Summary
Background
Important information to support healthcare quality improvement is often recorded in free text documents such as radiology reports. Natural language processing (NLP) methods may help extract this information, but these methods have rarely been applied outside the research laboratories where they were developed.
Objective
To implement and validate NLP tools to identify long bone fractures for pediatric emergency medicine quality improvement.
Methods
Using freely available statistical software packages, we implemented NLP methods to identify long bone fractures from radiology reports. A sample of 1,000 radiology reports was used to construct three candidate classification models. A test set of 500 reports was used to validate the model performance. Blinded manual review of radiology reports by two independent physicians provided the reference standard. Each radiology report was segmented and word stem and bigram features were constructed. Common English “stop words” and rare features were excluded. We used 10-fold cross-validation to select optimal configuration parameters for each model. Accuracy, recall, precision and the F1 score were calculated. The final model was compared to the use of diagnosis codes for the identification of patients with long bone fractures.
Results
There were 329 unique word stems and 344 bigrams in the training documents. A support vector machine classifier with Gaussian kernel performed best on the test set with accuracy=0.958, recall=0.969, precision=0.940, and F1 score=0.954. Optimal parameters for this model were cost=4 and gamma=0.005. The three classification models that we tested all performed better than diagnosis codes in terms of accuracy, precision, and F1 score (diagnosis code accuracy=0.932, recall=0.960, precision=0.896, and F1 score=0.927).
Conclusions
NLP methods using a corpus of 1,000 training documents accurately identified acute long bone fractures from radiology reports. Strategic use of straightforward NLP methods, implemented with freely available software, offers quality improvement teams new opportunities to extract information from narrative documents.
Keywords: Natural language processing, machine learning, quality improvement, pediatrics, emergency medicine
1. Background and Significance
Electronic health records (EHRs) provide opportunities for quality improvement and research that were previously infeasible. Unfortunately, improvements in healthcare quality have not been consistently observed after EHR implementation [1, 2]. One barrier to supporting quality improvement efforts is that significant amounts of information needed for these efforts are only available in a narrative format [3, 4]. Also, data entry in many coded fields, such as diagnoses, may be driven by administrative or billing activities, which may reduce their clinical accuracy [3, 5–7]. Changes in terminology over time, such as the transition from ICD-9 to ICD-10 in the United States, present additional challenges [8].
Radiology reports of diagnostic tests are one particularly rich source of clinical diagnostic information. Several researchers have described methods for extracting information from these reports using natural language processing (NLP) [9–11]. Unfortunately, despite the potential of NLP, clinical researchers and quality improvement teams have not broadly adopted these methods. This may be because reliable NLP methods are relatively new, and also in part due to a perception that NLP methods are complex and should not be used without highly specialized experts specifically trained in these methods. For example, sophisticated NLP systems typically require user input to optimize text processing steps such as correction of spelling errors, analyzing document structure, splitting sentences, word sense disambiguation, negation detection, and part-of-speech tagging [18]. At present, even the most sophisticated NLP systems require local adjustments or problem-specific adjustments to ensure information is extracted with sufficient accuracy. Also, the lack of NLP evaluation studies has been cited as an important barrier to implementation [12].
The Pediatric Emergency Care Applied Research Network (PECARN) has constructed a clinical registry of structured and narrative data from electronic health records at seven emergency departments associated with four health systems [13, 14]. Investigators implemented a quality improvement intervention addressing multiple clinical domains using audit and feedback reports derived from this registry. One quality improvement metric involved the pain management for children presenting the emergency department with acute long bone fractures. Concerns regarding the inclusivity of diagnosis code data for fractures resulted in a decision to pursue NLP as a method for more accurately identifying children with long bone fractures. In this manuscript, we describe our approach to identifying long bone fractures in radiology reports to support a multi-site quality improvement effort using NLP and machine learning tools that are widely available at no cost. To achieve our goals, we sought to use readily available software familiar to research teams with typical statistical skills to ensure local experts may remain involved to maintain the system over time.
2. Objective
Our objective was to implement and prospectively validate NLP methods for identifying long bone fractures in radiology reports to support a pediatric emergency medicine quality improvement project.
3. Methods
We developed NLP methods to identify long bone fractures using radiology reports for children treated in seven pediatric emergency departments associated with 4 health systems and using two distinct EHR vendors. The emergency departments involved with the registry include four pediatric emergency departments within academic children’s hospitals and three satellite community pediatric emergency departments each affiliated with one of the main emergency departments. The annual census of the sites ranged from 29,735 to 92,568 patient visits with 878,349 total patients visits over the two-year study period. As our intent was to use these methods in a quality improvement endeavor that would provide audit and clinician feedback regarding long bone fracture care, we required an NLP algorithm that identified long bone fractures with high precision (positive predictive value). Our pre-specified performance goals for the algorithm were a precision of at least 0.95 and recall (sensitivity) of at least 0.8.
3.1 Study population and setting
We included de-identified radiology reports for children aged 0 to 18 years receiving treatment at any of the seven pediatric emergency departments affiliated with the four health systems participating in the PECARN Registry between 1/1/2013 and 12/31/2014. The PECARN Registry data resides in a centralized database at the DCC. This registry contains data extracted from electronic health records and includes comprehensive emergency department disease, treatment, and outcome information [13, 14]. All narrative documents, including radiology reports, were de-identified using an automated process at each study location prior to submission to the DCC [15, 16].
Our study was conducted in two phases. In the first phase we developed NLP methods using a sample of radiology reports collected between 1/1/2013 and 7/31/2014. In the second phase we prospectively validated the ongoing performance of these methods using a sample of radiology reports collected between 8/1/2014 and 12/31/2014. The accuracy of coded encounter diagnoses for identifying long bone fractures was also assessed in this phase (relevant ICD-9 codes are listed in ► Table 1). All analyses were performed in R version 3.1.3 [17]. The following additional R packages were required: “glmnet” (regularized logistic regression), “e1071” (support vector machine), “randomForest” (random forest algorithm), “tm” (text mining), “SnowballC” (word stemming), and “RWeka” (multiple machine learning tools).
Table 1.
