Abstract
MicroRNA-221 and microRNA-222 (miR-221/222) have been identified as oncogenes and confirmed to be overexpressed in various types of cancer. However, the regulation mechanism of miR-221/222 in oral squamous cell carcinoma (OSCC) remains to be fully elucidated. Previously, an miR-221/222 sponge was successfully constructed and its effect on the downregulation of miR-221/222 expression was investigated. In the present study, the dual luciferase reporter assay revealed a phosphatase and tensin homolog (PTEN) deletion on chromosome 10 to be a target gene of miR-221/222. It was also demonstrated that miR-221/222 suppression by transfection with an miR-221/222 sponge in vitro resulted in upregulation of PTEN. Notably, the proliferation and invasiveness of the miR-221/222 sponge-transfected cells was significantly inhibited, while apoptosis was promoted, when determined by Cell Counting Kit-8, Transwell assays and flow cytometry. The results of the present study prove that miR-221/222 may downregulate the expression of PTEN in OSCC cells and function as oncogenes, providing a novel insight into the underlying mechanism of OSCC tumorigenesis. The present study suggests that upregulating the expression of PTEN by downregulation of miR-221/222 may be a potential treatment for OSCC.
Keywords: oral squamous cell carcinoma, microRNA-221/222, sponge, phosphatase and tensin homologue, apoptosis
Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNAs that negatively regulate gene expression by binding to the 3′untranslated region (3′UTR) of target mRNAs (1,2). There is emerging evidence that miRNAs function as oncogenes or tumor suppressors through interaction with a number of the signal transduction pathways involved in transformation and carcinogenesis (3–5). miRNA-221/222 (miR-221/222) are located in tandem on the X chromosome and have been reported as oncogenic miRNAs in multiple types of advanced cancer (6–9). A number of studies have observed overexpression of miR-221/222 in malignancies and the available data suggest that miR-221/222 may become suitable targets for cancer treatment (7–9).
The miRNA sponge method was introduced in 2007 by Ebert et al (10); it is used to create RNAs containing multiple tandem binding sites complementary to the miRNAs of interest and leads to continuous loss of miRNA function in cells and transgenic organisms. The miRNA sponge has proven to be a valuable tool for miRNA loss of function experimental systems (11). In a previous study, a miR-221/222 sponge was successfully constructed and its inhibitory effects on miR-221/222 in oral squamous cell carcinoma (OSCC) cells were investigated (12).
Phosphatase and tensin homolog (PTEN) is a tumor suppressor gene that is frequently mutated in many types of cancer (13–16). PTEN expression is downregulated in a wide range of malignancies, including glioblastoma, pancreatic cancer, colorectal carcinoma, breast cancer and OSCC (13,14,16–20). The phosphoinositide 3-kinase/protein kinase B (PI3K/Akt) signaling pathway is involved in multiple biological processes, including cellular apoptosis, cell cycle regulation, survival and proliferation (21). Previous studies have demonstrated that aberrant activation of the PI3K/Akt signaling pathway has a significant role in tumorigenesis and tumor metastasis (14). PTEN functions as a tumor suppressor by negatively regulating the PI3K/Akt signaling pathway (22). miRNAs, including miR-17-5p and miR29b, have been reported to regulate the expression of PTEN (19,23). However, little is known about the roles of miR-221/222 in the expression of PTEN in OSCC.
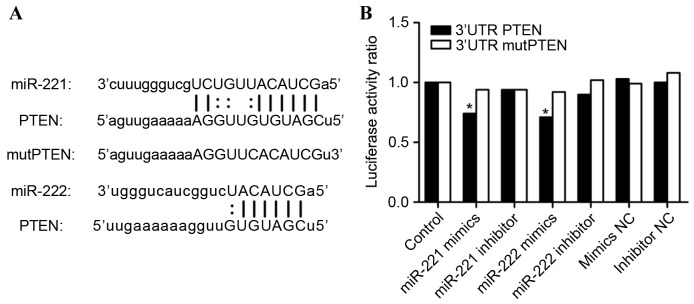
In the present study, bioinformatics analysis revealed that the PTEN gene may be a direct target of miR-221/222. Binding sites for miR-221/222 were identified in the 3′UTR of PTEN using miRanda (http://www.microrna.org/) and the result is shown in Fig. 1A. miR-221/222 were identified as potent regulators of PTEN. An miR-221/222 sponge was constructed in CAL27 and HSC6 OSCC cells to validate the induction of apoptosis, and reduction of cell proliferation and invasion, through miR-221/222 inhibition and the upregulation of PTEN expression.
Figure 1.
