Summary
Objective
Pharmacogenomic-guided dosing has the potential to improve patient outcomes but its implementation has been met with clinical challenges. Our objective was to develop and evaluate a clinical decision support system (CDSS) for pharmacogenomic-guided warfarin dosing designed for physicians and pharmacists.
Methods
Twelve physicians and pharmacists completed 6 prescribing tasks using simulated patient scenarios in two iterations (development and validation phases) of a newly developed pharmacogenomic-driven CDSS prototype. For each scenario, usability was measured via efficiency, recorded as time to task completion, and participants’ perceived satisfaction which were compared using Kruskal-Wallis and Mann Whitney U tests, respectively. Debrief interviews were conducted and qualitatively analyzed. Usability findings from the first (i.e. development) iteration were incorporated into the CDSS design for the second (i.e. validation) iteration.
Results
During the CDSS validation iteration, participants took more time to complete tasks with a median (IQR) of 183 (124–247) seconds versus 101 (73.5–197) seconds in the development iteration (p=0.01). This increase in time on task was due to the increase in time spent in the CDSS corresponding to several design changes. Efficiency differences that were observed between pharmacists and physicians in the development iteration were eliminated in the validation iteration. The increased use of the CDSS corresponded to a greater acceptance of CDSS recommended doses in the validation iteration (4% in the first iteration vs. 37.5% in the second iteration, p<0.001). Overall satisfaction did not change statistically between the iterations but the qualitative analysis revealed greater trust in the second prototype.
Conclusions
A pharmacogenomic-guided CDSS has been developed using warfarin as the test drug. The final CDSS prototype was trusted by prescribers and significantly increased the time using the tool and acceptance of the recommended doses. This study is an important step toward incorporating pharmacogenomics into CDSS design for clinical testing.
Keywords: Clinical decision support systems, pharmacogenetics, CPOE, computer-assisted drug therapy, warfarin
1. Background And Significance
Personalized medicine is an emerging field with the fundamental objective to predict a patient’s response in order to individualize drug therapy for improved medication effectiveness and safety. Pharmacogenomic-guided drug therapy is at the forefront of personalized medicine with over 150 United States Food and Drug Administration (FDA) approved drug labels incorporating pharmacogenomic information.[1] Nonetheless, the translation of pharmacogenomic testing into routine clinical practice has been slow due to several obstacles towards its implementation [2–4]. Currently, the routine use of clinical pharmacogenomics is limited to centers with resources to overcome these obstacles [5–9].
A major obstacle in the implementation of clinical pharmacogenomics is the rapid pace that science has identified genetic polymorphisms that can predict drug response. This large amount of data can be overwhelming for clinicians to decipher. Therefore, the implementation of personalized medicine has largely relied on mechanisms to collect and interpret large amounts of information through computerized systems, such as clinical decision support systems (CDSS); defined as any tool or system which provides filtered clinical and patient-related information to clinicians or patients in order to assist in patient care [10, 11]. However, there have been limited assessments of CDSSs on perspectives, usability, and user satisfaction with no data available in pharmacists despite their role in personalized medicine [12, 13].
Warfarin is a commonly prescribed drug with pharmacogenomic implications due to its narrow window for effectiveness and high incidence of bleeding complications [14]. The variability in warfarin response is partially due to genetic differences in the cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase complex subunit 1 (VKORC1) [15]. Pharmacogenomic tests may help clinicians manage warfarin more effectively and minimize the risk of serious bleeding [16, 17]. In August 2007, the United States FDA approved a change in warfarin labeling to incorporate information related to how CYP2C9 and VKORC1 genetic variability can impact appropriate warfarin dosing and response [18].
While a routine genotype-guided warfarin dosing service has been shown to be feasible from a procedural standpoint, there has been limited incorporation into clinical practice [19, 20]. An important setback is the fact that there are conflicting reports regarding the effectiveness of incorporating genetic testing into warfarin dosing [21, 22]. The aforementioned barrier in the complexity of the genetic output also presents clinical challenges given the large amount of data [17, 19, 23]. While less complex pharmacogenomics-guided CDSSs have been implemented, more complex genetic and clinical factors need further refinement. This is particularly true with warfarin due to additional factors, such as diet, concurrent medications, and medical indication that alter dosing requirements. These factors complicate prescribers’ decisions, especially on how to dose the medication and make it difficult to further assess the impact of genetic information [24].
While some CDSS assessments have applied pharmacogenomics, these were often limited in scope and generally did not recommend a specific dose [25, 26]. To develop CDSS interfaces, a number of methods have been identified. One such method is simulation, or scenario-based testing which allows for controlled testing of specific situations without risk of patient harm and can be used in situations where application is currently limited, such as pharmacogenomics [27]. The objective of this research was to develop a CDSS prototype to help pharmacists and physicians incorporate pharmacogenomic results into clinical decisions using warfarin as a model drug. Following development, scenario-based testing to assess the usability of the CDSS tool was performed. This paper examines the results of two iterations of CDSS tool development and testing.
2. Methods
2.1 Setting
This study was conducted at Wishard Hospital and in the Human-Computer Interaction and Simulation Laboratory at the Richard L. Roudebush Veterans Affairs (VA) Medical Center in Indianapolis, IN [28]. This research conforms to the Statement on reporting of evaluation studies in Health Informatics (STARE-HI) guidelines [29].
2.2 Prototype Design and Redesign
The prototype CDSS was developed as a computer-based application using the dosing algorithm provided by a federally funded website (warfarindosing.org) and designed to incorporate pharmacogenomics information into warfarin dosing [30, 31]. Two CDSS interface prototypes were developed (development and validation iterations) by a team which included a pharmacogenomics expert, a human factors engineer, informatics experts, and pharmacists. In the development iteration, clinicians were shown screenshots of the potential CDSS to obtain input prior to testing. During the validation iteration, the results from the development iteration were used to revise the interface. The CDSS was incorporated into a high fidelity prototype of the VA Computerized Patient Record System which is the VA version of an electronic health record (EHR), which is used by all healthcare providers, including pharmacists. The presentation of information within the EHR is the same regardless of the user.
While the prototypes incorporated the same information as warfarindosing.org, information was grouped and broken into tabs to reduce potential information overload. Additionally, the prototypes included previous INR measures with historical dosing, and a comment box to document additional information not captured by the algorithm which may impact warfarin dosing, such as a diet. In the prototype EHR system, the CDSS triggers when a prescriber places a new order or adjusts warfarin dosing for a patient. The CDSS has four tabs that can be accessed. First, the ‘Genetic Considerations’ tab displays the polymorphism results of genetic tests for CYP2C9 and VKORC1. Second, ‘Clinical Considerations’ has patient information such as age and warfarin indication. Third, the ‘INR/Dose History’ shows the dates and results of the patient’s International Normalized Ratio (INR) tests. Fourth, ‘Notes’ allows the prescriber to enter notes regarding the patient and/or their dosing. The mock EHR system was purposely designed with limited ordering functionality, and users could only make modifications to a current warfarin order or initiate a new warfarin order. The usability of an initial and revised version of the CDSS prototype was evaluated in two iterations approximately one year apart to allow for the interface redesign and the data collection was completed prior to publication of conflicting reports related to the effectiveness of pharmacogenomic-guided warfarin dosing in 2013 [21, 22]. Usability findings from the development iteration were used to inform an enhanced CDSS prototype for evaluation in the validation iteration (► Figure 1). ► Table 1 describes the modifications in detail.
