Abstract
Ubiquitin-conjugating enzyme E2T (UBE2T), a member of the ubiquitin-conjugating E2 family in the ubiquitin-proteasome pathway, has been reported to be overexpressed in certain tumor types and to have an important role in the Fanconi anemia pathway. In the present study, the expression of UBE2T and its association with bladder cancer were investigated; to the best of our knowledge, this has not been reported previously. Immunohistochemistry and western blot analysis demonstrated that UBE2T was significantly upregulated in bladder cancer tissues and cell lines compared with adjacent normal bladder tissues and a normal human urinary tract epithelial cell line, respectively. UBE2T was detectable in the nuclei and cytoplasm of cancer cells, exhibiting stronger expression in the nuclei. A UBE2T-siRNA-expressing lentivirus was constructed and used to infect human bladder cancer 5637 cells, in order to examine the role of UBE2T in bladder cancer cell growth in vitro. The knockdown of UBE2T significantly decreased bladder cancer cell proliferation and colony formation. Furthermore, UBE2T silencing induced cell cycle arrest at G2/M phase and increased cell apoptosis. Therefore, UBE2T serves an important role in the growth of bladder cancer cells, and may be considered as a potential biomarker and therapeutic target for bladder cancer.
Keywords: UBE2T, bladder cancer, siRNA, proliferation, apoptosis, cell cycle
Introduction
Bladder cancer is ranked as the second most common type of urinary system carcinoma; in the United States, ~74,000 new cases and ~16,000 mortalities occurred in 2015 (1). Although there are various treatments available, including radical cystectomy, transurethral resection of bladder tumor (TURBT), chemotherapy and radiotherapy, the therapeutic outcomes remain unsatisfactory. The 5-year survival rate of invasive bladder cancers in 2014 was 70.2%, and it has a high rate of recurrence (50–90%) (2). Due to the absence of more effective therapies for bladder cancer, further research into the underlying molecular mechanisms of its tumorigenesis and the development of novel treatments aimed at specific molecular targets are required.
Ubiquitin-conjugating enzyme E2T (UBE2T; also known as HSPC150) is a member of E2 the family in the ubiquitin-proteasome pathway, a complex protein degradation system that serves crucial roles in extensive biological processes, including cell cycle control, signal transduction and tumorigenesis (3). Certain ubiquitin-conjugating enzymes (E2s), such as UBC2/Rad6, UBC9 and UBE2C, have been reported to be closely associated with tumorigenesis (4–7). The E2s accept ubiquitin from the E1 complex and catalyze its covalent attachment to other proteins. UBE2T has been shown to bind the E3 ubiquitin-protein ligase Fanconi anemia complementation group L (FANCL), which is the Fanconi anaemia core complex, and to catalyze the monoubiquitination of Fanconi anemia complementation group D2, a key step in the DNA damage pathway (8,9). In addition, upregulation of UBE2T has been reported in certain cancer types (10,11). However little is known with regard to the association between UBE2T and bladder cancer.
In the present study, the expression of UBE2T was evaluated in human bladder cancer tissues and cell lines. In addition, the effects of lentivirus-mediated specific small interfering RNA (siRNA) knockdown of UBE2T on the proliferation, colony formation, apoptosis and cell cycle of bladder cancer cells were assessed. The findings revealed a potential oncogenic role for UBE2T in bladder cancer, and suggest that it may serve as a biomarker or therapeutic target for bladder cancer.
Materials and methods
Tissue samples
Tissue samples were obtained from 25 bladder cancer patients who underwent radical cystectomy or transurethral resection at the Department of Urology of Peking University First Hospital (Beijing, China). Tumor-adjacent normal bladder tissues were obtained from an area >2.0 cm away from the margin of the cancer tissue. The histological characteristics of the samples were evaluated by hematoxylin and eosin staining and confirmed by experienced urological pathologists. Fresh samples were fixed in 4% paraformaldehyde for 12–24 h and then paraffin-embedded for immunohistochemistry, or snapfrozen immediately following the resection and stored in liquid nitrogen for western blotting. Informed consent was obtained in all cases and protocols were approved by the Medical Ethics Committee of Peking University First Hospital.