PTEN was confirmed to be a target gene of miR-221/222. (A) Sequences of miR-221/222, the PTEN 3′UTR and the mutPTEN 3′UTR, including the binding sites of PTEN-miR-221/222. (B) Luciferase activity ratio of PTEN 3′UTR and mutPTEN 3′UTR luciferase constructs when transfected with miR-221/222 mimics, mimics NC, miR-221/222 inhibitor or inhibitor NC (*P<0.05). miR-221, microRNA-221/222; PTEN, phosphatase and tensin homolog; 3′UTR, 3′ untranslated region; mutPTEN, mutated PTEN; NC, negative control.
Materials and methods
Cells and cell culture
The 293T cell line was purchased from Land Unicomed (Guangzhou, China). The OSCC CAL27 cell line was purchased from American Type Culture Collection (ATCC; Manassas, VA, USA) and the OSCC HSC6 cell line was kindly provided by Dr J. Silvio Gutkind (NIH; Besthesda, MD). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.,), 100 IU/ml penicillin and 100 µg/ml streptomycin, and maintained at 37°C in a humidified 5% CO2 atmosphere.
Dual luciferase reporter assay
The 293T cells were cultured in 24-well plates, transfected with 0.5 µg of the psi-CHECK2-PTEN (Land Unicomed) or 0.5 µg of the psi-CHECK2-mutPTEN (Land Unicomed), and 20 µM of miR-221/222 inhibitor (Gene Pharma, Shanghai, China) or 20 µM of miR-221/222 mimics (Gene Pharma) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc. USA). Cells were lysed by Passive Lysis Buffer (Promega Corporation, Madison, WI, USA) and collected at 48 h post-transfection, and luciferase activity was detected using the Dual-Luciferase Reporter Assay system (Promega Corporation) according to the manufacturer's protocol.
Construction of miRNA sponge and transfection
An miR-221/222 sponge was constructed by inserting tandemly arrayed miRNA binding sites into the 3′UTR of a reporter gene encoding destabilized enhanced green fluorescent protein driven by the murine cytomegalovirus promoter. Binding sites contained 3 hsa-miR-221 and hsa-miR-222 antisense sequences complementary to miR-221/222. The miR-221/222 sponge and empty vector were transfected using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and the transfection medium was replaced with fresh DMEM containing 10% FBS 6 h later. Following treatment for 48 h, CAL27 and HSC6 cells were divided into 3 groups, including the control, scramble and miR-221/222 sponge, and used for subsequent analysis.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total miRNA and mRNA were extracted using the miRNeasy mini kit (Qiagen GmbH, Hilden, Germany) and the RNeasy Micro kit (Qiagen GmbH) according to the manufacturer's protocol. RT-qPCR was performed using the Primescript RT reagent kit (Takara, Otsu, Japan) with U6 and 18S rRNA as the loading controls. For the detection of miR-221/222 and PTEN, the SYBR Green PCR assay (SYBR® Premix Ex Taq™; Takara) was performed and the primer sequences used were as follows: miR-221 forward, ACACTCCAGCTGGGAGCTACATTGTCTGCTGG and reverse, CTCAACTGGTGTCGTGGA; miR-222 forward, ACACTCCAGCTGGGAGCTACATCTGGCTACTG and reverse, CTCAACTGGTGTCGTGGA; U6 forward, CTCGCTTCGGCAGCACA and reverse, AACGCTTCACGAATTTGCGT; PTEN forward, TTGTGGTCTGCCAGCTAAA and reverse, CGCTCTATACTGCAAATGCT; 18S rRNA forward, CCTGGATACCGCAGCTAGGA and reverse, GCGGCGCAATACGAATGCCCC. The PCR was performed in triplicate consisting of 40 cycles of a denaturation step at 95°C for 5 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec after a cycle of a pre-denaturation step at 95°C for 30 sec on an ABI PRISM® 7500 Sequence Detector (Applied Biosystems, Foster City, CA, USA). Relative expression levels were calculated using the 2−ΔΔCq method (24).