Complete list of ICD-9 codes for long bone fractures.
| ICD-9 Code | Description |
|---|---|
| 733.11 | Pathologic fracture of humerus |
| 733.12 | Pathologic fracture of radius or ulna* |
| 733.14 – 733.15 | Pathologic fracture of femur* |
| 733.16 | Pathologic fracture of tibia or fibula* |
| 733.93 | Stress fracture of tibia or fibula |
| 733.96 – 733.97 | Stress fracture of femur |
| 810.0 – 810.3 | Fracture of clavicle |
| 812.00 – 812.59 | Fracture of humerus |
| 813.00 – 813.93 | Fracture of radius or ulna |
| 818.0 – 818.1 | Ill-defined fractures of upper limb† |
| 819.0 – 819.1 | Multiple fractures of upper limbs† |
| 820.00 – 821.39 | Fracture of femur |
| 823.00 – 823.92 | Fracture of tibia or fibula |
| 824.0 – 824.9 | Fracture of ankle (malleolus) |
| 827.0 – 827.1 | Ill-defined fractures of lower limb† |
| 828.0 – 828.1 | Multiple fractures of lower limbs† |
* Pathologic fractures were not the focus of the pain management quality improvement project, but were included in our criteria for this NLP project. However, there were no occurrences of these codes in our test sample of radiology reports.
† We chose to include these ICD-9 codes in our criteria for this NLP project. It is possible these codes may be used in situations where no long bone fracture occurred. In our test sample of radiology reports there was one occurrence of the code 827.0, which was used to describe a fracture of the tibia and fibula.
3.2 NLP development phase
The following sections describe our methods for selecting a training sample of radiology reports, pre-processing the reports to construct feature vectors, developing the NLP models, and visualizing the behavior of these models. Resource constraints were important considerations throughout our project. These constraints were particularly important in the NLP development phase. We needed to invest effort in manually reviewing documents to establish a training corpus of reasonable size before any additional work was possible. We chose our initial training sample (N=1,000 training documents) based on what the project team considered reasonable and feasible with a plan to re-evaluate based on “learning curves,” which are discussed in the subsequent sections.
Selection of radiology reports enriched in long bone fractures
We used orders for ketamine—the preferred procedural sedation agent for long bone fracture reduction at all study sites—as a marker to identify visits more likely to be associated with long bone fractures where pain management was required. From these visits, we extracted a random sample of 500 plain film radiograph reports. Another sample of 500 reports was extracted from visits where there was no ketamine order. These 1000 reports, which included all types of plain film radiographs (e.g. extremity, chest, abdomen, etc), were manually reviewed and independently labeled by two clinician authors (RG and EA) to identify the subset where an acute long bone fracture was present. The reference standard definition of a radiology report positive for long bone fracture was the description of an acute long bone (clavicle, humerus, radius, ulna, femur, tibia, or fibula) fracture on any plain film radiograph examination. Reports related to healing fractures, fractures of other non-long bones, or those without mention of any long bone fractures were considered negative. When managing fractures in the pediatric emergency department setting, the treatment team must ultimately decide whether or not a child has a fracture. Consequently, reports with ambiguous phrases (e.g. “possible fracture,” and “fracture versus normal variant”) were reconciled by consensus of the two clinician reviewers as either positive or negative in a binary fashion based on their impression of whether fracture treatment was required based on the findings described in the radiology report.
Radiology report pre-processing
Our approach to converting radiology report text into a format (vector of numerical features) suitable as input for machine learning classifiers, involved a series of NLP steps derived from successful approaches described in prior manuscripts [9, 18]. We used a sequence of regular expressions—a text pattern matching technique—to segment each radiology report into the four sections: (1) clinical history, (2) description of comparison films, (3) findings, and (4) impression. These regular expressions, along with all the code developed for this project, are included in the online appendix. When present, we used text from the “findings” section for the NLP analysis. Text from the summary “impression” section was used for the NLP analysis only when there was no section describing the detailed study findings. We excluded text from the clinical history and comparison film from further analysis, because these sections frequently described previous fractures or clinical concerns for fracture and decreased the precision of the NLP algorithm.
Document normalization
To reduce feature scarcity and improve algorithm performance, we normalized radiology reports by replacing all specific anatomic references to long bones with the word stem “longbon.” We used regular expressions to identify terms such as “clavicle,” “clavicular”, “humerus,” “supracondylar” and “femur” that relate to long bones or specific regions of long bones. We were concerned that the numerous descriptions of hand and wrist bones in anatomic relation to forearm long bones would degrade the usefulness of long bone terms as features in our models. Therefore, we also normalized these terms by replacing them with the word stem “handbon.” We also replaced all sequences of numeric digits with the letter “N.” We then removed a limited set of English language “stop words” that occurred commonly in many radiology reports such as “the” and “a” [19]. All negation terms were retained (e.g. “no” and “not”).
Feature construction
Each document was tokenized into individual words with the RWeka library, and word stemming (reduction of inflection or derived words to root form) was performed with the Snowball algorithm [20, 21]. We included these word stems and bi-grams constructed from those stems (i.e. N-grams of length 2) as candidate features in our models. We created a binary feature matrix for each document where matrix element m,n = 1 or 0 indicated the presence or absence, respectively, of word or bigram n in document m. The resulting matrix had 9,908 features, which proved to be computationally intractable for fitting several of the models used in this project. We therefore excluded the 9,235 features that were present in less than 1% of documents to reduce dimensionality and improve computational efficiency during the model construction tasks. In the case of the random forest algorithm, model fitting with the reduced set of features was computationally 30-fold faster, which greatly facilitated cross-validation tasks. The remaining features that were present in at least 1% of documents were saved in a dictionary for use during the testing phases.