Fig. 1.
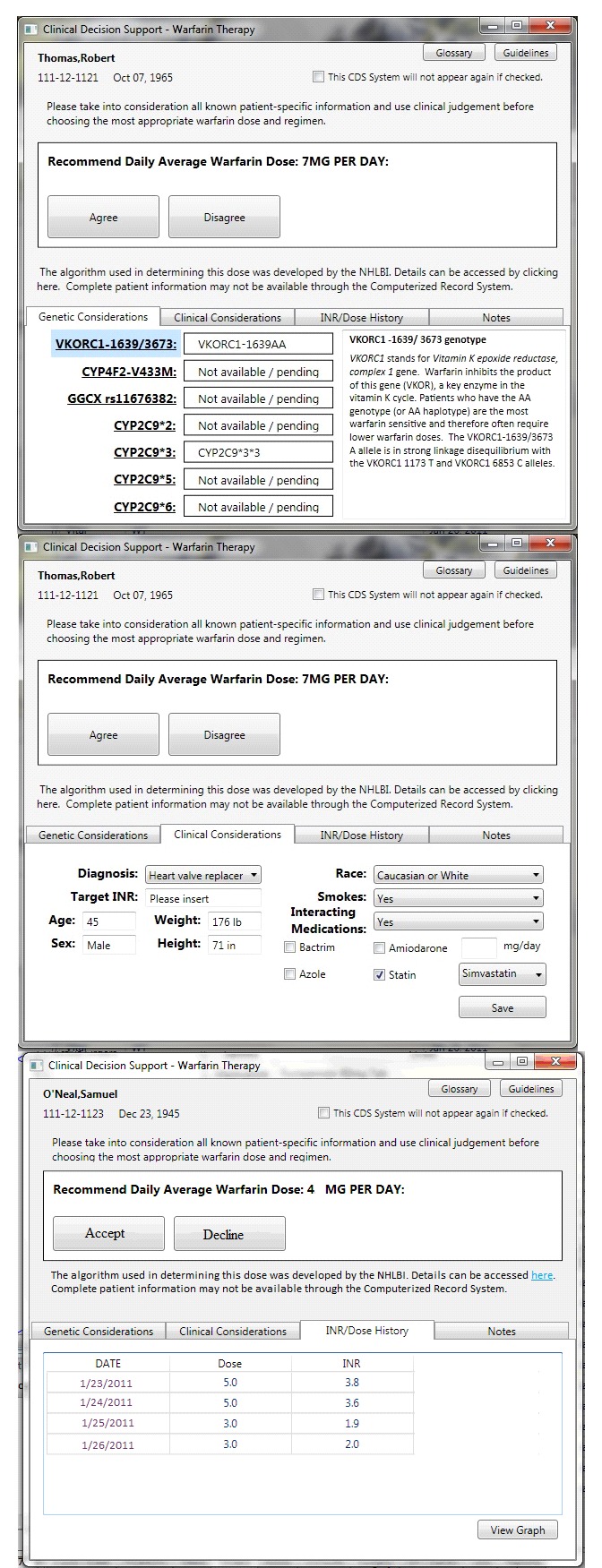
Initial view of the CDSS for fictitious patients upon activation during the validation iteration, showing A) the “Genetic Considerations” tab. B) The “Clinical Considerations” tab. C) The “INR/Dose History” tab. No changes were made to the “Notes” tab where prescribers could enter additional information from the development iteration.
Table 1.
Design Changes between Iterations and Rationale for Changes
| Initial Design Concern | Results from Testing of Initial Design in Development Phase | Design Change and Intended Goal | Results from Testing of Design Changes in Validation Phase |
|---|---|---|---|
| “Consider- ed”/”Not Considered” Buttons | Participants were confused about what other implications (legal liability- etc.) may be tied to “Consider- ed”/”Not Considered” | Change: Adjusted wording on buttons to “Accept”/”Decline” Goal: Reduce confusion and shift button choice to reflect clinical decision | No voiced confusion over “Accept”/”Decline” wording |
| Algorithm explanation “The recommended dose was estimated using clinical and genetic factors as listed in the Dosing Algorithm tab.” | 7 (87.5%) were not sure what the CDSS was using to calculate the dose |
Change: “The algorithm used in determining the dose was developed by the NHLBI. Details can be accessed here.” Goal: Improve transparency of the CDSS |
No participants voiced concerns over general content of the algorithm |
| No link to warfa- rindosing.org | Participants were not sure how to verify CDSS recommended doses |
Change: Link added to warfarindosing.org Goal: Improve transparency of the CDSS and the source of the algorithm |
3 (75%) of participants accessed the warfarindos- ing.org website from the provided link |
| Genotype explanations only found in the Glossary | Participants did not always see the Glossary or recognize it had the desired information |
Change: Titles in “Genetic Considerations” tab are hyperlinks to present explanations to the right of the results Goal: Improve understanding of the genetic results to facilitate clinical decision making |
3 (75%) of the participants expressed that having the genetic information presented was beneficial |
| Liver Disease presented as a yes/no drop down box in “Clinical Considerations” | Participants were unsure what constituted “Liver Disease” |
Change: Liver disease drop down box removed Goal: Reduce confusion and screen clutter |
1 (25%) participant asked if a patient had liver disease |
| Amiodarone presented as a yes/no dropdown box in “Clinical Considerations” | Participant confusion related to interacting medications and if amiodarone was the only one included in dose calculation |
Change: Amiodarone dropdown removed Goal: Reduce confusion related to interacting medications used in algorithm |
No participants voiced confusion over amiodarone |
| No additional interacting medications presented | 7 (87.5%) were uncertain which- if any- other medications were included in the algorithm |
Change: Interacting Medications yes/no dropdown box added; if “yes” selected then check boxes appear for algorithm included interacting medications (► Figure 1B) Goal: Improved transparency related to interacting mediations |
2 (50%) voiced confusion over other possible interacting medications which may be included or not included in the algorithm |
| No interacting medication details | Algorithm includes the amiodarone dose and specific statins in calculation |
Change: Add selection boxes to indicate the dose of amiodarone- and a dropdown list for selecting the specific statin Goal: Improved congruence between algorithm and CDSS content |
No comments regarding amiodarone dose or statins |
| Suggested dose presented on “INR/Dose History” tab | 1 (12.5%) was unsure of the significance of the column |
Change: Suggested dose removed Goal: Reduce confusion and screen clutter |
No participants voiced a need for suggested doses |
| INR presented before dose on “INR/ Dose History” tab | 1 (12.5%) felt prior dose is needed to provide context for INR results |
Change: Columns switched so dose is presented before INR Goal: Format information to reflect participant thought process |
No participants voiced confusion over order of information presentation |
CDSS: Clinical decision support system
NHLBI: National Heart, Lung, and Blood Institute
INR: International normalized ratio
2.3 Study Design
Six simulated patient scenarios representing inpatient care were used in both iterations.. The first three scenarios required the participants to initiate warfarin therapy, while the last three scenarios required a warfarin dose adjustment. All participants received the scenarios in the same order and were instructed that the scenarios were simulated but to complete them as they would in their current practice. The warfarin dosage recommendations from the CDSS were based on the scenarios clinical and pharmacogenomic factors in the algorithm.