Western blot analysis
Following 48 h of treatment with empty vector and miR-221/222 sponge, 3 groups of cells were washed with pre-chilled phosphate-buffered saline (PBS) 3 times and solubilized in RIPA buffer with 1% protease inhibitor (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and 10% phosphatase inhibitor (Roche Diagnostics GmbH, Mannheim, Germany). Cell lysates were centrifuged for 15 min at 14,000 × g and 4°C, and the protein concentration was subsequently measured using the Enhanced BCA protein assay kit (CWBIO, Beijing, China) according to the manufacturer's protocol. The proteins were diluted with 5X loading buffer (CWBIO, Beijing, China) and denatured at 99°C for 10 min. A total of 35 g of protein per group was separated on 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA) by electroblotting. The membranes were blocked with 5% skimmed milk at room temperature for 1 h, then washed in Tris-buffered saline and Tween 20 (TBST) and incubated with primary antibodies against PTEN (catalog no. 9188), phosphorylated Akt (pAkt; catalog no. 4060), Akt (catalog no. 4691) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; catalog no. 5174) (dilution, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA) overnight at 4°C. Subsequently, the blots were washed 3 times with TBST and incubated with Horseradish Peroxidase-conjugated AffiniPure Goat Anti-Rabbit IgG (1:3,000; Cell Signaling Technology, Inc.) for 1 h at room temperature. The membranes were detected by an enhanced chemiluminescence reagent kit (GE Healthcare Life Sciences, Chalfont, UK) and visualized using the AlphaView SA system (Protein Simple, San Jose, CA, USA). The specific protein bands were quantified by ImageJ 1.48 (National Institutes of Health, Bethesda, MD, USA) with the density of GAPDH used as the loading control.
Cell Counting Kit-8 (CCK8) assay
The viabilities of the control and miR-221/222 sponge transfected cells were evaluated using the CCK8 assay (Beyotime Institute of Biotechnology, Haimen, China). Respectively, CAL27 (4,000 cells/well) and HSC6 (3,000 cells/well) cell lines were seeded into 96 well plates 1 day prior to transfection. Following the transfection method previously mentioned, at time points 12, 24, 36, 48, 60 and 72 h, 10 µl CCK8 reagent per well was added and incubated for 2 h at 37°C in a humidified environment. Optical density values were measured at a wavelength of 450 nm. The data were derived from quadruplicate samples of at least three independent experiments.
Transwell® assay
The invasive ability of the CAL27 and HSC6 cell lines was evaluated by the Transwell assay, which measures the movement of cells across a Matrigel®-coated membrane. Transwell upper chambers were coated with 50 µl of 20% growth factor-reduced Matrigel prior to the experiment. The parental and transfected cells (CAL27, 1×105 cells/chamber; HSC6, 8×104 cells/chamber) were seeded into the upper chambers in 100 µl serum-free DMEM, while 600 µl DMEM supplemented with 10% FBS was added to the lower chambers. The cells were incubated at 37°C in a humidified 5% CO2 atmosphere for 24 h, and the cells and Matrigel in the upper chambers were subsequently discarded. Cells in the lower chamber were fixed with 4% paraformaldehyde solution for 20 min and then stained with 0.05% crystal violet for 15 min, followed by washing with PBS. Images were captured using an inverted microscope and the number of invasive cells was counted.
Flow cytometry
After transfection for 48 h, CAL27 and HSC6 cells were collected in the log phase of growth by centrifugation for 5 min at 1,000 × g and 25°C, and rinsed with PBS 3 times. The cell apoptosis assay was performed using an Annexin-V-FLUOS Staining kit (Roche Diagnostics GmbH). Cells were resuspended in incubation buffer at a density of 106/ml. Subsequently, 5 µl Annexin V-fluorescein isothiocyanate and 5 µl propidium iodide (PI) were added to the cell suspension according to the manufacturer's protocol. The cells were incubated for 15 min at room temperature in the dark. The stained cells were analyzed by an FC500 flow cytometer (Beckman Coulter, Fullerton, CA, USA).
Statistical analysis
All data are presented as the mean ± SD. The results were determined by Student's t-test using SPSS version 15.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-221/222 direct targeting of the PTEN mRNA 3′UTR
To validate whether PTEN is a direct target of miR-221/222, PTEN 3′UTR and mutPTEN 3′UTR luciferase constructs were transfected into into 293T cells with miR-221/222 mimics, mimic negative control (NC), miR-221/222 inhibitor or inhibitor NC. Luciferase activity was detected using a Dual-Luciferase Reporter Assay System. As can be observed in Fig. 1B, the luciferase activity of 293T cells transfected with miR-221/222 and PTEN 3′UTR was reduced when compared with all other treatment groups.