Model construction
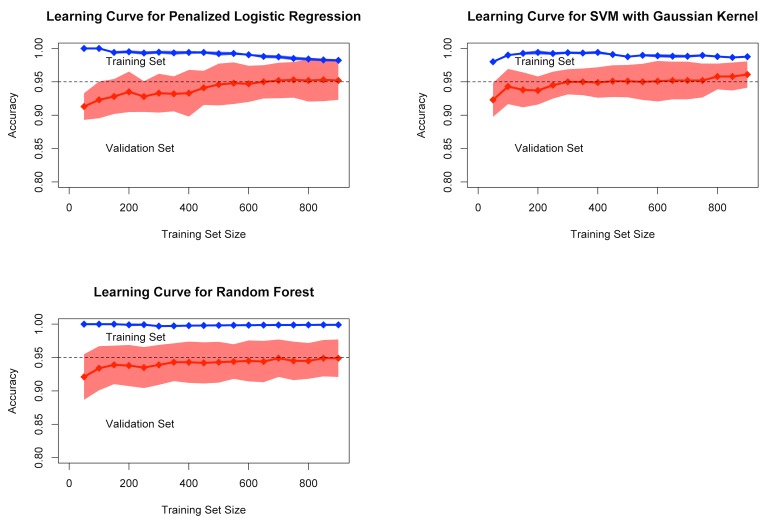
We evaluated the performance of three machine learning classifier models: one with a linear decision boundary (regularized logistic regression), and two with non-linear decision boundaries (support vector machine with a Gaussian kernel, and random forests) [22, 23]. Using the documents in the training corpus, optimal model configuration parameters (e.g. regularization constant) were selected using available cross-validation functions in R (“cv.glmnet” for the regularized logistic regression, “tune.svm” for the support vector machine, and “tuneRF” for the random forest model). To ensure the models were appropriately fit to the training data, learning curves were plotted for each model with model accuracy, (true positives + true negatives) / number documents, as the outcome. To construct these plots, we first stratified the training corpus by the document label determined from manual chart review (fracture present vs. absent). Within these strata we then partitioned the training data into 10 groups of equal size (100 documents each) and constructed 10 learning curves per model using each group as a hold out validation set for one of the learning curves. In each learning curve, documents from the other nine groups were sequentially added. With each addition of training documents, model coefficients were fit to the subgroup of training data, and the accuracy of the model was measured against both the documents used to train the model as well as the 100 held-out validation documents. Consistently high accuracy on the training samples in the learning curves with poor accuracy on the validation samples implies high variance (i.e. the model is “over-fit” to the training data). Poor accuracy on the training samples typically indicates bias (i.e. the model is “under-fit”). We also inspected the slope of the learning curves to determine whether expanding our training corpus beyond the initial sample of 1,000 manually reviewed documents was likely to yield higher accuracy.
Model visualization
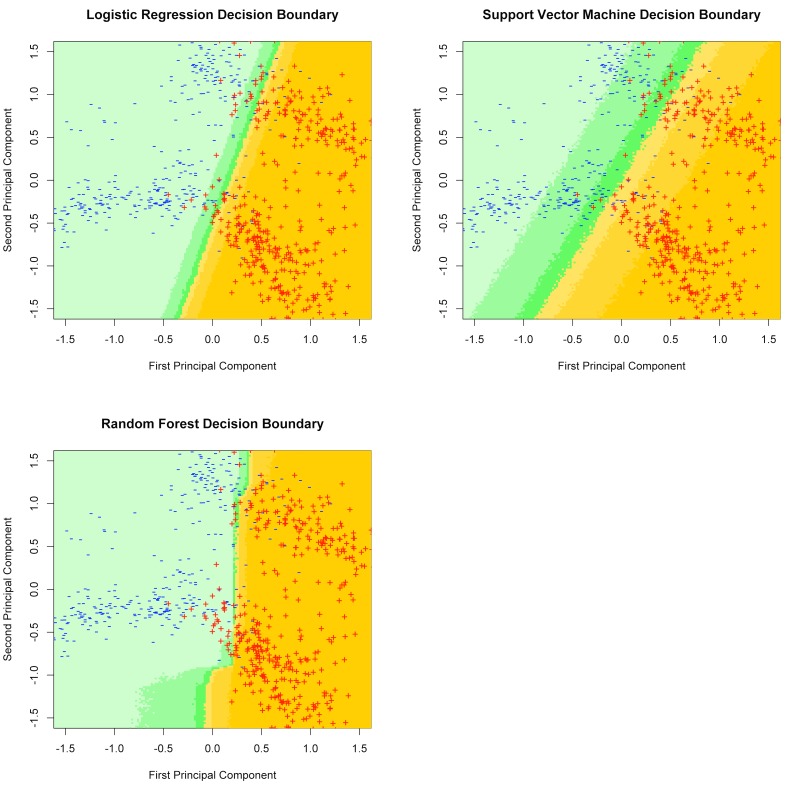
For the purposes of illustration and to understand potential differences between the models, we visualized the decision boundary where a document would transition from being classified as positive for a long bone fracture to negative. To construct these figures we transformed the matrix of features using principal component analysis. Each component was normalized to have a mean of zero and variance of one. We then fit each of the models to the transformed data and visualized the predicted proportion of positive documents occurring at each point along the first two principal coordinates, assuming independent normal distributions for the remaining components.
3.4 NLP testing phase
After selecting optimal features and model parameters for the three models, a new set of 500 radiology reports was extracted from the time period after the model was constructed (8/1/2014 to 12/31/2014). Similar to our approach in developing the training corpus, we ensured adequate numbers of positive fractures were available in the test corpus of radiology reports by selecting 250 reports from encounters where ketamine was used, and 250 from encounters where it was not used. The same authors who reviewed the training documents (RG and EA) independently reviewed the test set reports to identify the presence of acute long bone fractures. The Kappa statistic for inter-rater agreement was calculated based on the authors’ initial review, and disagreements were resolved by consensus to establish the reference standard. These new reports were pre-processed using the same steps described previously with the exception that the feature matrix was constructed using the dictionary of word stems and bi-grams selected for the training documents. Consequently, novel features in the test set that were not seen in the training set did not contribute to classification. The models were then used to predict whether an acute long bone fracture was documented. Recall, precision and F1 score were measured for each model. We used bootstrap sampling (10,000 iterations) to estimate confidence intervals for each performance statistic, and to calculate the two-tailed p-value for the difference between each model’s performance and the performance of diagnosis codes.
3.5 Sensitivity analyses
We performed two sensitivity analyses to better understand the performance of the machine learning classification models. For our first sensitivity analysis, we calculated accuracy, recall, precision and F1 score for the classification models within each of the four healthcare systems participating in this project. Site level analysis (N=7 sites) was not feasible due to small numbers of radiology reports describing long bone fractures available in the test corpus from some of the smaller pediatric emergency departments.