Cell culture
SV-HUC-1 normal human urinary tract epithelial cells were obtained from ATCC (Manassas, VA, USA) and cultured in Hyclone F-12K medium (GE Healthcare Life Sciences, Logan, UT, USA). The bladder cancer cell lines T24, 5637, BIU-87 and EJ were cultured in RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences), and J82 cells were cultured in high-glucose Dulbecco's modified Eagle's medium (Hyclone; GE Healthcare Life Sciences). All bladder cancer cell lines were obtained from the Institute of Urology of Peking University (Beijing, China). All media contained 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), penicillin G (100 U/ml) and streptomycin (100 µg/ml) (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). Cells were maintained as a monolayer culture at 37°C in a humidified atmosphere containing 5% CO2.
Immunohistochemistry
After fixing with 4% formalin and embedding in paraffin wax, tissues were cut into 5-µm sections using a microtome. The sections were deparaffinized in xylene and rehydrated with graded concentrations of alcohol. Subsequently, the slides were treated with 3% H2O2 to block the endogenous peroxidase activity and heated (95°C) for 2.5 min in citrate buffer (10 mmol/l; pH 6.0) for antigen retrieval. To reduce nonspecific binding, 10% normal goat serum was applied. Subsequently, the slides were incubated with primary rabbit anti-human UBE2T polyclonal antibody (#bs-18089R; Bioss, Inc., Woburn, MA, USA; dilution, 1:500) at 4°C overnight, and a PowerVision™ two-step histostaining reagent and 3,3-diaminobenzidine tetrahydrochloride substrate kit (ZSGB-Bio, Bejing, China) were used to visualize the localization of the antigen, according to the manufacturer's instructions.
Western blotting
Total protein from bladder cancer samples and cell lines were prepared with ice-cold radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck Millipore), quantified using BCA protein assay reagent (Pierce Chemical Co., Rockford, IL, USA), and separated by SDS-PAGE. Following electrophoresis, proteins in the gel were transferred to polyvinylidene difluoride membranes (Merck Millipore). After blocking for 1 h with 5% non-fat milk, the membranes were incubated overnight at 4°C with a rabbit monoclonal antibody against UBE2T (#ab179802; Abcam, Cambridge, MA, USA; dilution, 1:500) and rabbit polyclonal antibody against GAPDH (#sc-25778; Santa Criz Biotechnology, Inc., Santa Cruz, CA, USA; dilution, 1:1,000), followed by incubation with a horseradish peroxidase-labeled goat anti-rabbit IgG secondary antibody (#sc-2004/sc-2005; Santa Cruz Biotechnology, Inc.; dilution, 1:5,000) at room temperature for 1 h. Signals were detected by application of ECL Western Blotting Detection Reagent (GE Healthcare Life Sciences) and visualized using a G:BOX Chemi Gel Documentation System (Syngene, Frederick, MD, USA).
Lentivirus vector construction for RNA interference
The sequence encoding the human UBE2T gene (NM_014176) was obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The lentivirus vector system was composed of the vector pGCSIL-GFP, which stably expresses the siRNA and a marker (GFP-RFP fusion protein), pHelper1.0 (gag/pol element) and Helper2.0 (VSV-G element); pHelper1.0 and pHelper2.0 contain essential virus packaging elements. The most effective double-stranded UBE2T-targeted siRNA sequence, PscSI14026 (5′-GTACACAACTCAACACAGAAA-3′), was synthesized and cloned into the pGCSIL-GFP vector by GeneChem Corporation (Shanghai, China). Psc-NC (5′-TTCTCCGAACGTGTCACGT-3′), which shows no homology to any known human genes, was used to generate the negative control (NC) lentiviral vectors. Cancer cells were plated and cultured in six-well plates (5×104 cells/well) until 60% confluent, and then appropriate volumes of the lentiviruses were added to the cells, according to the multiplicity of infection value (number of lentiviruses per number of cells) recommended by the manufacturer. The interference efficiency of the vectors was determined by western blot analysis.