Patient scenarios were designed to test various conditions in which the prescriber may order warfarin, including comorbidities, concurrent medications, and genetic test results. Scenarios were designed to mimic patients the participants may encounter in practice without being overly complex (i.e. they did not include excessive superfluous disease states or medication allergies that complicate prescribing). Because warfarin is the only medication they could modify in the system, they were asked to verbalize any other changes they would make to the patient’s medications or laboratory tests they would order to help explain their warfarin prescribing choice. Upon choosing a warfarin dose to order, prescribers moved into the ordering component of the EHR and were presented with a drug-drug interaction alert to resolve before completing the order for each scenario. The drug-drug interaction alert is a part of the current EHR system. The purpose of the drug-drug interactions was to simulate clinical practice to assess usability of the CDSS under situations that were not covered in the algorithm.
2.4 Participant Selection and Recruitment
Participants were recruited from an integrated, university-based medical campus that utilizes an EHR system. A total of 12 unique participants were recruited for the study. Eight participants were recruited for the first iteration of testing and four for the second iteration [32-34]. Practitioners were eligible if they were currently a practicing physician or clinical pharmacist with training or an active practice in the fields of cardiology, anticoagulation, internal medicine or ambulatory care. Residents and fellows were eligible for study participation, but students were not eligible. Participants were eligible even if they had not prescribed warfarin in their current practice to simulate new prescribers. Participants were offered a $50 gift card as compensation. The study was approved by the Indiana University-Purdue University of Indianapolis (IUPUI) Institutional Review Board and each participant provided written informed consent.
2.5 Data Collection and Outcome Measures
One study investigator moderated the usability tests with a standardized introductory script. The purpose of the introductory script was to ensure that users had a consistent baseline understanding of the EHR system and understood the study procedures. Participants used a desktop computer with the EHR and CDSS and an adjacent monitor was used for usability questionnaire administration. As participants completed simulated patient scenarios, their computer screen actions as well as verbal and non-verbal responses were recorded via a webcam and Morae® Software (TechSmith; Okemos MI). Each participant completed a demographic questionnaire. The participant was allowed to review the electronic health record and prescribe or adjust warfarin therapy for the patient. Once they had determined a dose and ordered it, a debrief interview was conducted and the researcher asked about features of the CDSS they found helpful or confusing, and aspects of the CDSS they would like to see changed to assist with prescribing. This procedure was repeated for the remaining scenarios, such that each participant completed all six scenarios. Sessions were designed to take about 30 minutes, but participants could use more time if needed. After completing the scenarios, participants completed the validated 19-item Computer System Usability Questionnaire (CSUQ) which assesses user satisfaction with the system usability on a seven point Likert type scale from strongly disagree to strongly agree [35].
2.5.1 Usability
Usability was assessed through efficiency and satisfaction. Efficiency was assessed in two ways: (1) Time on task – measured from the time the participant clicked to start each scenario to the final signing of the warfarin order (2) Time spent with the CDSS tool – measured from the first time the participant first accessed the CDSS to the final signing of the warfarin order. These definitions were to distinguish time participants spent on the entire scenario including familiarizing themselves with the patient case versus time spent on the using the CDSS prescribing task. Satisfaction was measured via the CSUQ with a 7-point Likert scale (1=Strongly Agree, 7=Strongly Disagree). Debrief interviews were also conducted after each scenario and at the conclusion of the six-scenario session.
2.5.2 Prescribing
Prescribing was assessed to evaluate participant confidence in the CDSS. The frequency of prescribing a standard starting dose of 4mg or 5mg was compared to the frequency of prescribing doses recommended by the CDSS.[36] A dose was considered to be in agreement with the CDSS if the dose ordered is the closest possible dose to the one recommended by the CDSS. For example, if the CDSS recommended a dose of 2.3 mg, and a participant ordered 2mg, it would be considered to agree with the CDSS.
2.6 Data Analyses
Time was manually extracted from Morae® recordings for analysis and quantitative data were analyzed using SPSS v.22. Participant demographics, efficiency, and satisfaction are represented using median and interquartile range (IQR). Mann-Whitney U tests were performed to assess differences in satisfaction between iterations. The Kruskal-Wallis test was used to assess efficiency between iterations and participant groups. The summation of efficiency measures from all scenarios were tested for normality with a Kolmogorov-Smirnov test and compared using the Student’s t-test or non-parametric equivalent. The acceptance of the CDSS recommended dose between iterations was assessed by the Fisher’s exact test. A one-sample t-test was used to compare the average dose prescribed for each scenario versus the CDSS recommended dose in each iteration following a Kolmogorov-Smirnov test for normality. In any instance where normality was not confirmed, a non-parametric equivalent test was used. Alpha was set to 0.05 for all tests.
Video and audio were collected to provide a record of the users’ actions and verbalizations for the qualitative analysis. The recordings were analyzed for confusion and difficulty in meeting task goals. Conclusions were drawn from erroneous assumptions or actions, statements of confusion, time required for tasks, and self-reported measures of satisfaction with the software by completing a usability questionnaire. Participant comments during the scenarios and debrief interview were extracted by a study investigator from the Morae® recordings. These comments were reviewed for reoccurring themes related to strengths and weaknesses that warranted design modifications. Comments from the participants in the development iteration were used to determine CDSS interface changes to be incorporated in the validation iteration.
3.0 Results
3.1 Participant Characteristics
Twelve volunteers completed the study: n=8 (four pharmacists and four physicians) for the development iteration and n=4 (two pharmacists and two physicians) for the validation iteration (► Table 2). The participants represented a range of inpatient and outpatient practices and had been practicing longer than five years. Although most physicians in the study routinely prescribed warfarin in clinical practice, three participants indicated that they have never prescribed warfarin. The participant characteristics between the iterations was not significantly different.
Table 2.