As a second sensitivity analysis, we sought to determine whether the pre-processing steps were truly necessary. For this analysis we repeated the process of building the three machine learning models, but without segmenting the documents to extract the findings or impression portion of the document as described previously in the radiology report pre-processing section. We also omitted the step of replacing references to specific long bones and hand bones as described in the section on document normalization. We compared model performance on the test set both re-using the model configuration parameters established during the original model construction tasks, as well as after re-tuning these parameters using the features that resulted from omitting the pre-processing steps.
4. Results
Manual review of 1,000 de-identified radiology reports in the training corpus identified 454 (45.4%) with acute long bone fractures. After pre-processing the text and constructing features, there were 329 word stems and 344 bigrams in at least 1% of the radiology reports. The most frequent examples are shown in ► Table 2. For our logistic regression model (glmnet), the optimal value for the regularization parameter lambda was 0.01, for the support vector machine with Gaussian kernel (SVM) the optimal parameters were gamma=0.005 and cost=4, and for random forests the model default parameters were optimal (ntree=500 and mtry=25, which is the square root of the number of features). Measures of variable importance for selected terms based on the regularized logistic regression and random forest models are shown in ► Table 3. As shown, some variables that were highly predictive in the logistic regression model had relatively low importance in the random forest model.
Table 2.
Top ten most frequent word stems (as constructed by the Snowball algorithm) and bigrams (sequential pairs of word stems) in the 1000 training documents. The percent of documents with at least one occurrence of each term is reported by category of document (acute long bone fracture present vs. absent). Note that some of the most frequent terms do not specifically relate to fractures.
| Percent of Documents | Percent of Documents | ||||
|---|---|---|---|---|---|
| Word Stem | Fracture | No Fracture | Bigram | Fracture | No Fracture |
| fractur | 96% | 41% | soft tissu | 39% | 37% |
| longbon* | 98% | 14% | distal longbon* | 54% | 3% |
| normal | 22% | 66% | tissu swell | 27% | 14% |
| distal | 76% | 10% | longbon longbon* | 37% | 3% |
| tissu | 39% | 37% | longbon fractur* | 32% | 1% |
| soft | 39% | 37% | fractur distal | 28% | 1% |
| align | 38% | 13% | fractur fragment | 24% | 1% |
| seen | 22% | 25% | pleural effus | 0% | 20% |
| left | 22% | 22% | displac distal | 19% | 1% |
| displac | 43% | 4% | joint effus | 8% | 8% |
*The term “longbon” was introduced during the normalization process to replace references to specific long bones (e.g. tibia, fibula) or specific portions of long bones (e.g. olecranon).
Table 3.
Measures of importance for the word stems and bigrams. The ten terms with the most positive and negative coefficients from the regularized logistic regression model are shown alongside the Gini importance measure from the random forest model, which approximates the permutation importance of the variable.
| Word Stem or Bigram | Coefficient (Regularized Logistic Regression) | Gini Importance (Random Forest) | Percent of Documents |
|---|---|---|---|
| longbon* | 3.00 | 61.76 | 52.1% |
| fractur | 1.79 | 18.71 | 66.0% |
| close reduct | 1.44 | 0.52 | 2.1% |
| cast | 1.06 | 7.75 | 12.4% |
| distal | 0.89 | 32.41 | 40.1% |
| distal forearm | 0.79 | 0.70 | 1.3% |
| through | 0.72 | 3.28 | 8.6% |
| metaphysi | 0.72 | 1.49 | 4.4% |
| angul | 0.66 | 16.78 | 19.4% |
| buckl | 0.62 | 1.95 | 4.0% |
| left elbow | 0.58 | 0.16 | 1.2% |
| fractur disloc | -0.61 | 1.57 | 5.8% |
| normal | -0.73 | 11.73 | 45.9% |
| handbon* | -0.73 | 2.36 | 5.9% |
| injuri | -0.77 | 0.51 | 1.4% |
| no visibl | -1.03 | 2.50 | 8.0% |
| heal | -1.22 | 0.73 | 1.8% |
| acut fractur | -1.30 | 0.58 | 2.0% |
| Nth* | -1.31 | 0.55 | 1.2% |
| no fractur | -1.75 | 3.41 | 7.3% |
| proxim handbon* | -2.31 | 1.31 | 1.7% |
*The terms “longbon,” “handbon,” and “Nth” were introduced during the text normalization process.
4.1 Learning curves and cross-validation accuracy
The learning curves for all three models with these parameters showed that good predictive performance was achieved on the validation set with only 400 to 500 documents for training. There were only slight increases in accuracy as additional documents were added to the training set (► Figure 1). In 10-fold cross-validation using all available training documents, the models all achieved similar, high accuracy. Across the 10 folds of cross-validation, the glmnet model had a mean accuracy of 0.952 (SD= .029), SVM had a mean accuracy of 0.961 (SD=.020), and the random forest model had a mean accuracy of 0.949 (SD=.028).
Fig. 1.
Learning curves showing performance on the training set (blue) and validation set (red). Mean accuracy and standard deviation (shaded area) are shown for 10-fold cross-validation for each of the three models.
4.2 Visualization of decision boundary
After plotting documents from the training set in principal component coordinates, a linear decision boundary was visible (► Figure 2). The shaded colors represent the probability that a document mapping to a particular location in the first two principal component coordinates would be classified as a positive document by each model. In these conceptual visualizations that used a dataset with lower dimensionality, the decision boundaries for the linear model (glmnet), and non-linear models (SVM and random forests) were remarkably similar. However, it is possible that model behaviors may differ in a higher dimensional space.
Fig. 2.
Illustration of decision boundaries for each model in principal component coordinates. Color shading indicates the proportion of documents projecting to each area in principal coordinates that are coded as “positive for long bone fracture” by each model.
