ABSTRACT
The recent Zaire Ebola virus (EBOV) outbreak in West Africa illustrates clearly the need for additional studies with humans and animals to elucidate the ecology of Ebola viruses (EBVs). In this study, we developed a serological assay based on the Luminex technology. Nine recombinant proteins representing different viral regions (nucleoprotein [NP], 40-kDa viral protein [VP40], and glycoprotein [GP]) from four of the five EBV lineages were used. Samples from 94 survivors of the EBOV outbreak in Guinea and negative samples from 108 patients in France were used to calculate test performance for EBOV detection and cross-reaction with other Ebola virus lineages. For EBOV antibody detection, sensitivities of 95.7%, 96.8%, and 92.5% and specificities of 94.4%, 95.4%, and 96.3% for NP, GP, and VP40, respectively, were observed. All EBOV-negative samples that presented a reaction, except for one, interacted with a single antigen, whereas almost all samples from EBOV survivors were simultaneously reactive with NP and GP (90/94) or with NP, GP, and VP40 (87/94). Considering as positive for past EBOV infection only samples that reacted with EBOV NP and GP, sensitivity was 95.7% and specificity increased to 99.1%. Comparing results with commercial EBOV NP and GP enzyme-linked immunosorbent assays (ELISAs; Alpha Diagnostic, San Antonio, TX), lower sensitivity (92.5%) and high specificity (100%) were observed with the same positivity criteria. Samples from EBOV survivors cross-reacted with GP from Sudan Ebola virus (GP-SUDV) (81.9%), GP from Bundibugyo Ebola virus (GP-BDBV) (51.1%), GP from Reston Ebola virus (GP-RESTV) (9.6%), VP40-SUDV (76.6%), and VP40-BDBV (38.3%). Overall, we developed a sensitive and specific high-throughput serological assay, and defined an algorithm, for epidemiological surveys with humans.
KEYWORDS: Ebola virus, serology, Luminex, recombinant proteins
INTRODUCTION
Since 1976 and the first formal identification of Ebola virus (EBV) disease in Yambuku, in the Democratic Republic of Congo (DRC), a total of 25 outbreaks have been reported today among humans in Africa, with a fatality rate between 25% and 90% (1). Twenty-three of the 25 Ebola virus disease outbreaks occurred in Central Africa, while two occurred in West Africa, including the most devastating outbreak in Guinea/Liberia/Sierra Leone between 2013 and 2016, infecting more than 28,000 persons and causing 11,308 deaths (2, 3). For unknown reasons, no Ebola virus disease epidemics were detected between the 1979 outbreak in DRC and the 1994 outbreak in Côte D'Ivoire. However, since 1999, the frequency of outbreaks has increased (1, 2, 4). There are currently five species of Ebola viruses: Zaire Ebola virus (EBOV) documented in the epidemics in DRC, Gabon, Congo, and Uganda and in the 2014 epidemic in West Africa; Sudan Ebola virus (SUDV) in Sudan, Tai Forest Ebola virus (TAFV) in Côte d'Ivoire, Bundibugyo Ebola virus (BDBV) in Uganda, and Reston Ebola virus (RESTV), found to infect macaques in Asia but with no evidence of lethality in humans (1, 5). Interestingly, phylogenetic analysis of Ebola virus sequences of the 2014 outbreak in West Africa showed that the virus species responsible for the disease is EBOV and was related to the strains of the outbreaks in Central Africa, some 2,000 km away from Guinea. Moreover, the viral strain identified in the recent outbreak in West Africa diverged from Central African strains 10 years ago.
Each Ebola outbreak is the result of a zoonotic transmission, and EBOV genetic material has been amplified from chimpanzee, gorilla, and antelope carcasses and from three fruit bat species in areas where human outbreaks occurred. Moreover, EBOV antibodies have also been detected in other nonhuman primate species and additional insectivorous and frugivorous bat species in areas where no Ebola outbreaks have been documented yet (6). Many uncertainties still remain regarding the ecology of Ebola viruses, how they are maintained between outbreaks in wildlife, and the role of wildlife, because EBOV can also cause disease in certain species, especially chimpanzees and gorillas (7, 8). On the other hand, we cannot rule out that more EBV outbreaks have occurred but were not recognized and thus not reported, since they are mainly located in isolated and remote forest areas, with poor health infrastructure and poor knowledge on the disease. A recent report on a possible filovirus outbreak in 1956 in DRC is in line with this hypothesis (9), and several studies reported the presence of antibodies (sometimes with high levels) in human populations in Central Africa (10).