Participant Demographics for Development and Validation Iterations
| Development (n=8) | Validation (n=4) | |
|---|---|---|
| Demographic | n (%) | n (%) |
| Primary practice | Critical care Ambulatory care Geriatrics Internal medicine Infectious disease | Pediatric hematology/oncology Clinical pharmacology Hospital Internal Medicine |
| Sex (male) | 6(75) | 4(100) |
| Physicians | 4(50) | 2(50) |
| Length of practice (years)* | 8.4 (3.1, 10.5) | 7.8 (5.5, 18.5) |
| Self-reported frequency of prescribing warfarin in clinical practice | ||
| Never | 1 (12.5) | 2(50) |
| Less than once a month | 3 (37.5) | 1(25) |
| Once a month | 2(25) | 0 |
| 2-3 times a month | 2(25) | 1(25) |
| Had prior experience with VA EHR | 7 (87.5) | 3(75) |
*Presented as median time (interquartile range)
VA: Veteran’s Affairs
EHR: Electronic health record
3.2 Efficiency
The time to complete each scenario for both the development and validation iteration ranged for individual tasks from 24 seconds to 9.45 minutes. The time to complete tasks requiring warfarin initiation were not statistically different than the dose adjustment tasks in either iteration. The median (IQR) time to complete warfarin initiation versus dose adjustment tasks were 98.5 (55–181) versus 102 (76.3–250.3) seconds, respectively, in the development iteration (p=0.20) and 137 (98.8–247.5) versus 189 (163.3–241.3), respectively, in the validation iteration (p=0.40).
The efficiency measures for the time on task for the sum of all scenarios were statistically longer in the validation phase versus the development phase with median (IQR) of 183 (124–247) versus 101 (73.5–197) seconds, respectively, p=0.01. The increased time to complete the task in the validation phase corresponded to a statistically significant increase in the combined time in the CDSS tool from all scenarios which increased from a median (IQR) of 78 (64–139) seconds in the development phase to 127 (83.5–192) seconds in the validation phase (p=0.02). There was not a statistically significant increase in any individual scenarios with an overall p=0.13 for time in task and p=0.35 for the time in tool from the ANOVA, as displayed in ► Figure 2A and ► Figure 2B, respectively.
Fig 2.
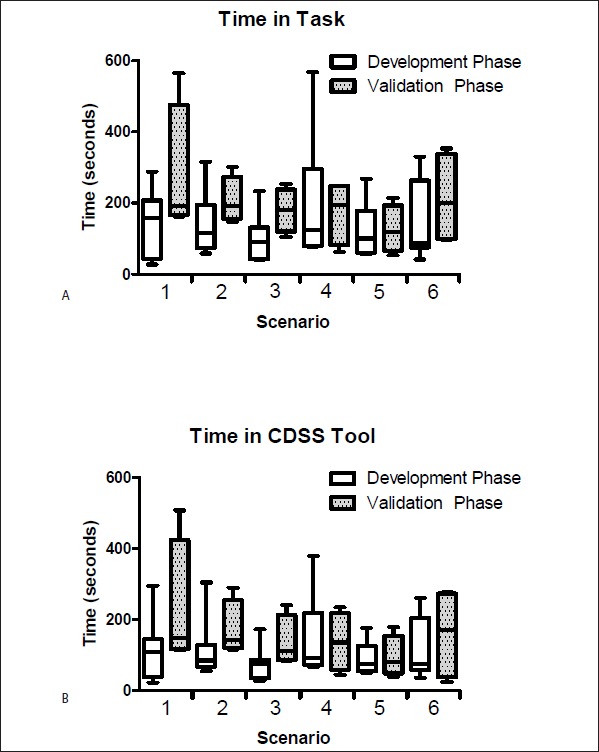
A) Time to complete a task for each individual scenario for all participants in the development (n=8) and validation (n=4) iterations. B) Time spent in the CDSS tool for all participants in the development and validation iterations for each individual scenario. The first 3 tasks were initiating a patient on warfarin; the remaining 3 were adjusting a warfarin dose. Data are presented in seconds as medians with interquartile ranges representing the boundaries of the boxes and whiskers representing the minimum and maximum observed times.
Pharmacists took longer than physicians to complete all tasks (p<0.001) and spent more time in the CDSS tool (p<0.001) for the combined scenarios in the development iteration. However in the validation phase, there was no difference between the time to complete a task (p=0.60) or time in the CDSS tool (p=0.42) between pharmacists and physicians, as displayed in ► Figure 3.
Fig 3.
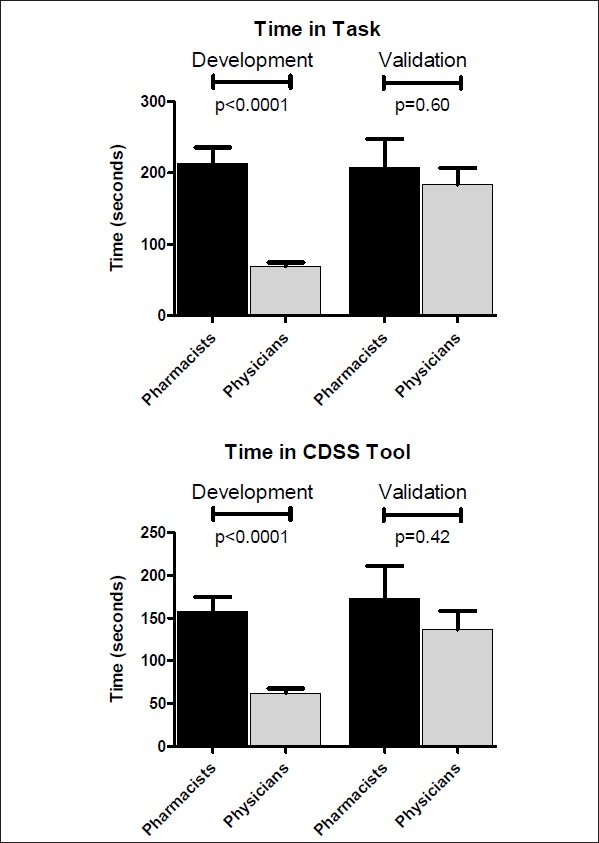
A) Time to complete the tasks for the combined scenarios by pharmacists (n=24 scenarios) and physicians (n=24 scenarios) in the development phase and by pharmacists (n=12 scenarios) and physicians (n=12 scenarios) in the validation phase. B) Time spent in the CDSS tool for the combined scenarios by pharmacists (n=24 scenarios) and physicians (n=24 scenarios) in the development phase and by pharmacists (n=12 scenarios) and physicians (n=12 scenarios) in the validation phase. Pharmacists spent significantly more time to complete the tasks and in the CDSS tool than physicians in the development but there was no statistical difference in the validation phase for time spent on task or in the CDSS tool. Data are presented as mean ± standard error of the mean given all data presented were normally distributed.
3.3 Prescribing
During the development iteration there were 48 opportunities (8 prescribers with 6 scenarios each) for prescribers to order warfarin. Of those opportunities, prescribers ordered the dose recommended by the CDSS twice (4%). Conversely, during the validation iteration, there were 24 opportunities to prescribe warfarin (4 prescribers X 6 scenarios), and prescribers ordered the CDSS recommended dose 9 (37.5%) times, p<0.001 versus frequency in the development iteration. The validation phase significantly improved the accuracy of doses prescribed relative to those of the CDSS recommended dose, as displayed in ► Table 3.