4.3 Test set performance
The prospectively collected test set of 500 documents contained 225 (45%) positive reports describing long bone fractures. There were only three disagreements between the two reviewers (kappa=0.988). After discussion between the reviewers, all three reports were coded as positive by consensus. Using the results of our manual review as the reference standard, ICD-9 CM diagnosis codes for these patient records identified presence of a long bone fracture in radiology reports with 93.2% accuracy (recall 0.96, precision 0.896, F1 score 0.927). All three NLP models achieved high levels of accuracy, recall, and precision on the prospectively collected test set. The performance statistics of our NLP models in this analysis were not different from the performance of diagnosis codes at traditional levels of statistical significance (See ► Table 4, model performance with pre-processing). Recall (sensitivity) for diagnosis codes was slightly higher than regularized logistic regression (recall 0.951), but was lower than the other models. Overall SVM performed best with an accuracy of 95.8% (recall 0.969, precision 0.94, and F1 score 0.954). Salient examples of radiology reports with the most positive or negative coefficients in the support vector machine are included in the online appendix.
Table 4.
Performance statistics and 95% confidence intervals of the three models with and without pre-processing (document segmentation and targeted word replacement for long bones and hand bones) on the 500 test documents compared to the performance of ICD-9 coded emergency department diagnoses.
| Accuracy | Recall | Precision | F1 Score | |
|---|---|---|---|---|
| ICD-9 Codes | 0.932 [0.904, 0.950] |
0.960 [0.927, 0.981] |
0.896 [0.854, 0.931] |
0.927 [0.899, 0.949] |
| Model performance with pre-processing | ||||
| Logistic Regression | 0.950 [0.926, 0.966] |
0.951 [0.916, 0.974] |
0.939 [0.900, 0.965]† |
0.945 [0.920, 0.964] |
| Support Vector Machine | 0.958 [0.934, 0.972]† |
0.969 [0.938, 0.987] |
0.940 [0.903, 0.966]† |
0.954 [0.931, 0.971]† |
| Random Forest | 0.950 [0.926, 0.964] |
0.973 [0.945, 0.991] |
0.920 [0.881, 0.950] |
0.946 [0.922, 0.964] |
| Model performance without pre-processing | ||||
| Logistic Regression | 0.954 [0.930, 0.968] |
0.951 [0.916, 0.974] |
0.947 [0.911, 0.971]* |
0.949 [0.924, 0.967] |
| Support Vector Machine | 0.960 [0.938, 0.974]* |
0.960 [0.928, 0.981] |
0.952 [0.916, 0.974]* |
0.956 [0.933, 0.972]* |
| Random Forest | 0.958 [0.936, 0.972]† |
0.978 [0.950, 0.991] |
0.932 [0.892, 0.960]† |
0.954 [0.931, 0.971]† |
*Difference in performance compared to coded diagnoses was statistically significant with p<0.05
†Difference in performance compared to coded diagnoses approached statistical significance with p<0.1, but 0.05
4.4 Sensitivity analyses
Omitting the pre-processing steps related to document segmentation and targeted word replacement for bone names yielded a small, but unexpected improvement in performance. This improvement was sufficient for several of the performance statistics in this sensitivity analysis to be superior to the performance of diagnosis codes with two-tailed p-values < 0.05 (► Table 4, model performance without pre-processing). Retuning the configuration parameters for each model in this sensitivity analysis had a negligible effect on classification performance (data not shown).
Performance of the three classification models as well as the accuracy of coded diagnoses was generally similar across the four health systems with the exception that precision was lower for all the models at one of the health systems (► Table 5). The precision of coded diagnoses at this same health system was also lower than for the other health systems. Excluding this health system from the analysis improved the precision of the best performing classification model (SVM) across the remaining three health systems to 0.962 and the precision of diagnosis codes to 0.908. Manual review of the documents incorrectly labeled as positive by the classification models at the health system with lowest precision revealed that most of the false positive radiology reports contained the phrase “fracture or dislocation visualized” in the context of negation (e.g. “No pelvis or hip fracture or dislocation visualized”). This phrase structure, which separates the negation term “no” from both the words “fracture” and “dislocation,” did not occur in the radiology reports from any of the other health systems. Other reasons for false positive reports that occurred across all four health systems included hand bone fractures that were described in relation to forearm bones (e.g. “fracture line extending from the ulnar metaphysis of the proximal phalanx”), and the presence of findings that may suggest a fracture (e.g. “irregularity at the medial condylar surface of the distal humerus.”)
Table 5.
Performance statistics of the three classification models as well as diagnosis codes within each of the four health systems.
| Regularized Logistic Regression | Support Vector Machine | Random Forest | Coded Diagnosis | |
|---|---|---|---|---|
| Health System 1 | ||||
| Accuracy | 0.967 | 0.967 | 0.967 | 0.942 |
| Recall | 0.962 | 0.942 | 1.000 | 0.962 |
| Precision | 0.962 | 0.980 | 0.929 | 0.909 |
| F1 score | 0.962 | 0.961 | 0.963 | 0.935 |
| Health System 2 | ||||
| Accuracy | 0.967 | 0.967 | 0.967 | 0.940 |
| Recall | 0.985 | 0.985 | 0.985 | 0.985 |
| Precision | 0.941 | 0.941 | 0.941 | 0.889 |
| F1 score | 0.962 | 0.962 | 0.962 | 0.934 |
| Health System 3 | ||||
| Accuracy | 0.936 | 0.960 | 0.928 | 0.928 |
| Recall | 0.910 | 0.955 | 0.925 | 0.940 |
| Precision | 0.968 | 0.970 | 0.939 | 0.926 |
| F1 score | 0.938 | 0.962 | 0.932 | 0.933 |
| Health System 4 | ||||
| Accuracy | 0.923 | 0.933 | 0.904 | 0.913 |
| Recall | 0.951 | 1.000 | 1.000 | 0.951 |
| Precision | 0.867 | 0.854 | 0.804 | 0.848 |
| F1 score | 0.907 | 0.921 | 0.891 | 0.897 |
5. Discussion
Using widely available free NLP and machine learning software and a corpus of 1,000 training documents, we successfully constructed predictive models to identify radiology reports that describe acute long bone fractures with high recall (sensitivity) and precision (positive predictive value) in a prospective validation experiment. The classification models performed well within each of the four healthcare systems with the exception of one health system where the performance was somewhat lower. Although in our primary analyses the performance of these models was not superior to ICD-9 diagnosis codes at traditional levels of statistical significance, the estimated performance more closely achieved our pre-specified performance criteria (recall 0.8 and precision 0.95). Interestingly, statistical significance was achieved for several performance characteristics in our sensitivity analysis, which involved less pre-processing of the original text. It is possible that the clinical history information that we thought would be distracting (e.g. “rule out fracture”) actually contained important predictors of long bone fracture (e.g. “motor vehicle accident”). Further study is required in larger cohorts to determine whether or not clinical history information, combined with the actual radiograph findings, truly yields better performance.