It is thus important to conduct large-scale retrospective and prospective serological surveys on human populations and wildlife to identify unreported epidemics of Ebola virus disease in areas where conditions of Ebola virus circulation are fulfilled and to elucidate the ecology of the virus. However, such studies require reliable serological tests to detect antibodies to the different Ebola virus lineages. Current methods of detection of antibodies to Ebola virus antigens in humans or nonhuman primates rely on various methodologies (11) and include enzyme-linked immunosorbent assay (ELISA) using whole-viral-lysate antigens (12, 13), synthetic peptides (14), or recombinant proteins (15, 16), and some studies also used Western blotting of whole viral lysates (6, 17). Various anti-Ebola virus antibody prevalences were reported using these methods. For example, a survey conducted in the Republic of Congo, where an immunofluorescence method was used, reported anti-Ebola virus IgG antibodies in 2.5% and 4% of urban and rural human populations, respectively (18). Another study in Gabon that used whole viral lysates to coat ELISA plates reported the presence of IgG to Zaire Ebola virus antigens in 1.3% to 21% of samples tested, depending on the gender and the geographic location (17). Recently, a study reported an IgG prevalence of 18.7% in pygmies from DRC, with an age-related increase, reaching 35% in those aged more than 60 years (10). These methods, although operational, are also time- and bench work-consuming and do not allow a high-throughput screening of samples available for some at minute quantities against all the available Ebola virus lineages. The specificities of some of these methods are also questionable due to the very high seroprevalence rates reported. There is thus a need for alternative methods and screening algorithms. The Multiple Analyte Profiling technology (xMAP; Luminex Corp., Austin, TX) is a flow cytometry-based system (19) that enables simultaneous detection of up to 100 analytes in a single well of a 96-well flat-bottom plate, limiting the volumes of scarce biological samples. We have previously used this technology to detect the prevalence of a wide diversity of simian immunodeficiency virus (SIV) lineages in wild African monkeys (20). In this study, we developed a Luminex-based assay for the simultaneous detection of antibodies to four of five species of the virus in humans and animals. Our data show that the novel assay is as sensitive as and more specific, more accurate, and more cost-effective than a commercial ELISA for the detection of EBOV IgG in human plasma.
RESULTS
Protein coupling conditions and working assay dilution.
To set up the Ebola virus xMAP assay, we used recombinant proteins of the glycoprotein (GP) region for four Ebola virus lineages, 2 representing the Zaire lineage (EBOV) and one for the Sudan (SUDV), Bundibudiyo (BDBV) and Reston (RESTV) lineages. We also included 3 recombinant proteins of the 40-kDa protein (VP40) region derived from EBOV, SUDV, and BDBV lineages, together with one nucleoprotein (NP) recombinant protein from EBOV lineage. We first titrated each recombinant protein from 1.25 μg/1.25 × 106 beads to 5 μg/1.25 × 106 beads. For each coupling condition, an assay was run on the BioPlex200 platform with an EBOV survivor's plasma diluted 1/200. We chose this dilution in this preliminary experiment to be under conditions similar to those of a commercial ELISA used for comparison purposes (see below). A negative plasma sample was tested in parallel. A signal-to-noise ratio was then calculated for each coupling condition. The one that gave the best signal-to-noise ratio and which was cost-effective was 2 μg of protein/1.25 × 106 beads. We used this protein quantity for all subsequent experiments for all 9 recombinant proteins.
Next, to determine the dilution to test plasma samples, we serially diluted from 1/40 to 1/1,000 six Ebola virus-negative plasma samples collected from patients living in France and tested them on GP and NP from EBOV (GP-EBOV and NP-EBOV, respectively) coupled to beads. Results of this titration (Table 1) showed that the median fluorescence intensities (MFIs) varied, on average, from 908 to 85 for NP and from 1,724 to 107 for GP, when diluted from 1/40 to 1/1,000, respectively. We next titrated a sample (MP1745) collected from a survivor of the 2014 Ebola outbreak in Guinea. We tested this sample diluted from 1/100 to 1/1,600 on the two GP and NP recombinants. On NP-EBOV, the average MFI from a triplicate varied from 15,834 to 11,496 for the dilutions 1/100 to 1/1,600, respectively. On GP-coupled beads, the average MFI varied from 12,051 to 2,960 for the Kissidougou/Makona strain and from 12,964 to 3,645 for the Mayinga strain for the dilutions of 1/100 to 1/1,600, respectively. Sudan and Bundibugyo recombinant glycoproteins also significantly cross-reacted with this sample (Table 2). From these two experiments, we concluded that plasma samples should be tested diluted 1/800 or less to exclude false-positive reactions, while signals from positive samples remained very high at dilutions between 1/800 and 1/1,600. We thus chose to test human samples at a final dilution of 1/1,000 in assay buffer.