miR-221/222 and PTEN expression in CAL27 and HSC6 cells
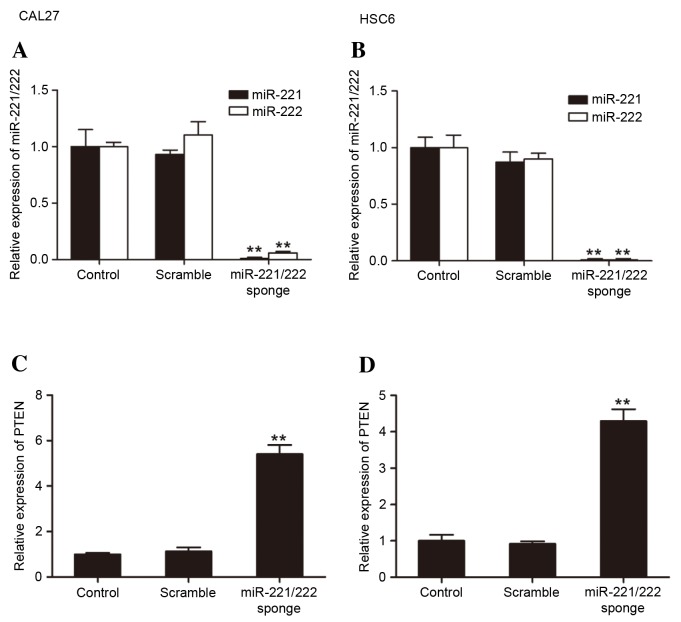
The present study analyzed the expression of miR-221/222 and PTEN by RT-qPCR. miR-221/222 sponge transfection significantly reduced the miR-221/222 levels compared with the control groups (CAL27: miR-221 vs. control, P=0.0063; miR-222 vs. control, P=0.0090; HSC6: miR-221 vs. control, P=0.00072; miR-222 vs. control, P=0.00058) (Fig. 2A and B). The bioinformatics analysis predicted that miR-221/222 would regulate the expression of PTEN. The results of the present study showed that in miR-221/222 sponge groups, a notable upregulation of PTEN was observed in CAL27 and HSC6 cells (Fig. 2C and D).
Figure 2.
RT-qPCR analysis of miR-221/222 and PTEN expression in CAL27 and HSC6 cells transfected with miR-221/222 sponge. RT-qPCR results showed significant downregulation of miR-221/222 following transfection with miR-221/222 sponge in (A) CAL27 and (B) HSC6 cells compared with the control groups (**P<0.01). The level of PTEN expression in the miR-221/222 sponge group was significantly increased compared with that of the control and scramble groups in (C) CAL27 and (D) HSC6 cells (**P<0.01). RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR-221/222, microRNA-221/222; PTEN, phosphatase and tensin homolog.
Determination of PTEN and pAkt expression in CAL27 and HSC6 cells
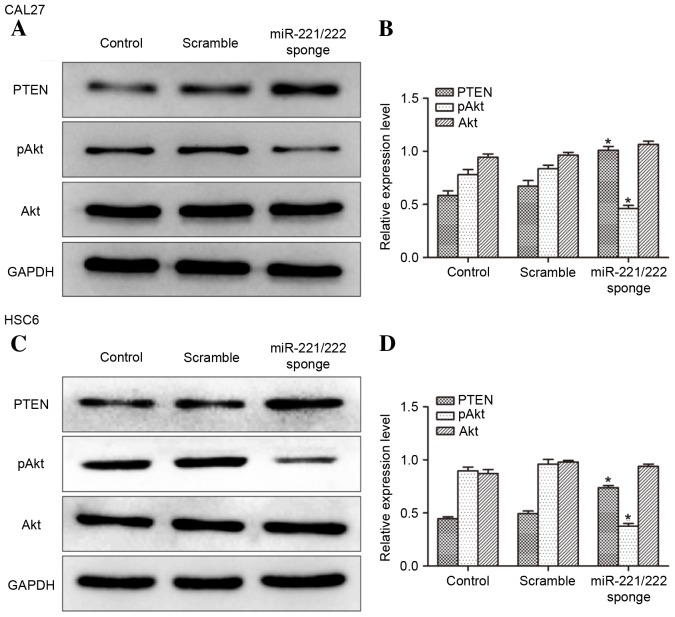
To investigate the impact of miR-221/222 on protein expression (Fig. 3) western blot analysis was performed. It was observed that the level of PTEN expression was increased and the expression of pAkt was decreased in miR-221/222 sponge transfected CAL27 and HSC6 cells compared with controls. The data suggest that the expression of PTEN in CAL27 and HSC6 cells was negatively regulated by miR-221/222.
Figure 3.