5.1 Effectiveness of simple NLP methods
Unlike many NLP pipelines, the approach we used was comparatively simple and did not require dictionaries of coded medical terms for named entity recognition, negation detection algorithms, or sentiment analysis. Furthermore, the learning curves for all the models we tested achieved maximum performance with fewer than 500 documents, suggesting these simple NLP approaches may provide value even with a smaller corpus of documents than was used in our study. In sensitivity analyses the classification models performed similarly well even with further simplification of our NLP pipeline. Specifically, document segmentation to extract the radiology “findings” section, and targeted word replacement related to the names of specific bones was not necessary to achieve satisfactory performance from the classification models.
These results are consistent with recent research. For example, Jung et al. found that comparatively simple NLP pipelines performed well at information extraction tasks related to pharmacovigilance [32]. Yadav et al. achieved a high degree of accuracy identifying orbital fractures using a decision tree model and raw text word counts as input features [9]. Sevenster et al. extracted measurement information with extremely high accuracy using text pattern matching techniques (recall and precision both > 0.99) [24]. Recently, one innovative team embedded NLP related specifically to long bone fractures in a real-time system within a single health system to improve the quality of decision-making by radiologists (e.g. to recommend additional imaging tests) [25]. Unlike our machine-learning approach, their system used pattern-matching techniques (regular expressions) combined with manually developed rules to extract specific anatomic location information. Our studies had similar accuracy, but differed in our technical approach and clinical objectives.
Nadkarni et al. hypothesized that NLP software may soon be available as a commodity [18]. Towards that goal, sophisticated and freely available NLP packages such as the Apache clinical Text Analysis and Knowledge Extraction System (cTAKES) offer excellent performance [26]. Unsupervised NLP methods that support information extraction tasks without the need for large corpuses of manually annotated text are also becoming available [27]. Hassanpour and Langlotz have recently developed an information extraction approach that extracts the majority of clinically significant information from radiology reports with high accuracy (recall 0.84 and precision 0.87) [28]. Unfortunately these packages have not yet become “commodities” and are typically used only by teams with robust programming talent. Given that significant amounts of important health information are recorded in free text within the EHR, the availability of accessible, user-friendly NLP libraries may vastly increase the amount of clinical information available for quality improvement and research activities.
5.2 Maintenance considerations
Our NLP pipeline is embedded within a quality improvement project that generates monthly feedback reports to pediatric emergency medicine providers regarding pain management of an acute injury. NLP pipelines to support projects of this nature must be simple to run repeatedly over time and must be easy to maintain. Formal evaluation of the maintenance process after implementation for our NLP system was beyond the scope of this study. However, changes in documentation style (e.g. as newly trained radiologists join the workforce or as documentation templates change) and terminology over time—aka “concept drift”—are very likely [29, 30]. We also anticipate the arrival of new study sites that may require additional system modifications to achieve the high level of accuracy required to support quality improvement activities related to the management of long bone fractures. During the progression of our study, we learned several lessons that inform maintenance tasks that will require ongoing vigilance by experts on the project team. These tasks included: (a) periodic assembly and hand labeling of additional documents enriched in radiology reports positive for long bone fractures; (b) manual verification that documentation segmentation continues to function correctly; (c) review our hand-curated synonyms for anatomic terms related to long bones and hand bones to make sure they remain appropriate; (d) periodic re-evaluation of the model output to ensure accuracy meets acceptable thresholds for all study sites; and (e) periodic review of diagnosis code accuracy to assess whether NLP is still required (i.e. in the event that diagnosis code accuracy improves over time, which is of particular interest at our study sites due to the recent adoption of ICD-10 codes).
At present the availability of NLP experts remains inadequate to directly and indefinitely support the ongoing maintenance of information extractions pipelines such as ours. Until such time that truly generalizable information extraction pipelines are available (and continuously maintained) as a commodity product for typical research and quality improvement teams, we feel the construction of a “manual of operations” to support the ongoing maintenance of our NLP pipeline is essential. We do not perceive the need for such ongoing maintenance as indication of a failure of the NLP methods. Instead, this is merely one of numerous components in the ongoing maintenance plan required during the life cycle of any project.
The notion that one can “push a button” to extract information from text is unrealistic. Consequently there is a need for local experts to participate in the development and ongoing maintenance of NLP systems that use machine learning approaches [31]. To ensure readily available members of the project team could adequately maintain our system, we used NLP methods available as standard modules in the R statistical software. These modules are readily accessible, free of charge, and straightforward to use by teams that are familiar with statistical software, even by those who have never previously used NLP. Although we ultimately chose to implement the highest performing model for our quality improvement project (support vector machine with Gaussian kernel), the logistic regression model performed nearly as well. Logistic regression with regularization (penalization of high coefficients to avoid over-fitting) is particularly appealing because of its ease of use and familiarity among most research and quality improvement teams. Additional research is required to monitor the accuracy of NLP over time and to measure the burden of maintenance.