Quantitative reverse transcription (RT)-polymerase chain reaction (PCR)
Total RNA was isolated from cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 2 µg RNA was reverse-transcribed into cDNA using M-MLV reverse transcriptase (Promega Cororation, Madison, WI, USA) and oligo(dT) 15 (Promega Corporation) as a primer. RT was performed under the following conditions: 42°C for 1 h and 75°C for 15 min. Quantitative PCR was performed using SYBR Green PCR Master Mix (Toyobo Co., Ltd., Osaka, Japan) in a final volume of 10 µl in the 7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer sequences were as follows: Forward, 5′-ATCCCTCAACATCGCAACTGT-3′ and reverse, 5′-CAGCCTCTGGTAGATTATCAAGC-3′ for UBE2T; forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′ for GAPDH. GAPDH served as the quantitative internal control. PCR was performed under the following conditions: 94°C for 30 sec, followed by 45 cycles of 94°C for 5 sec and 60°C for 30 sec. The expression of UBE2T mRNA was normalized to GAPDH according to the ΔΔCq method (12).
Cell growth assay
Cell growth was measured via multiparametric high-content screening (HCS). Briefly, human bladder cancer 5637 cells that had been infected with NC lentivirus or UBE2T-siRNA lentivirus were seeded at a density of 2,000 cells per well in 96-well plates, and then incubated at 37°C with 5% CO2 for 5 days. Plates were processed with the ArrayScan™ HCS software (Cellomics Inc.) for analysis each day. The system is a computerized, automated fluorescence-imaging microscope that automatically identifies stained cells and reports the intensity and distribution of fluorescence in each individual cell. Images were acquired for each fluorescence channel, using suitable filters and a 20x objective lens. In each well, >800 cells were analyzed.
BrdU incorporation assay
DNA synthesis in proliferating cells was determined by a BrdU incorporation assay, using a BrdU kit (#11647229001; Roche Diagnostics, Basel, Switzerland) following the manufacturer's instructions. The assay was performed in triplicate and repeated three times.
Colony formation assay
Cells infected with UBE2T-siRNA lentivirus or NC lentivirus were seeded in 6-well plates at a density of 800 cells/well and cultured at 37°C for 14 days. Medium was replaced every 2–3 days. Cells were then washed twice with PBS, fixed with 4% paraformaldehyde, stained with Giemsa for 10 min and washed three times with double distilled H2O. Colonies were photographed and counted under a microscope (Leica DM IL; Leica Microsystems, Wetzlar, Germany).
Cell cycle analysis
Cells infected with UBE2T-siRNA lentivirus or NC lentivirus were collected, washed twice with ice-cold PBS, and fixed with 70% ice-cold ethanol. Following fixation overnight and subsequent rehydration in PBS for 30 min at 4°C, the samples were stained for 30 min in darkness with 50 µg/ml propidium iodide (#P4170; Sigma-Aldrich; Merck Millipore) containing 125 U/ml protease-free RNase, and then analyzed using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Cell cycle analysis was conducted using ModFit 2.0 software (BD Biosciences). The assay was performed in triplicate.
Cell apoptosis analysis
Cell apoptosis was assayed by staining with Annexin V-allophycocyanin (eBioscience Annexin V Apoptosis Detection Kit APC; #88-8007; Affymetrix, Inc., Santa Clara, CA, USA) and detected using a FACSCalibur flow cytometer. The assay was performed in triplicate.
Statistical analysis
SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA) was employed for statistical analyses. All values in the text and figures are expressed as the mean ± standard deviation of these observations. P<0.05 was considered to indicate statistical significance.