Expression of PTEN, Akt and pAkt demonstrated by western blotting. The expression of PTEN, Akt and pAkt in (A and B) CAL27 and (C and D) HSC6 cells was measured by western blotting following transfection of empty vector or miR-221/222 sponge. An increase in the expression of PTEN was observed in the CAL27 and HSC6 cells of the miR-221/222 sponge group, while the expression of pAkt decreased (*P<0.05 vs. control). PTEN, phosphatase and tensin homolog; Akt, protein kinase B; pAkt, phosphorylated Akt; miR-221/222, microRNA-221/222; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
miR-221/222 sponge inhibits CAL27 and HSC6 cell proliferation
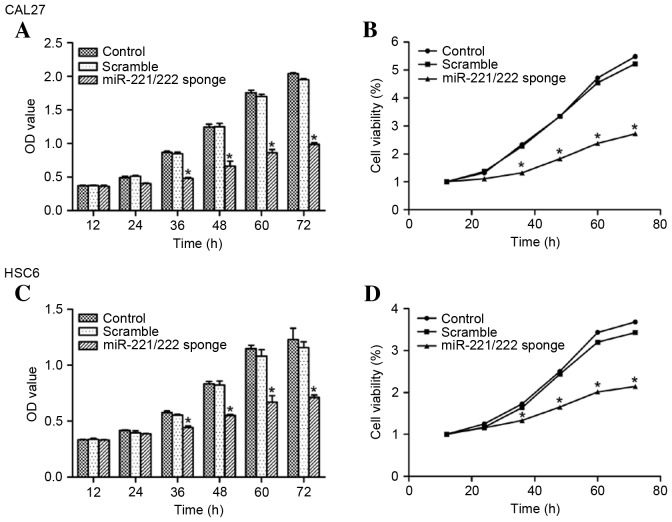
The effect of miR-221/222 on the growth of CAL27 and HSC6 cells was determined using a CCK8 assay. Compared with the control group, the miR-221/222 sponge inhibited CAL27 and HSC6 cell growth (P<0.05 vs. control) (Fig. 4).
Figure 4.
miR-221/222 influences oral squamous cell carcinoma cell growth. Cell counting Kit-8 assay showed that in (A and B) CAL27 and (C and D) HSC6 cells, transfection with an miR-221/222 sponge inhibited cell proliferation significantly compared with the control groups (*P<0.05). There were no significant differences between the control group and the scramble group (P>0.05). miR-221/222, microRNA-221/222; OD, optical density.
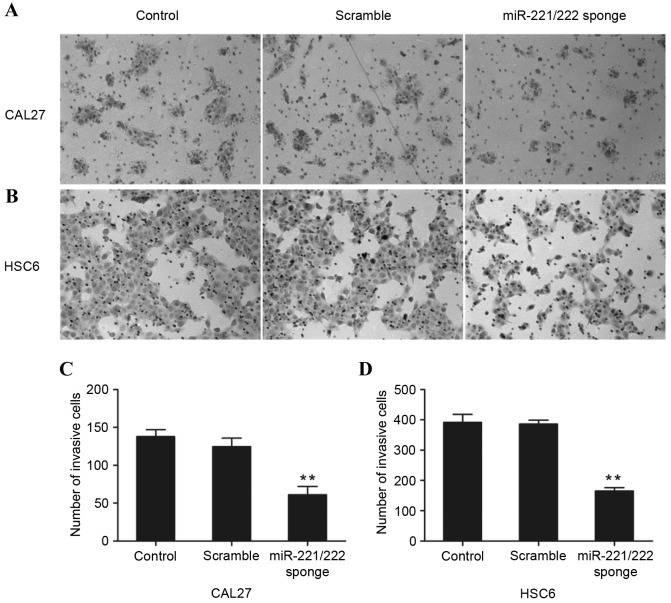
miR-221/222 sponge induces CAL27 and HSC6 cell invasion
The role of miR-221/222 on cell invasion was further assessed by a Transwell assay. As shown in Fig. 5, the invasive ability of CAL27 and HSC6 cells that were transfected with the miR-221/222 sponge was significantly reduced when compared with the control groups (P<0.01, P=0.0087 and P=0.00063 vs. control, respectively).
Figure 5.
Effect of the miR-221/222 sponge on the invasive ability of oral squamous cell carcinoma cells was investigated by Transwell assay. Invasive ability decreased in (A) CAL27 and (B) HSC6 cells when transfected with the miR-221/222 sponge compared with the control group (**P<0.01 vs. control). Cell counts of invasion (C) CAL27 and (D) HSC cells were observed in 5 random fields (magnification, ×100). miR-221/222, microRNA-221/222.
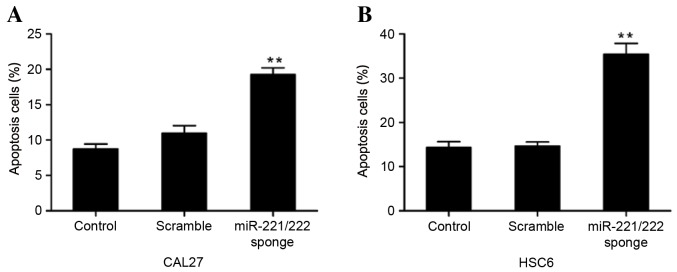
miR-221/222 sponge induces CAL27 and HSC6 cell apoptosis
To examine the effect of the miR-221/222 sponge on CAL27 and HSC6 cell apoptosis, Annexin V-PI analysis was performed. The results showed that the number of apoptotic cells was significantly increased following treatment with the miR-221/222 sponge, compared with the untreated groups, suggesting that miR-221/222 sponge treatment induced apoptosis (P=0.00024 and P=0.00003 vs. control, respectively) (Fig. 6).
