway reconstruction with multi-shell HARDI and FOD-based analysis. NeuroImage. 2016;125:767–779. doi: 10.1016/j.neuroimage.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tournier JD, Calamante F, Connelly A. Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions; Proc. 18th Annual Meeting of the Intl. Soc. Mag. Reson. Med.(ISMRM).2010. p. 1670. [Google Scholar]
- 16.Benson NC, Butt OH, Datta R, Radoeva PD, Brainard DH, Aguirre GK. The retinotopic organization of striate cortex is well predicted by surface topology. Current Biology. 2012;22(21):2081–2085. doi: 10.1016/j.cub.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]