Results
UBE2T is highly expressed in bladder cancer tissues and cell lines
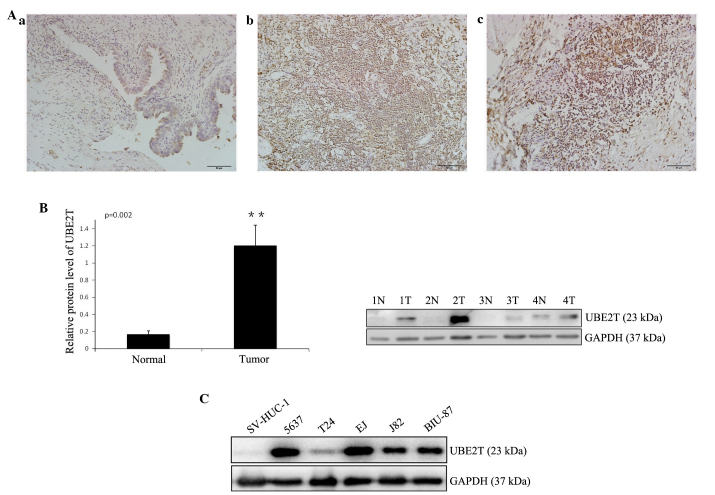
In order to study the expression of UBE2T in bladder cancer tissues, immunohistochemistry was performed on 25 pairs of bladder cancer and adjacent normal tissues. Weak staining was detected in normal bladder tissue, while strong staining of UBE2T was observed in almost all cancer specimens (Fig. 1A). UBE2T protein could be detected in the nuclei and cytoplasm of cancer cells, but was stronger in the nuclei, in concordance with a previous report (11). A western blot analysus was performed to assess UBE2T protein levels in 12 bladder cancer samples and 5 cancer cell lines (5637, T24, J82, EJ and BIU-87). The results revealed that UBE2T expression was higher in bladder cancer tissues (Fig. 1B) and cell lines (Fig. 1C) compared with tumor-adjacent normal bladder tissues and SV-HUC-1 normal human urinary tract epithelial cells. Of the bladder cancer cell lines, 5637 exhibited the highest UBE2T expression, and was therefore selected for further functional characterization.
Figure 1.
Increased expression of UBE2T in human bladder cancer tissues and cell lines. (A) Representative immunohistochemical staining of UBE2T in normal control tissues and bladder cancer tissues (magnification, ×200; scale bar, 50 µm): (a) Negative or weak UBE2T expression in normal bladder tissues; (b) strong expression of UBE2T in transitional cell carcinoma of the bladder; (c) strong expression of UBE2T in squamous cell carcinoma of the bladder. (B) Representative western blots showing the expression of UBE2T in bladder cancer tissues (tumor/T) and tumor-adjacent bladder tissues (normal/N). The relative UBE2T protein expression is expressed as the mean ± standard deviation from 12 cases, with GAPDH serving as a loading control. **P<0.01. (C) Western blots showing the expression of UBE2T in human bladder cancer cell lines. The normal human urinary tract epithelial cell line SV-HUC-1 was used as negative control. GAPDH served as the loading control. The results represent at least three separate experiments. UBE2T, ubiquitin-conjugating enzyme E2T.
Lentivirus-mediated knock-down of UBE2T in 5637 cells
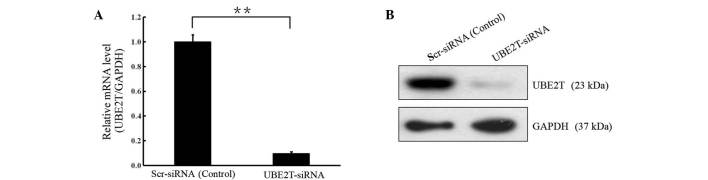
In order to investigate the biological function of UBE2T in bladder cancer, lentivirus-mediated knockdown of UBE2T was performed in cells of the 5637 bladder cancer cell line. UBE2T mRNA and protein levels were assessed by RT-qPCR and western blot analyses. As shown in Fig. 2, the mRNA and protein levels of UBE2T were reduced in cells infected with UBE2T-siRNA compared with the cells infected with NC siRNA (P<0.01), indicating effective knockdown of the target sequence.
Figure 2.