5.3 Limitations
This NLP project was narrowly focused on extracting a single type of information – presence of an acute long bone fracture – from radiology reports completed in the pediatric emergency departments of four academic health systems. Our system does not extract information regarding which long bone was fractured. Additionally, we purposefully enriched our study cohort with documents describing fractures that required procedural sedation for reduction. Consequently less painful fractures, such as small avulsion fractures, were likely under-represented in our corpus of study documents, which may limit our NLP pipeline’s ability to identify these fracture types. We attempted to mitigate this limitation by including an equal number of radiology reports from ED visits where no procedural sedation occurred. Due to our use of an enriched cohort of documents the performance of our NLP pipeline will likely have different performance characteristics than would be observed in a completely random cohort. Because there are differences in radiology documentation style at other health systems or for different types of clinical problems, the relatively simple NLP pipeline we developed may perform differently in other health systems, and could be less successful at extracting other types of information. Even within our own experiment the observed performance of the pipeline varied across the four health systems. We observed a few occurrences of complex phrase structures at one particular health system, which would likely require more sophisticated algorithms (e.g. ConText or cTAKES) to improve accuracy beyond that of our simpler approach [26, 32]. Notably, the accuracy of ICD-9 diagnosis codes also varied across these health systems. Also, our study was not designed to test performance over time; it is possible that the performance of this algorithm could degrade over time as radiology documentation styles change, and it is possible in the future that newer diagnosis coding systems such as ICD-10 may outperform our NLP pipeline. Two clinicians who were not radiologists manually review the radiology reports to establish our reference standard. We thought this approach was most appropriate for our study because the decision to treat a child’s fracture is made by members of the treatment team who are not radiologists. However, it is possible that radiologists would have coded the reports differently.
6. Conclusions
Standard NLP methods packaged in freely available software can be used to construct a simple pipeline that accurately identifies acute long bone fractures from narrative radiology reports. In the context of a project to improve the quality of pain management for children with long bone fractures, the estimated performance of NLP more closely achieved our pre-specified criteria than ICD-9 coded diagnosis. This NLP performance was achieved without using more sophisticated tools such as medical dictionaries, negation detection algorithms, or sentiment analysis. Strategic use of NLP methods offers the potential to make use of unstructured narrative documents in quality improvement and clinical research efforts.
7. Knowledge Assessment
Question 1. What phenomenon is indicated by consistently high accuracy on learning curves for training data coupled with poor accuracy on validation samples?
Feature bias
Data variance
Sample independence
Sample dependence
Preferred Answer: B. Data variance
Data variance describes situations where predictive models are “over-fit” to the available training data. In this situation the model easily fits or “memorizes” the training data and produces high accuracy when used to perform predictions on the training data. Unfortunately, in this situation the model is not adequately generalized to accurately classify new validation samples that were not present in the training set. This occurs when the model has sufficient degrees of freedom (i.e. learnable parameters) to be sensitive to variance in the data that may arise because the data generating process contains a random component or if the sampled data set does not reflect the true population distribution. Many additional techniques are often applied to avoid over fitting including cross-validation, regularization, early stopping, pruning, and Bayesian priors.
Feature bias refers to situations where predictive models are “under-fit” to the available data and yields poor accuracy for both the training and validation samples. Sample dependence vs. independence refers to situations where the class of some samples may depend on other samples, which can reduce the effective sample size (e.g. if members of a sample are highly correlated or similar), but this issue does not typically cause the pattern of accuracy described in the question.
Question 2. Which of the following is the most likely barrier to using electronic health record (EHR) data in quality improvement efforts?
Vendors rarely provide tools to extract data
The amount of data in the EHR is unmanageable
Necessary information is often in free text format
Patient consent is required to use EHR data
Preferred Answer: C. Necessary information is often in free text format
There are significant amounts of codified information in electronic health records that are produced by order entry and billing activities. This codified data may be sufficient to determine some cohorts or outcomes related to quality improvement. However, crucial information to assure accuracy may reside in the free text documentation.
Although there are challenges in extracting and using electronic health record data, vendors typically provide tools or services related to data extraction as part of their product portfolio. The amount of data can be overwhelming, but there exist many tools and robust databases to help manage large EHR datasets. Although patient consent is often required for participation in research, it is typically not required for quality improvement activities or may not be required for research use of de-identified data from the electronic health record.
8. Clinical Relevance Statement
Important information to support healthcare quality improvement is often recorded in free text documents. Natural language processing may help extract free text information, but these methods have rarely been applied beyond the laboratories where they were developed. As part of an effort to improve the quality of pain management for children with long bone fractures, we successfully implemented simple NLP methods using freely available software to identify acute long bone fractures from radiology reports with high accuracy.
Acknowledgements
We wish to acknowledge Evaline A Alessandrini, MD, MSCE, Lalit Bajaj, MD, MPH, and Marc H Gorelick, MD, MSCE for their work establishing the PECARN Registry. We also acknowledge Mar-lena Kittick, MPH for her tireless assistance coordinating PECARN Registry activities and preparing this manuscript.
Funding Statement
Funding This project work was supported by the Agency for Healthcare Research and Quality (AHRQ) grant R01HS020270. The PECARN infrastructure was supported by the Health Resources and Services Administration (HRSA), the Maternal and Child Health Bureau (MCHB), and the Emergency Medical Services for Children (EMSC) Network Development Demonstration Program under cooperative agreements U03MC00008, U03MC00001, U03MC00003, U03MC00006, U03MC00007, U03MC22684, and U03MC22685. This information or content and conclusions are those of the author and should not be construed as the official position or policy of, nor should any endorsements be inferred by HRSA, HHS or the U.S. Government.
Footnotes
Conflicts of Interest
The authors have no conflicts of interest relevant to the work described in this article.
Protection of Human Subjects
The Institutional Review Boards of all sites and the Data Coordinating Center (DCC) approved this project and waived the requirement for consent from individual children/families.