Table 3.
Doses recommended by CDSS versus actual dose ordered by participants
| Development Iteration (n=8) |
Validation Iteration (n=4) |
|||
|---|---|---|---|---|
| Scenario | Task Type | Recommended Dose per day (mg) | Dose Prescribed | Dose Prescribed |
| 1 | Initiation | 7.0 | 5.5 (5.0, 6.4) | 7.0 (6.0, 7.0) |
| 2 | Initiation | 9.0 | 5.0 (4.8.5.0)* | 4.0 (3.0, 6.0) |
| 3 | Initiation | 7.0 | 5.0 (5.0, 5.0)* | 6.0 (4.4, 7.0) |
| 4 | Adjustment | 4.3 | 4.0 (3.0, 4.0)* | 3.0 (2.8, 3.1)* |
| 5 | Adjustment | 1.6 | 1.6 (1.0, 2.0) | 0.80 (0.0, 1.7) |
| 6 | Adjustment | 2.9 | 3.5 (3.0, 4.5)* | 2.9 (2.7, 3.7) |
Data presented as median (IQR)
* p<0.05 versus the CDSS recommended dose
The most commonly prescribed initial dose in the development iteration was the standard starting doses of 4 mg or 5 mg in 18/24 (75%) opportunities. In all of those opportunities, the CDSS had recommended a starting dose greater than 5 mg. In the 12 opportunities (4 participants X 3 scenarios) in the validation iteration to initiate a patient on warfarin, prescribers ordered a 5 mg dose twice (12.5%) versus the aforementioned 75% in development iteration, p<0.001. The CDSS recommended dose was ordered six times for the initiation of warfarin (i.e. 50% vs. 4.2% in development iteration, p=0.03).
Prescribers were more likely to check the CDSS recommendation against the warfarindosing.org website, after a link to the website was included, and then accept the CDSS dose (75% in the validation iteration vs. 0% in the development iteration, p<0.001). One prescriber errantly ordered a dose of 12.5mg, a dose with potential for adverse events, due to the erroneous pre-programmed default dose on the ordering screen being 12.5 mg in the development iteration. During the validation iteration, the default dose was set to the recommended CDSS dose and no doses were ordered in error.
3.4 Satisfaction
Participant responses for CSUQ items were generally positive for both the development and validation iterations with no significant differences between the development and validation iteration (► Table 4). The details of the CSUQ questionnaire are provided in ► Table 4. A qualitative analysis was performed to better discern both positive and negative aspects of the developed CDSS and are discussed in sections 3.4.1–3.4.3.
Table 4.
Post-Session Computer System Usability Questionnaire (CSUQ) for Development and Validation Iterations
| Score | Development (n=8) | Validation (n=4) | |
|---|---|---|---|
| Median (IQR) | Median (IQR) | p value | |
| Overall satisfaction (items 1-19*) | 2.0 (2.0, 4.0) | 3.0 (2.0, 4.0) | 0.29 |
| System usefulness (items 1-8) | 2.0 (2.0, 3.0) | 3.0 (2.0, 3.0) | 0.61 |
| Information quality (items 9-15) | 3.0 (2.0, 5.0) | 4.0 (3.0, 5.0) | 0.14 |
| Interface quality (items 16-18) | 2.0 (2.0, 3.3) | 2.5 (2.0, 3.3) | 1.0 |
* all questions on the CSUQ use 1=Strongly Agree, 2=Agree, 3=Somewhat Agree, 4=Neither Agree nor Disagree, 5=Somewhat Disagree, 6=Disagree, and 7=Strongly Disagree.
3.4.1 Positive Aspects of the CDSS
Participants in the development iteration stated they liked having all the relevant information in one location without “having to flip back and forth through the patient chart”. Two participants in the validation iteration commented that the CDSS was “user friendly”. When participants had access to the warfarindosing.org website through the CDSS, one participant commented of the CDSS, “this is much simpler than the website, which I like”. However participants in both iterations expressed a concern about ordering a dose greater than the often standard starting dose of 5mg for a patient initiating warfarin. One participant stated “I’ve never started a patient with 7[mg] before, usually just do 5[mg]”. In both iterations of the study, some participants verbalized a trust in the CDSS, “at this point I am just trusting the computer” or that its presence validated the use of a dose other than what they would normally prescribe.
3.4.2 Confusing Aspects of the CDSS
Participants in both the development (100%) and validation (75%) iterations expressed some initial confusion related to the genetic information, with one participant in the development iteration stating “it would be nice to have a preamble on what the polymorphisms do”. One physician specifically stated that the usual method (starting at 4–5mg and waiting for INRs) works so the physician did not want enhanced information on the genetic information.
There was general confusion in both iterations regarding drug-drug interactions with warfarin. The validation iteration was revised to display the drug-interactions that were included in the algorithm for the recommended dose calculation. The algorithm included amiodarone, sulfamethoxazole/trimethoprim, azole antibiotics, and statins, but the scenarios intentionally included other medications which interact with warfarin. This created confusion as participants were unsure if the algorithm included all the possible interacting medications. When presented with a warfarin drug-drug interaction alert for medications not included in the CDSS, nine participants were initially unsure whether or not to go back and adjust the dose. Because participants could not clearly discern what was used in the algorithm, they expressed confusion and some initial mistrust of the algorithm in both iterations until they identified that the interacting medications listed in the CDSS were the only ones included in the algorithm. In some cases, this confusion resulted in participants exiting the ordering process and returning to the CDSS to reevaluate and determine if the dose needed to be changed.
3.4.3 Suggested CDSS Design Changes
Across both iterations, participants expressed several potential CDSS changes. Participants felt the explanation of the genetic polymorphisms should include more actionable information (i.e. ‘decrease dose of warfarin by 50%’, rather than ‘slow metabolizer’) as all participants expressed having no exposure to pharmacogenomics-guided warfarin dosing prior to the study. They felt this information would assist in determining the accuracy of the dose recommended by the CDSS. Eight participants also expressed a desire for all recommended doses to be orderable amounts. Currently the CDSS recommends fractions of a milligram; limiting recommended doses to those manufactured doses available at the healthcare institution would save the prescriber from rounding the dose inappropriately or ordering a dose that could not be accurately dispensed to the patient. Three participants (1 in the development iteration, and 2 in the validation iteration), wanted the option to order INRs or to hold warfarin doses via the CDSS interface for patients with a high INR. They felt it would be beneficial to manage warfarin completely within the CDSS rather than having to order laboratory tests elsewhere within the EHR. Four participants across the two iterations wanted a link to the exact algorithm rather than the warfarindosing.org website so they could do the math themselves if desired, and felt that having access to the algorithm itself would help with CDSS transparency.