Lentivirus-mediated knockdown of UBE2T in 5637 cells. (A) Reverse transcription-quantitative polymerase chain reaction and (B) western blotting revealed that UBE2T mRNA and protein levels were decreased significantly following UBE2T knockdown (**P<0.01). GAPDH served as the loading control. UBE2T, ubiquitin-conjugating enzyme E2T; siRNA, small interfering RNA.
Knockdown of UBE2T inhibits the proliferation of 5637 cells
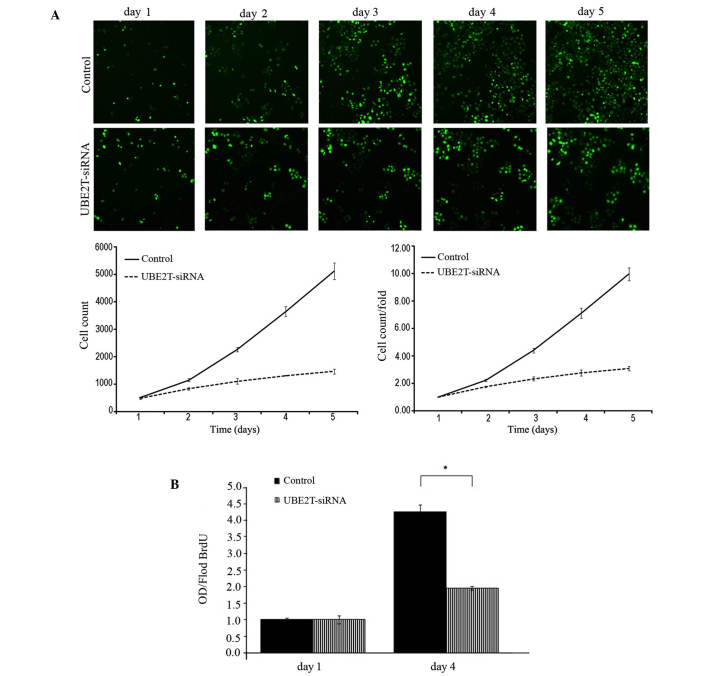
Following infection with lentiviral-mediated siRNA, GFP-expressing 5637 cells were monitored every day for 5 days by Cellomics and 5-day cell proliferation curves were drawn. Cell growth rate was defined as the cell count of Nth day/cell count of 1st day, where N = 2, 3, 4 or 5. The results revealed that downregulation of UBE2T decreased the total number of cells and slowed the cell growth rate (Table I and Fig. 3A). Furthermore, DNA synthesis was analyzed by BrdU incorporation assay on the 1st and 4th days in the 5637 cells. The results demonstrated decreased DNA synthesis in the UBE2T-siRNA lentivirus-infected group, indicating that cell proliferation was significantly reduced over the course of 4 days (P=0.0127; Fig. 3B). Collectively, these results indicate that the UBE2T gene may be closely associated with the proliferation of 5637 cells.
Table I.
Cell numbers and growth rate as counted by cellomics.
| Cell count | Fold change in cell count | |||
|---|---|---|---|---|
| Time | Control | UBE2T-siRNA | Control | UBE2T-siRNA |
| Day 1 | 513.3±13.05 | 473.0±36.29 | 1.00±0.00 | 1.00±0.00 |
| Day 2 | 1,143.3±58.79 | 826.3±39.70 | 2.23±0.09 | 1.75±0.06 |
| Day 3 | 2,258.3±76.54 | 1,103.3±97.11 | 4.40±0.17 | 2.33±0.14 |
| Day 4 | 3,646.3±176.19 | 1,302.3±14.50 | 7.11±0.37 | 2.76±0.22 |
| Day 5 | 5,113.0±295.13 | 1,457.3±93.22 | 9.96±0.47 | 3.09±0.15 |
Fold change in cell count was defined as cell count of Nth day / cell count of 1st day, where N=2, 3, 4 or 5.
Figure 3.