Figure 6.
Percentage of apoptotic oral squamous cell carcinoma cells following treatment with a miR-221/222 sponge. The apoptosis of (A) CAL27 and (B) HSC6 cells was analyzed by flow cytometry at 48 h post-treatment with the empty vector and miR-221/222 sponge. Annexin V-propidium iodide analysis showed that the percentage of apoptotic cells was significantly increased compared with the control and scramble groups (**P<0.01). miR-221/222, microRNA-221/222.
Discussion
In previous years, increasing evidence has highlighted the importance of miRNAs in the development and progression of various types of cancer, including OSCC (25). miRNAs are considered to be novel molecular targets in the diagnosis and treatment of human carcinoma (1). Abnormal expression of miR-221/222 has been observed and implicated in multiple neoplasms, but to the best of our knowledge the equivalent research into OSCC has not been performed (7,8,26).
PTEN was demonstrated to be a direct target gene of miR-221/222, and the binding sites for miR-221/222 in the PTEN 3′UTR were predicted by bioinformatic analysis. Furthermore, the results of the Dual-Luciferase Reporter Assay confirmed that PTEN was a target gene of miR-221/222. To investigate further, it was necessary to remove miR-221/222 function. This could be achieved through an antisense oligonucleotide inhibitor, genetic knockout or miRNA sponge (10,11,27). In the present study, a miR-221/222 sponge was constructed in order to downregulate the expression of miR-221/222. Results from RT-qPCR confirmed that transfection with the miR-221/222 sponge significantly reduced the miR-221/222 levels compared with the control groups. miRNA sponge technology was initially introduced and verified by Ebert et al (10) through the creation of RNAs containing multiple binding sites complementary to the miRNA of interest, leading to the continuous loss of miRNA function (28,29). The sponge is usable in a long-term miRNA knockout experiment, while maintaining an efficiency comparable with knockout by antisense oligonucleotide inhibitors (10). It is more convenient to use the sponge rather than perform a gene knockout to build miRNA silencing cells or animal models. Additionally, a sponge with common sequences could block a whole miRNA family, which has the same seed regions (10,12). Moreover, a sponge can be constructed to work on multiple miRNAs synchronously by connecting different inhibiting sequences of relevant miRNAs in series (10,12). It should, however, be noted that the sponge may exhibit inefficient inhibition at excessively high concentrations of miRNA, as the sponge reaches maximum saturation (11). In previous studies, an miR-221/222 sponge was successfully constructed and its effect in OSCC cells was investigated (12). In addition to this, the results of the present study demonstrated that the miR-221/222 sponge satisfactorarily inhibited the expression of miR-221/222.
In the present study, it was identified that miR-221/222 regulates OSCC cell proliferation, invasive ability and apoptosis. The results of the present study show that CAL27 and HSC6 cell proliferation and invasive ability was inhibited, and apoptosis was promoted, when miR-221/222 expression was suppressed. Yang et al (26) studied the miR-221/222 expression in OSCC and demonstrated that upregulating the expression of miR-221/222 increased the growth and tumorigenesis of OSCC cells. Jiang et al (30) demonstrated that miR-222 targets the p53 upregulated modulator of apoptosis expression, impacting OSCC cell growth, apoptosis and invasion, which is consistent with the results of the present study. The RT-qPCR and western blotting data suggest that the expression of PTEN in CAL27 and HSC6 is negatively correlated with miR-221/222 levels. The results demonstrated that PTEN is a crucial functional target of miR-221/222 in OSCC cells.
Previous studies have observed abnormal expression of PTEN in OSCC and that its expression levels were correlated with the degree of carcinoma differentiation (13,14,31). In the present study, it was observed that knockdown of miR-221/222 in CAL27 and HSC6 cells resulted in upregulation of PTEN and downregulation of pAkt. PTEN has been identified as a tumor suppressor and negative regulator of the PI3K/Akt signaling pathway. The PI3K/Akt signaling pathway has been identified as an oncogenic pathway and regulator of numerous cellular processes, including proliferation, cell cycle, migration, invasion and apoptosis (32,33). The results of the present study suggest that OSCC cell proliferation, invasion and apoptosis are associated with expression of PTEN. Activated PTEN dephosphorylates phosphatidylinositol 3,4,5-trisphosphate to give phosphatidylinositol 4,5-bisphosphate, and inhibits the phosphorylation status of Akt, which results in decreased oncogenic activity (34). Akt is a key molecule in the PI3K signaling pathway, and the activity of Akt is tightly regulated by its phosphorylation status (33). pAkt is activated in various types of OSCC and has been confirmed to promote cell proliferation, invasion, migration and reduce apoptosis (23,35). Research has shown that loss of PTEN function may be a factor leading to Akt signaling pathway activation and is consistent with the results of the present study (14,36).