References
- 1.Black AD, Car J, Pagliari C, Anandan C, Cresswell K, Bokun T, McKinstry B, Procter R, Majeed A, Sheikh A. The impact of eHealth on the quality and safety of health care: a systematic overview. PLoS medicine 2011; 8(1): e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Poelgeest R, Heida J-P, Pettit L, de Leeuw RJ, Schrijvers G. The Association between eHealth Capabilities and the Quality and Safety of Health Care in the Netherlands: Comparison of HIMSS Analytics EMRAM data with Elsevier’s “The Best Hospitals” data. Journal of Medical Systems 2015; 39(9): 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PRO, Bernstam EV, Lehmann HP, Hripcsak G, Hartzog TH, Cimino JJ, Saltz JH. Caveats for the Use of Operational Electronic Health Record Data in Comparative Effectiveness Research 2013; 51: S30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devine EB, Capurro D, van Eaton E, Alfonso-Cristancho R, Devlin A, Yanez ND, Yetisgen-Yildiz M, Flum DR, Tarczy-Hornoch P. Preparing Electronic Clinical Data for Quality Improvement and Comparative Effectiveness Research: The SCOAP CERTAIN Automation and Validation Project. EGEMS; (Washington, DC: ) 2013; 1(1): 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roane TE, Patel V, Hardin H, Knoblich M. Discrepancies identified with the use of prescription claims and diagnostic billing data following a comprehensive medication review. Journal of managed care pharmacy JMCP 2014; 20(2):165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quan H, Parsons GA, Ghali WA. Validity of procedure codes in International Classification of Diseases, 9th revision, clinical modification administrative data 2004; 42(8):801–9. [DOI] [PubMed] [Google Scholar]
- 7.Heintzman J, Bailey SR, Hoopes MJ, Le T, Gold R, O‘Malley JP, Cowburn S, Marino M, Krist A, DeVoe JE. Agreement of Medicaid claims and electronic health records for assessing preventive care quality among adults. The Oxford University Press 2014; 21(4):720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krive J, Patel M, Gehm L, Mackey M, Kulstad E, Li JJ, Lussier YA, Boyd AD. The complexity and challenges of the International Classification of Diseases, Ninth Revision, Clinical Modification to International Classification of Diseases, 10th Revision, Clinical Modification transition in EDs. The American journal of emergency medicine 2015; 33(5):713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yadav K, Sarioglu E, Smith M, Choi H-A. Automated outcome classification of emergency department computed tomography imaging reports. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine 2013; 20(8):848–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedlin J, Mahoui M, Jones J, Jamieson P. Knowledge Discovery and Data Mining of Free Text Radiology Reports. IEEE 2011; 89–96. [Google Scholar]
- 11.Womack JA, Scotch M, Gibert C, Chapman W, Yin M, Justice AC, Brandt C. A comparison of two approaches to text processing: facilitating chart reviews of radiology reports in electronic medical records. Perspectives in health information management / AHIMA, American Health Information Management Association 2010; 7: 1a. [PMC free article] [PubMed] [Google Scholar]
- 12.Hersh W. Evaluation of biomedical text-mining systems: lessons learned from information retrieval. Briefings in bioinformatics 2005; 6(4):344–56. [DOI] [PubMed] [Google Scholar]
- 13.Alpern ER, Alessandrini EA, Casper TC, Bajaj L, Gorelick MH, Gerber JS, Funai T, Grundmeier RW, Enriquez R, Dean JM, Campos DA, Deakyne SJ, Bell J, Hayes KL, Kittick M, Chamberlain JM. Benchmarks in Pediatric Emergency Medicine Performance Measures Derived from an Multicenter Electronic Health Record Registry. Platform Presentation at the Pediatric Academic Societies; San Diego, CA: 2015. [Google Scholar]
- 14.Deakyne SJ, Grundmeier RW, Campos DA, Hayes KL, Cao J, Enriquez R, Bell J, Fahim C, Casper TC, Funai T, Scheid B, Kittick M, Dean JM, Alessandrini EA, Bajaj L, Gorelick MH, Chamberlain JM, Alpern ER. Building a Pediatric Emergency Care Electronic Medical Registry. Poster Session at the Pediatric Academic Societies; San Diego, CA: 2015. [Google Scholar]
- 15.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng CK, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. Lippincott Williams & Wilkins; 2000; 101(23):E215–20. [DOI] [PubMed] [Google Scholar]
- 16.Neamatullah I, Douglass MM, Lehman L-WH, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody GB, Mark RG, Clifford GD. Automated de-identification of free-text medical records. BioMed Central Ltd 2008; 8(1): 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Core Team. R: A Language and Environment for Statistical Computing [Internet]. 3rd ed Vienna, Austria: R Foundation for Statistical Computing; Available from: http://www.R-project.org/ [Google Scholar]
- 18.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. The Oxford University Press 2011; 18(5):544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinerer I, Hornik K. Package “tm” [Internet]. cran.r-project.org 2015. [cited 2015 Sep 10] pp. 34–5. Available from: https://cran.r-project.org/web/packages/tm/tm.pdf.
- 20.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14(3):130–7. [Google Scholar]
- 21.Bouchet-Valat M. Package “SnowballC” [Internet]. cran.r-project.org 2014. [cited 2015 Sep 10]. Available from: https://cran.r-project.org/web/packages/SnowballC/SnowballC.pdf.
- 22.Cortes C, Vapnik V. Support-Vector Networks. Kluwer Academic Publishers 1995; 20(3):273–97. [Google Scholar]
- 23.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010; 33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 24.Sevenster M, Buurman J, Liu P, Peters JF, Chang PJ. Natural Language Processing Techniques for Extracting and Categorizing Finding Measurements in Narrative Radiology Reports. 2015;6(3):600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Do BH, Wu AS, Maley J, Biswal S. Automatic retrieval of bone fracture knowledge using natural language processing. J Digit Imaging 2013; 26(4):709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pathak J, Bailey KR, Beebe CE, Bethard S, Carrell DC, Chen PJ, Dligach D, Endle CM, Hart LA, Haug PJ, Huff SM, Kaggal VC, Li D, Liu H, Marchant K, Masanz J, Miller T, Oniki TA, Palmer M, Peterson KJ, Rea S, Savova GK, Stancl CR, Sohn S, Solbrig HR, Suesse DB, Tao C, Taylor DP, Westberg L, Wu S, Zhuo N, Chute CG. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. The Oxford University Press 2013; 20(e2):e341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasko TA, Denny JC, Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PloS one. Public Library of Science 2013; 8(6): e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artificial Intelligence In Medicine 2016; 66: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widmer G, Kubat M. Learning in the Presence of Concept Drift and Hidden Contexts. Machine Learning. Kluwer Academic Publishers-Plenum Publishers 1996; 23(1):69–101. [Google Scholar]
- 30.Learning under Concept Drift: an Overview. Vol. cs.AI, arXiv.org 2010.
- 31.Holzinger A. Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Inf. Berlin: Springer; 2016; 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports. JBI 2009; 42(5):839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]