Lentivirus-mediated knockdown of UBE2T inhibited the proliferation of 5637 cells. (A) UBE2T knockdown attenuated 5637 cell growth potential in vitro. Cell growth was measured via multiparametric high-content screening every day for five days. Data are presented as the mean ± standard deviation. (B) The DNA synthesis rate was slowed in response to UBE2T downregulation, as analyzed by BrdU incorporation assay on the 1st and 4th days. (Control vs. UBE2T-siRNA, *P<0.05). UBE2T, ubiquitin-conjugating enzyme E2T; siRNA, small interfering RNA.
Knockdown of UBE2T induced cell cycle arrest in 5637 cells
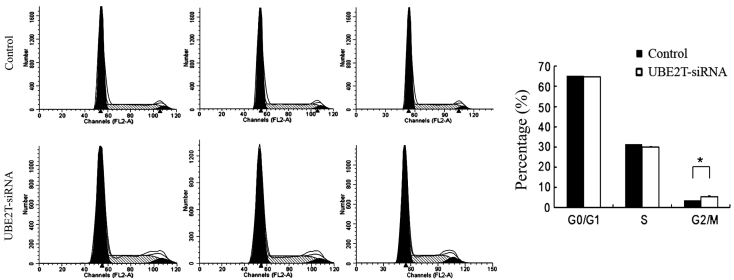
Alterations in cell proliferative are usually caused by changes in the cell cycle or apoptosis. In order to investigate whether the downregulation of UBE2T has an effect on the cell cycle of 5637 cells, flow cytometry was employed. As shown in Fig. 4, the control group exhibited the following distribution: G0/G1 phase, 65.5%; S phase, 31.64%; and G2/M phase, 3.66%. By contrast, in the UBE2T-siRNA group, the proportions were 64.58% in G1 phase, 30.83% in S phase, and 5.79% G2/M phase. Compared with the control group, the UBE2T-siRNA group demonstrated a significant increase in the percentage of cells in G2/M phase (3.66±0.34% vs. 5.79±0.55%; P=0.0185), suggesting that cells were arrested in G2/M phase following UBE2T gene silencing and that the UBE2T gene is significantly correlated with the cell cycle of 5637 cells.
Figure 4.
Knockdown of UBE2T expression induced G2/M phase arrest. The cell cycle of 5637 cells was analyzed by propidium iodide staining and flow cytometry. Compared with the control, UBE2T-siRNA cultures showed a significant increase in cells in G2/M phase (*P<0.05). UBE2T, ubiquitin-conjugating enzyme E2T; siRNA, small interfering RNA.
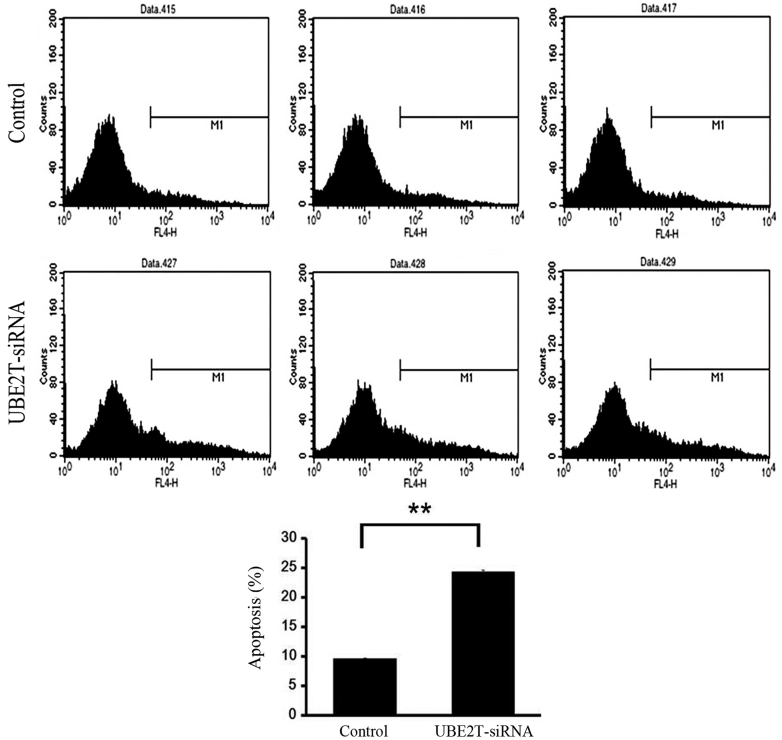
Knockdown of UBE2T induces apoptosis of 5637 cells
Flow cytometry was used to investigate the effect of UBE2T on apoptosis of 5637 cells infected with NC lentivirus or UBE2T-siRNA lentivirus. As shown in Fig. 5, the percentage of cells undergoing apoptosis was significantly increased in the UBE2T-siRNA group compared with NC group (24.46±0.39% vs. 9.51±0.01%; P=0.0001). This result affirmed that UBE2T may be associated with the apoptosis of bladder cancer cells.