4. Discussion
In this study, we iteratively developed and evaluated a CDSS using pharmacogenomic-guided warfarin dosing algorithms and incorporated participant feedback through user-centered design. The final CDSS tool improved the variation in efficiency measures between pharmacists and physicians while improving accuracy of prescribing relative to the dose recommended by the algorithm without altering user satisfaction. The optimization of CDSS to guide clinical pharmacogenomics for physicians and pharmacists may accelerate the translation of science from research to practice to overcome a major obstacle in personalized medicine [37, 38].
Pharmacists took longer to complete scenarios than physicians during the development iteration, but not in the validation iteration. Pharmacists used more time than physicians during the development iteration, but the time needed for pharmacists to complete the tasks did not vary widely between the development and validation iterations. Prior literature has found that pharmacists spend more time on some tasks than physicians, including medication reconciliation in which pharmacists spent twice as long as physicians to reconcile medications. [39] In that investigation, pharmacists spent more time reviewing the patient profile before beginning the task of medication reconciliation, similar to what was observed in the development iteration of the present study. However there were no differences between pharmacists and physicians during the validation iteration, with the actions and comments of physicians more closely reflecting those of pharmacists in both iterations. This suggests the design may have been the reason for different time to completion between pharmacists and physicians rather than true differences between the groups. During the validation iteration, physicians’ interests more closely aligned with the pharmacists: physicians demonstrated more interest in understanding the genetics and how to apply the patient’s test results. This may help explain why the time to complete the scenario results were similar to pharmacists with the enhanced CDSS prototype. These findings, combined with more frequent prescribing of the CDSS recommended dose, suggests progress towards a more universal CDSS design for warfarin pharmacogenomics that supports decision-making for both physicians and pharmacists. No CDSS design differences between pharmacists and physicians were identified.
Interestingly, there was a difference between how pharmacists and physicians perceived the CDSS should be used in the development iteration that was alleviated with modifications for the validation iteration. During the development iteration, pharmacists were interested in the CDSS tool, but they were doubtful that it could incorporate all the data they had access to in the EHR and perceived a limited benefit to adoption. However, they were interested in learning more about pharmacogenomics and how to apply the information presented. These data suggest that further assessments of pharmacists’ perspectives on pharmacogenomic-guided CDSS and differences from physicians is warranted. Our results of initial skepticism are similar to those reported in a study that implemented a personal digital assistant mechanism to support warfarin dosing [40]. In that study, some viewed the dosing assistant as a tool for providing better, safer care, while others with more experience prescribing warfarin felt the intervention had limited positive impact.
It has been suggested that underutilization or ignoring of CDSS interventions could be due, at least in part, to gaps in the logic presentation [41]. Participants in the development iteration were reluctant to trust non-standard warfarin doses when they had no access to the original algorithm or the algorithm website. While the participants in the validation iteration more commonly verbalized trust of the CDSS recommendations, they still felt it would be best to have access to the original algorithm in order to double-check the calculations themselves. CDSS modifications included efforts to also improve transparency for interacting medications. While there were fewer participant concerns about the interacting medications in the validation iteration, there were still a number of questions related to which interacting medications were accounted for in the algorithm, and future work may elucidate the best way to present interacting medications.
Another indication of greater participant confidence during the validation iteration was the reduction in the variation of doses prescribed in the validation iteration. Prescribers in the development iteration were more likely to initiate patients on the standard dose of 4 or 5 mg than those in the validation iteration. This standard dosing has been demonstrated to overestimate the necessary dose in 18% of patients, while another study has shown that the algorithm used in the current study over-predicted the dose in 7% of patients [42]. Prescribers in the validation phase more commonly ordered the dose recommended by the CDSS for both initiating and adjusting warfarin doses. The frequency of ordering the CDSS recommended dose was similar to another study which utilized warfarin pharmacogenomics to provide dosing recommendations [43].
There was one instance during the development iteration where a prescriber unknowingly ordered a dose of 12.5mg, a potentially fatal dose, but this did not occur during the validation iteration. It should be noted, however, that while a small number of participants can identify a large number of problems with an interface, rare events and ‘critical usability incidents’, such as an accidental fatal warfarin dose, may require a greater number of participants. CDSS design modifications of the validation iteration may not completely eliminate the potential for critical usability incidents to occur [44, 45]. By making design changes to the prototype CDSS to increase usability, prescribing variability decreased and more closely aligned with the algorithm’s recommendations, which could further reduce the potential for a fatal adverse drug event. Participant confusion and ordering errors may further be reduced by providing recommended doses which are orderable rather than fractions of a milligram.
4.1 Limitations
This study focused on developing and assessing a user-interface for pharmacogenomic-guided CDSSs. While this is a barrier to the clinical implementation, there are other unique challenges, particularly for institutions that have already implemented these CDSSs. These barriers include discretely representing pharmacogenomic data in the EHR and managing genetic knowledge. These obstacles were not assessed in the current study.
Between the development and validation iterations, there was an overall increase in time required to complete tasks and use of the CDSS tool. While the time to complete the scenarios was less in the development iteration, poorly designed systems can decrease time required in a similar manner to well-designed CDSS.[46] For example, decreased time using the tool may occur if individuals dismiss the CDSS tool as invalid and move on to the next task. Indeed the increased time to complete tasks in the validation phase corresponded to increased time in the CDSS tool.
This study focused on physicians and pharmacists, but other prescribers such as nurse practitioners may respond differently or illuminate other potential advantages or disadvantages of the CDSS tool. All participants were aware that the patient scenarios were simulated and while they were instructed to treat the patients as though they were real, the prescribers may not have taken the scenarios as seriously as if the scenarios were actual patients. Additionally, while participants were at a distance from the study investigator, there was no barrier between them, which may have altered their responses. The scenarios were all inpatient cases and prescribers may have treated the scenarios differently if the patients represented outpatient cases since inpatients can be monitored more closely for adverse events and warfarin more easily adjusted. Given the iterative design of the study, each iteration had a relatively small sample size limiting the application of inferential statistics. Thus, user satisfaction was largely assessed by qualitative methods.
4.2 Future Work
The results of the validation iteration show promise in the wider application of the CDSS to include genetic testing. Although warfarin was chosen as the probe for the CDSS, the tool may be applicable to other pharmacogenomics-guided drugs with appropriate alterations. Pilot testing of the CDSS in a clinical setting would allow for field observations to evaluate how the tool is used by prescribers during patient care. It would also provide a greater opportunity to identify potential unintended consequences associated with the CDSS design and inform additional design improvements. Research is also needed to assess if such a CDSS is effective in reducing the number of bleeding events which occur while stabilizing patients on warfarin and if the time to goal drug concentrations or INR can be reduced.