In the present study, it was demonstrated that PTEN is a crucial functional target of miR-221/222 in OSCC cells. When miR-221/222 is inhibited by the miR-221/222 sponge, the results suggest that miR-221/222 has a regulatory function in OSCC cell proliferation and invasion. Through the targeting of PTEN, the PI3K/Akt signaling pathway may also be involved. The results of the present study indicate a potential novel therapy for OSCC.
Acknowledgements
The present study was supported by the National Natural Sciences Foundation of China (grant nos. 81272554 and 81472526), the Guangdong Natural Sciences Foundation (grant no. S2011020003247) and the Guangdong Sciences and Technology Project (grant nos. 2011B050400030 and 2012B031800387).
References
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebrahimi F, Gopalan V, Smith RA, Lam AK. miR-126 in human cancers: Clinical roles and current perspectives. Exp Mol Pathol. 2014;96:98–107. doi: 10.1016/j.yexmp.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 3.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Han L, Zhang A, Wang G, Jia Z, Yang Y, Yue X, Pu P, Shen C, Kang C. Adenovirus-mediated shRNAs for co-repression of miR-221 and miR-222 expression and function in glioblastoma cells. Oncol Rep. 2011;25:97–105. [PubMed] [Google Scholar]
- 5.Mittal SP, Mathai J, Kulkarni AP, Pal JK, Chattopadhyay S. miR-320a regulates erythroid differentiation through MAR binding protein SMAR1. Int J Biochem Cell Biol. 2013;45:2519–2529. doi: 10.1016/j.biocel.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by Targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, Kang C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37:1621–1626. doi: 10.3892/ijo_00000816. [DOI] [PubMed] [Google Scholar]
- 8.Quintavalle C, Garofalo M, Zanca C, Romano G, Iaboni M, del Basso De Caro M, Martinez-Montero JC, Incoronato M, Nuovo G, Croce CM, Condorelli G. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPµ. Oncogene. 2011;31:858–868. doi: 10.1038/onc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jikuzono T, Kawamoto M, Yoshitake H, Kikuchi K, Akasu H, Ishikawa H, Hirokawa M, Miyauchi A, Tsuchiya S, Shimizu K, Takizawa T. The miR-221/222 cluster, miR-10b and miR-92a are highly upregulated in metastatic minimally invasive follicular thyroid carcinoma. Int J Oncol. 2013;42:1858–1868. doi: 10.3892/ijo.2013.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou LJ, Zhao W, Jiang FF, Liu ZF, Ou Yang Y, Yu DS. Construction and validation of microRNA-221/222 sponge vector. Chinese Journal of Stomatological Research (Electronic Version) 2015:106–112. [Google Scholar]
- 13.Rahmani A, Alzohairy M, Babiker AY, Rizvi MA, Elkarimahmad HG. Clinicopathological significance of PTEN and bcl2 expressions in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2012;5:965–971. [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Wu Q, Liu Z, Luo X, Fan Y, Liu Y, Zhang Y, Hua S, Fu Q, Zhao M, et al. Downregulation of FAP suppresses cell proliferation and metastasis through PTEN/PI3K/AKT and Ras-ERK signaling in oral squamous cell carcinoma. Cell Death Dis. 2014;5:e1155. doi: 10.1038/cddis.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorr C, Janik C, Weg M, Been RA, Bader J, Kang R, Ng B, Foran L, Landman SR, O'Sullivan MG, et al. Transposon mutagenesis screen identifies potential lung cancer drivers and CUL3 as a tumor suppressor. Mol Cancer Res. 2015;13:1238–1247. doi: 10.1158/1541-7786.MCR-14-0674-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin PC, Lin JK, Lin HH, Lan YT, Lin CC, Yang SH, Chen WS, Liang WY, Jiang JK, Chang SC. A comprehensive analysis of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) loss in colorectal cancer. World J Surg Oncol. 2015;13:186. doi: 10.1186/s12957-015-0601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ, Kim JH, Yin J, Yoo H, Lee SH, Park JB. Silencing of MicroRNA-21 confers radio-sensitivity through Inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7:e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. 2013;3:465–477. [PMC free article] [PubMed] [Google Scholar]