Figure 5.
Knockdown of UBE2T increased cell apoptosis. Cell death was determined by Annexin V and flow cytometric analysis. UBE2T-siRNA cultures showed a significant increase in apoptosis compared with the control (**P<0.01). UBE2T, ubiquitin-conjugating enzyme E2T; siRNA, small interfering RNA.
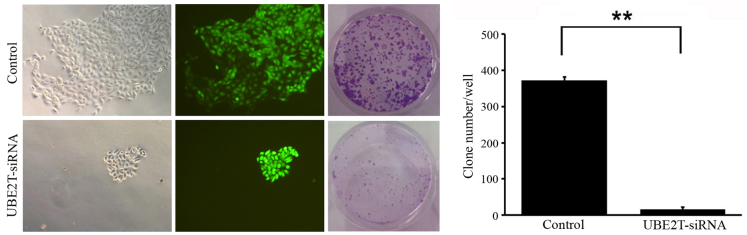
Knockdown of UBE2T decreases colony formation of 5637 cells
To determine the effect of UBE2T knockdown on bladder cancer cell tumorigenesis in vitro, colony formation assays were performed. The results revealed that UBE2T knockdown in 5637 cells caused a substantial reduction in colony formation compared with the NC cells. The number of colonies in UBE2T-siRNA lentivirus-infected cells was significantly lower than that in the NC group (14±15 vs. 372±61; P=0.009; Fig. 6). These results suggest that the expression of UBE2T may be associated with the progress of malignant changes.
Figure 6.
Knockdown of UBE2T resulted in lower bladder cancer cell colony formation efficiency, as assessed by colony formation assay (magnification, ×100). The number of colonies in UBE2T-siRNA lentivirus-infected cells was statistically less than that of the negative control group (**P<0.01).
Discussion
Although extensive information is available regarding bladder cancer at the genetic and molecular levels, differing clinical courses and the limited value of established prognostic markers have compelled researchers to investigate the mechanisms of tumorigenesis in order to identify novel molecular components for use in bladder cancer diagnosis as well as therapy. Tumorigenesis is considered a multistep and complex process, which includes genomic instability and mutation, evasion of apoptosis, resistance to cell death, and dysregulation of cellular energetic processes (13). The ubiquitin-proteasome pathway serves an important role in cell proliferation, apoptosis, the cell cycle and DNA repair (14,15). For the ubiquitin-proteasome pathway, there are 2 E1 enzymes, ~40 E2 enzymes, and >600 E3 ligases, allowing for thousands of combinations of E2-E3 complexes (9). UBE2T, as an important member of the E2 family, is gaining increasing attention due to its role in the process of tumorigenesis and its potential prognostic and therapeutic value (9–11).