4.3 Conclusions
A CDSS tool has been developed to help integrate pharmacogenomic information into prescribing decisions using warfarin as a model drug. Iterative usability testing informed a CDSS design that enhanced physician and pharmacist confidence in the recommended warfarin doses. Study findings suggest that condensing all relevant information from the patient profile into one location may aid prescriber decision-making. While prescribers were more willing to accept the CDSS recommended doses after the design changes, there was still confusion around how to apply the CYP2C9 and VKORC1 genetic results particularly in the presence of drug-interactions. The use of pharmacogenomic information to guide prescribing is increasing. Improving the usability of CDSS systems for physicians and pharmacists may aid adoption and facilitate safer prescribing. The CDSS prototype we developed can serve as a starting point for designing more advanced CDSS tools to incorporate new genetic tests into practice.
Acknowledgments
We would like to acknowledge Josh Williams and Lara Schmidt for their contributions to the prototype development.
Funding Statement
Funding: This work was supported by a Lilly seed grant to the Purdue University College of Pharmacy. Drs. Russ was supported in part by a VA HSR&D Research Career Development Award (CDA 11–214) and Dr. Overholser was supported in part by K08HL095655 (BRO) from the National Heart, Lung, and Blood Institute, National Institutes of Health (Bethesda, MD). Views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veterans Affairs or the U.S. government.
Footnotes
Clinical Relevance Statement
A clinical decision support system for pharmacogenomic-driven drug dosing has been developed for clinical evaluation. Disparity between physicians and pharmacists in the utilization of the clinical decision support system was eliminated by design modifications. Improving the usability of CDSS systems for physicians and pharmacists may aid adoption of clinical pharmacogenomics and facilitate safer prescribing.
Conflict of Interest
The authors declare that they have no conflicts of interest in the research.
Human Subjects Protections
The study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects, and was reviewed by the Indiana University-Purdue University of Indianapolis (IUPUI) Institutional Review Board.
References
- 1.Food and Drug Administration: Table of pharmacogenomics biomarkers in drug labeling. [cited 2014 Dec 1]. Available from: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ ucm083378.htm.
- 2.Arwood MJ, Chumnumwat S, Cavallari LH, Nutescu EA, Duarte JD. Implementing Pharmacogenomics at Your Institution: Establishment and Overcoming Implementation Challenges. Clin Transl Sci. 2016; doi: 10.1111/cts.12404. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, Staley BJ, Dong HJ, Allan RW, Liu JF, Cooper-Dehoff RM, Anderson RD, Conlon M, Clare-Salzler MJ, Nelson DR, Johnson JA. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet 2014; 166C(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy KD, Decker BS, Carpenter JS, Flockhart DA, Dexter PR, Desta Z, Skaar TC. Prerequisites to implementing a pharmacogenomics program in a large health-care system. Clin Pharmacol Ther 2014; 96: 307-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottesman O, Scott SA, Ellis SB, Overby CL, Ludtke A, Hulot JS, Hall J, Chatani K, Myers K, Kannry JL, Bottinger EP. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 2013; 94: 214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics 2013; 14: 723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O‘Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, Cox NJ, Ratain MJ. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 2012; 92: 446-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Jirjis JN, Roden DM. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 2012; 92: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuldiner AR, Palmer K, Pakyz RE, Alestock TD, Maloney KA, O‘Neill C, Bhatty S, Schub J, Overby CL, Horenstein RB, Pollin TI, Kelemen MD, Beitelshees AL, Robinson SW, Blitzer MG, McArdle PF, Brown L, Jeng LJ, Zhao RY, Ambulos N, Vesely MR. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am J Med Genet C Semin Med Genet 2014; 166C(1):76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teich JM, Osheroff JA, Pifer EA, Jenders RACDS Expert Review Panel. Clinical decision support in electronic prescribing: recommendations and an action plan: report of the joint clinical decision support workgroup. J Am Med Inform Assoc 2005; 12: 365-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan B. Evaluating informatics applications- clinical decision support systems literature review. I Int J Med Inform 2001; 64: 15–37. [DOI] [PubMed] [Google Scholar]
- 12.Devine EB, Lee CJ, Overby CL, Abernethy N, McCune J, Smith JW, Tarczy-Hornoch P. Usability evaluation of pharmacogenomics clinical decision support aids and clinical knowledge resources in a computerized provider order entry system: a mixed methods approach. Int J Med Inform 2014; 83(7): 473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura AA, Shirts BH, Salama J, Smith JW, Devine B, Tarczy-Hornoch P. Physician perspectives of CYP2C19 and clopidogrel drug-gene interaction active clinical decision support alerts. Int J Med Inform 2016; 86: 117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budnitz DS, Pollock DA, Weidenbach KN, Mendelsohn AB, Schroeder TJ, Annest JL. National surveil-lance of emergency department visits for outpatient adverse drug events. JAMA 2006; 296: 1858-1866. [DOI] [PubMed] [Google Scholar]
- 15.Food and Drug Administration FDA clears genetic lab test for warfarin sensitivity. Sep 17, 2007. [cited 2011 Feb 9]. Available from: http://www.fda.gov/newsevents/newsroom/pressannouncements/2007/ucm108984.htm.
- 16.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A Genome-Wide Association Study Confirms VKORC1, CYP2C9, and CYP4F2 as Principal Genetic Determinants of Warfarin Dose. PLoS Genetics 2009; 5(3):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swen J, Wilting I, de Goede AD, Grandia L, Mulder H, Touw DJ, de Boer A, Conemans JM, Egberts TC, Klungel OH, Koopmans R, van der Weide J, Wilffert B, Guchelaar HJ, Deneer VH. Pharmacogenetics: From Bench to Byte. Clin Pharmacol Ther 2008; 83: 781-787. [DOI] [PubMed] [Google Scholar]
- 18.Food and Drug Administration: FDA approves updated warfarin (Coumadin) prescribing information: new genetic information may help providers improve initial dosing estimates of the anticoagulant for individual patients. [cited 2011 Feb 9]. Available from: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2007/ucm108967.htm.