- 19.Fang L, Li H, Wang L, Hu J, Jin T, Wang J, Yang BB. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974–2987. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beg S, Siraj AK, Prabhakaran S, Jehan Z, Ajarim D, Al-Dayel F, Tulbah A, Al-Kuraya KS. Loss of PTEN expression is associated with aggressive behavior and poor prognosis in Middle Eastern triple-negative breast cancer. Breast Cancer Res Treat. 2015;151:541–553. doi: 10.1007/s10549-015-3430-3. [DOI] [PubMed] [Google Scholar]
- 21.Sun MM, Zhang MZ, Chen Y, Li SL, Zhang W, Ya GW, Chen KS. Effect of PTEN antisense oligonucleotide on oesophageal squamous cell carcinoma cell lines. J Int Med Res. 2012;40:2098–2108. doi: 10.1177/030006051204000607. [DOI] [PubMed] [Google Scholar]
- 22.Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ, Li GX. Anti-miRNA-221 sensitizes human colorectal carcinoma cells to radiation by upregulating PTEN. World J Gastroentero. 2013;19:9307–9317. doi: 10.3748/wjg.v19.i48.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia LF, Huang YP, Zheng YF, Lyu MY, Wei SB, Meng Z, Gan YH. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN-AKT signaling pathway by targeting Sp1. Oral Oncol. 2014;50:1062–1071. doi: 10.1016/j.oraloncology.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Ni Z, Xu X, Xiao J. MiR-32 Functions as a tumor suppressor and directly targets EZH2 in human oral squamous cell carcinoma. Med Sci Monit. 2014;20:2527–2535. doi: 10.12659/MSM.892636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Shen WG, Liu CJ, Chen YW, Lu HH, Tsai MM, Lin SC. miR-221 and miR-222 expression increased the growth and tumorigenesis of oral carcinoma cells. J Oral Pathol Med. 2011;40:560–566. doi: 10.1111/j.1600-0714.2010.01005.x. [DOI] [PubMed] [Google Scholar]
- 27.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 28.Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, Halsema N, Kroesen BJ, van den Berg A. Generation of miRNA sponge constructs. Methods. 2012;58:113–117. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MK, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen BJ, van den Berg A. Rapid generation of MicroRNA Sponges for MicroRNA Inhibition. PLoS One. 2012;7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang F, Zhao W, Zhou L, Zhang L, Liu Z, Yu D. miR-222 regulates the cell biological behavior of oral squamous cell carcinoma by targeting PUMA. Oncol Rep. 2014;31:1255–1262. doi: 10.3892/or.2014.2985. [DOI] [PubMed] [Google Scholar]
- 31.Kurasawa Y, Shiiba M, Nakamura M, Fushimi K, Ishigami T, Bukawa H, Yokoe H, Uzawa K, Tanzawa H. PTEN expression and methylation status in oral squamous cell carcinoma. Oncol Rep. 2008;19:1429–1434. [PubMed] [Google Scholar]
- 32.Cohen Y, Goldenberg-Cohen N, Shalmon B, Shani T, Oren S, Amariglio N, Dratviman-Storobinsky O, Shnaiderman-Shapiro A, Yahalom R, Kaplan I, Hirshberg A. Mutational analysis of PTEN/PIK3CA/AKT pathway in oral squamous cell carcinoma. Oral Oncol. 2011;47:946–950. doi: 10.1016/j.oraloncology.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Liu YZ, Wu K, Huang J, Liu Y, Wang X, Meng ZJ, Yuan SX, Wang DX, Luo JY, Zuo GW, et al. The PTEN/PI3K/Akt and Wnt/β-catenin signaling pathways are involved in the inhibitory effect of resveratrol on human colon cancer cell proliferation. Int J Oncol. 2014;45:104–112. doi: 10.3892/ijo.2014.2392. [DOI] [PubMed] [Google Scholar]
- 34.Giudice FS, Squarize CH. The determinants of head and neck cancer: Unmasking the PI3K pathway mutations. J Carcinog Mutagen. 2013 doi: 10.4172/2157-2518.S5-003. (Suppl 5): pii: 003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie Q, Yan Y, Huang Z, Zhong X, Huang L. MicroRNA-221 targeting PI3-K/Akt signaling axis induces cell proliferation and BCNU resistance in human glioblastoma. Neuropathology. 2014;34:455–464. doi: 10.1111/neup.12129. [DOI] [PubMed] [Google Scholar]
- 36.Gao F, Huang W, Zhang Y, Tang S, Zheng L, Ma F, Wang Y, Tang H, Li X. Hes1 promotes cell proliferation and migration by activating Bmi-1 and PTEN/Akt/GSK3β pathway in human colon cancer. Oncotarget. 2015;6:38667–38680. doi: 10.18632/oncotarget.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]