The effect of UBE2T and its interaction with the E3 ligase FANCL in the Fanconi anemia pathway, which is required for the repair of damaged DNA, have been well studied in recent years (9,16–19). Furthermore, the location of the UBE2T gene, 1q32.1, is amplified in breast cancer, liver tumors and cervical tumors (20). Ueki et al (10) demonstrated that UBE2T is overexpressed in breast cancer and is critical in the development and/or progression of breast cancer due to its interaction with and regulation of the BRCA1/BARD1 complex (10). Hao et al (11) also reported that UBE2T is highly expressed in lung cancer tissues and cell lines. It is well known that hypoxia is one of the various conditions of tumor microenvironment (21). Hypoxia has been demonstrated to downregulate UBE2T expression, which correlates with an increased sensitivity to crosslinking agents consistent with a defective Fanconi anemia pathway (9). UBE2T has also been described as a target gene promoted by the E2F transcription factors, which are known to serve a key role in the timely expression of genes required for cell cycle progression and proliferation (22). Considering these facts, UBE2T may interact with certain key molecules that are significantly involved in cell cycle regulation or tumorigenesis. In addition, pathways of which UBE2T is a component, for example the Fanconi anemia pathway and BRCA1 pathway, may also be exploited as potential targets for tumor diagnosis and treatment.
In summary, the present study confirmed that UBE2T is overexpressed in bladder cancers and that knockdown of UBE2T with lentivirus-mediated specific siRNA significantly suppresses the proliferation and colony formation of 5637 bladder cancer cells, as well as inducing cell cycle arrest and increasing the rate of apoptosis. Future studies should expand the number of bladder cancer specimens to assess whether UBE2T levels are correlated with tumor grade, pathological stage or lymph node metastasis. In addition, the mechanisms of UBE2T in regulating bladder cancer tumorigenesis and progression, and the potential associations between UBE2T and downstream molecules (such as BRCA1 and Fanconi anemia pathway factors) also require further studies. Such studies regarding the role of UBE2T in cell cycle regulation and tumorigenesis will undoubtedly provide novel insights into potential bladder cancer therapies.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (nos. 81172419 and 81402083).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21314. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 3.Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009;19:1905–1911. doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shekhar MP, Lyakhovich A, Visscher DW, Heng H, Kondrat N. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 2002;62:2115–2124. [PubMed] [Google Scholar]
- 5.Dong M, Pang X, Xu Y, Wen F, Zhang Y. Ubiquitin-conjugating enzyme 9 promotes epithelial ovarian cancer cell proliferation in vitro. Int J Mol Sci. 2013;14:11061–11071. doi: 10.3390/ijms140611061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuliang S, Lei C, Guangwu J, Changjie L. Involvement of ubiquitin-conjugating enzyme E2C in proliferation and invasion of prostate carcinoma cells. Oncol Res. 2013;21:121–127. doi: 10.3727/096504013X13832473329953. [DOI] [PubMed] [Google Scholar]
- 7.Xie C, Powell C, Yao M, Wu J, Dong Q. Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int J Biochem Cell Biol. 2014;47:113–117. doi: 10.1016/j.biocel.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Hodson C, Purkiss A, Miles JA, Walden H. Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3-E2 selection. Structure. 2014;22:337–344. doi: 10.1016/j.str.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaekers CH, van den Beucken T, Meng A, Kassam S, Thoms J, Bristow RG, Wouters BG. Hypoxia disrupts the Fanconi anemia pathway and sensitizes cells to chemotherapy through regulation of UBE2T. Radiother Oncol. 2011;101:190–197. doi: 10.1016/j.radonc.2011.05.059. [DOI] [PubMed] [Google Scholar]
- 10.Ueki T, Park JH, Nishidate T, Kijima K, Hirata K, Nakamura Y, Katagiri T. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009;69:8752–8760. doi: 10.1158/0008-5472.CAN-09-1809. [DOI] [PubMed] [Google Scholar]
- 11.Hao J, Xu A, Xie X, Hao J, Tian T, Gao S, Xiao X, He D. Elevated expression of UBE2T in lung cancer tumors and cell lines. Tumour Biol. 2008;29:195–203. doi: 10.1159/000148187. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama KI, Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 15.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 16.Longerich S, Kwon Y, Tsai MS, Hlaing AS, Kupfer GM, Sung P. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014;42:5657–5670. doi: 10.1093/nar/gku198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 2012;40:4553–4561. doi: 10.1093/nar/gks053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: A basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421–8430. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 20.Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–4753. doi: 10.1038/sj.onc.1208641. [DOI] [PubMed] [Google Scholar]
- 21.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 22.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]