- 19.Baudhuin LM, Langman LJ, O‘Kane DJ. Translation of pharmacogenetics into clinically relevant testing modalities. Clin Pharmacol Ther 2007; 82: 373-376. [DOI] [PubMed] [Google Scholar]
- 20.Nutescu EA, Drozda K, Bress AP, Galanter WL, Stevenson J, Stamos TD, Desai AA, Duarte JD, Gordeuk V, Peace D, Kadkol SS, Dodge C, Saraf S, Garofalo J, Krishnan JA, Garcia JG, Cavallari LH. Feasibility of implementing a comprehensive warfarin pharmacogenetics service. Pharmacotherapy 2013; 33: 1156-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, Kesteven P, Christersson C, Wahlström B, Stafberg C, Zhang JE, Leathart JB, Kohnke H, Maitland-van der Zee AH, Williamson PR, Daly AK, Avery P, Kamali F, Wadelius M; EU-PACT Group. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 2013; 369(24): 2294-2303. [DOI] [PubMed] [Google Scholar]
- 22.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, Rosenberg YD, Eby CS, Madigan RA, McBane RB, Abdel-Rahman SZ, Stevens SM, Yale S, Mohler ER, 3rd, Fang MC, Shah V, Horenstein RB, Limdi NA, Muldowney JA, 3rd, Gujral J, Delafontaine P, Desnick RJ, Ortel TL, Billett HH, Pendleton RC, Geller NL, Halperin JL, Goldhaber SZ, Caldwell MD, Califf RM, Ellenberg JH; COAG Investigators. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 2013; 369(24): 2283-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmadian L, van Engen-Verheul M, Bakhshi-Raiez F, Peek N, Cornet R, de Keizer NF. The role of standardized data and terminological systems in computerized clinical decision support systems: Literature review and survey. Int J Med Inform 2011; 80: 81–93. [DOI] [PubMed] [Google Scholar]
- 24.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, Kahn SF, May HT, Samuelson KM, Muhlestein JB, Carlquist JF; Couma-Gen Investigators. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation 2007; 116: 2563-2570. [DOI] [PubMed] [Google Scholar]
- 25.Manzi SF, Fusaro VA, Chadwick L, Brownstein C, Clinton C, Mandl KD, Wolf WA, Hawkins JB. Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration- experience from a pediatric tertiary care facility. J Am Med Inform Assoc 2016. Jun 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hicks JK, Stowe D, Willner MA, Wai M, Daly T, Gordon SM, Lashner BA, Parikh S, White R, Teng K, Moss T, Erwin A, Chalmers J, Eng C, Knoer S. Implementation of clinical pharmacogenomics within a large health system: from electronic health record decision support to consultation services. Pharmacotherapy 2016. Jun 17. [DOI] [PubMed] [Google Scholar]
- 27.Kushniruk AW, Patel VL. Cognitive and usability engineering methods for the evaluation of clinical information systems. J Biomed Inform 2004; 37(1):56–76. [DOI] [PubMed] [Google Scholar]
- 28.Russ AL, Weiner M, Russell SA, Baker DA, Fahner WJ, Saleem JJ. Design and implementation of a hospital-based usability laboratory: insights from a Department of Veterans Affairs laboratory for health information technology. Jt Comm J Qual Patient Saf 2012; 38: 531-540. [DOI] [PubMed] [Google Scholar]
- 29.Talmon J, Ammenwerth E, Brender J, de Keizer N, Nykänen P, Rigby M. STARE-HI--Statement on reporting of evaluation studies in Health Informatics. Int J Med Inform 2009; 78: 1–9. [DOI] [PubMed] [Google Scholar]
- 30.Lenzini P, Wadelius M, Kimmel S, Anderson JL, Jorgensen AL, Pirmohamed M, Caldwell MD, Limdi N, Burmester JK, Dowd MB, Angchaisuksiri P, Bass AR, Chen J, Eriksson N, Rane A, Lindh JD, Carlquist JF, Horne BD, Grice G, Milligan PE, Eby C, Shin J, Kim H, Kurnik D, Stein CM, McMillin G, Pendleton RC, Berg RL, Deloukas P, Gage BF. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin Pharmacol Ther 2010; 87: 572-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.International Warfarin Pharmacogenetics Consortium Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen J, Landauer T. A mathematical model of the finding of usability problems. Proceedings of the INTERACT,93 and CHI,93 conference on Human factors in computing systems, Amsterdam, The Netherlands. 1993. p. 206–13. [Google Scholar]
- 33.Nielsen J. Ch 2: What is Usability? In: Usability Engineering. San Francisco, CA: Morgan Kaufmann Publishers; 1993: 23–48. [Google Scholar]
- 34.Nielsen J. Usability 101: Introduction to Usability. Jan 4, 2012 [cited 2013 Feb 5]; Available from: http://www.nngroup.com/articles/usability-101-introduction-to-usability/.
- 35.Lewis JR. IBM computer usability satisfaction questionnaires: psychometric evaluation and instructions for use. International Journal of Human-Computer Interaction 1995; 7: 57–78. [Google Scholar]
- 36.ECRI Institute Warfarin therapy management. May 12, 2011 [cited 2013 Aug 9]; Available from: http://www.guideline.gov/content.aspx?id=24513.
- 37.Crews KR, Cross SJ, McCormick JN, Baker DK, Molinelli AR, Mullins R, Relling MV, Hoffman JM. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am J Health Syst Pharm 2011; 68: 143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weitzel KW, Alexander M, Bernhardt BA, Calman N, Carey DJ, Cavallari LH, Field JR, Hauser D, Junkins HA, Levin PA, Levy K, Madden EB, Manolio TA, Odgis J, Orlando LA, Pyeritz R, Wu RR, Shuldiner AR, Bottinger EP, Denny JC, Dexter PR, Flockhart DA, Horowitz CR, Johnson JA, Kimmel SE, Levy MA, Pollin TI, Ginsburg GS; IGNITE Network. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics 2016; 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kushniruk AW, Santos SL, Pourakis G, Nebeker JR, Boockvar KS. Cognitive analysis of a medication reconciliation tool: applying laboratory and naturalistic approaches to system evaluation. Stud Health Technol Inform 2011; 164: 203-207. [PubMed] [Google Scholar]
- 40.Kravitz RL, Neufeld JED, Hogarth MA, Paterniti DA, Dager W, White RH. From insight to implementation: lessons from a multi-site trial of a PDA-based warfarin dose calculator. Henriksen K, Battles JB, Marks ES, Lewin DI, editors. Advances in Patient Safety: From Research to Implementation. Rockville, MD: Agency for Healthcare Research and Quality; 2005. [PubMed] [Google Scholar]
- 41.Horsky J, Schiff GD, Johnston D, Mercincavage L, Bell D, Middleton B. Interface design principles for usable decision support: a targeted review of best practices for clinical prescribing interventions. J Biomed Inform 2012; 45: 1202-1216. [DOI] [PubMed] [Google Scholar]
- 42.Shaw PB, Donovan JL, Tran MT, Lemon SC, Burgwinkle P, Gore J. Accuracy assessment of pharmacogenetically predictive warfarin dosing algorithms in patients of an academic medical center anticoagulation clinic. J Thromb Thrombolysis 2010; 30: 220-225. [DOI] [PubMed] [Google Scholar]
- 43.Nutescu EA, Drozda K, Bress AP. Feasibility of Implementing a Comprehensive Warfarin Pharmacogenetics Service. Pharmacotherapy 2013; 33(11): 1156-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen J. How many test users in a usability study? June 4, 2012 [cited 2013 Aug 6]; Available from: http://www.nngroup.com/articles/how-many-test-users/.
- 45.Barnum C. The ‘magic number 5’ is it enough for web testing. Information Design Journal and Document Design 2003; 11(2/3): 160-170. [Google Scholar]
- 46.Smith-Jackson TL, Wogalter MS. Methods and Procedures in Warnings Research. Wogalter MS, editor. Handbook of Warnings. Mahwah, NJ: Lawrence Erlbaum Associates; 2006. [Google Scholar]