TABLE 1.
Titration of Ebola virus-negative plasma samples on Zaire Ebola virus recombinant NP and GP
| Plasma dilution | NP-Mayinga MFI (n = 6) |
GP-Mayinga MFI (n = 6) |
||
|---|---|---|---|---|
| Mean | SD | Mean MFI | SD | |
| 40 | 909 | 582 | 1,724 | 942 |
| 80 | 542 | 364 | 1,048 | 537 |
| 100 | 137 | 83 | 606 | 443 |
| 160 | 327 | 216 | 540 | 339 |
| 200 | 241 | 162 | 518 | 401 |
| 320 | 201 | 133 | 347 | 250 |
| 400 | 140 | 96 | 271 | 195 |
| 800 | 92 | 75 | 119 | 85 |
| 1,000 | 86 | 76 | 107 | 38 |
TABLE 2.
Titration of an Ebola virus survivor's plasma sample on Zaire Ebola virus recombinant NP and GP
| Protein | Value (MFI/100 beads) at indicated plasma dilution |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 100 |
200 |
400 |
800 |
1,600 |
||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| NP-Mayinga (n = 3) | 15,835 | 804 | 15,035 | 831 | 15,771 | 276 | 13,300 | 870 | 11,496 | 331 |
| GP-Mayinga (n = 3) | 12,964 | 762 | 11,026 | 398 | 9,424 | 337 | 6,163 | 71 | 3,645 | 151 |
| GP-Kissidougou-Makona (n = 3) | 12,051 | 357 | 9,908 | 386 | 8,239 | 477 | 5,331 | 178 | 2,960 | 91 |
| GP-SUDV (n = 3) | 8,291 | 434 | 6,231 | 100 | 4,773 | 267 | 2,909 | 42 | 1,546 | 71 |
| GP-BDBV (n = 3) | 5,575 | 191 | 3,818 | 51 | 2,657 | 104 | 1,499 | 35 | 694 | 12 |
| GP-RESTV (n = 3) | 491 | 26 | 284 | 4 | 174 | 11 | 93 | 4 | 44 | 2 |
Repeatability of the assay.
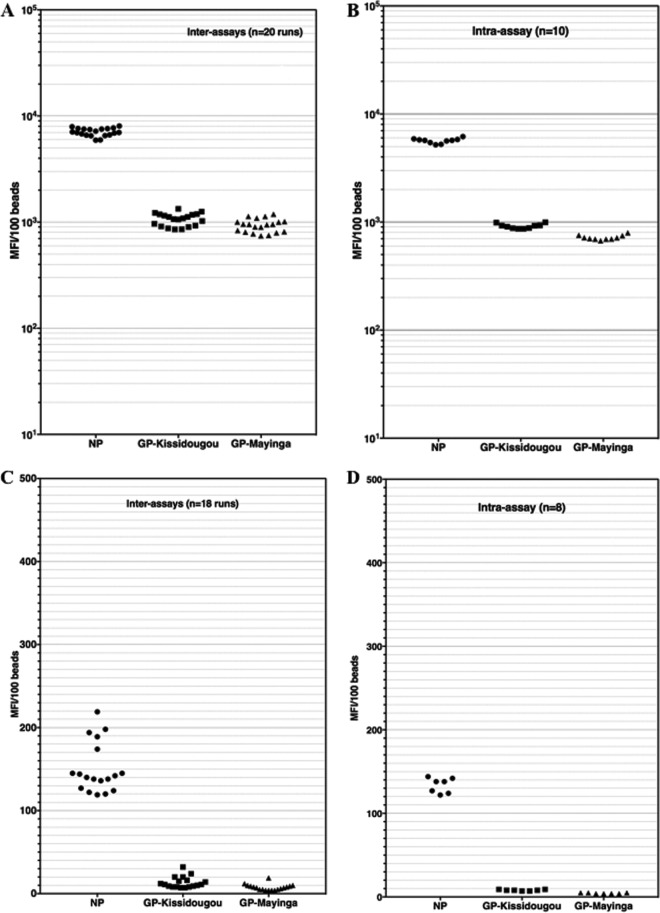
We next evaluated the repeatability of the test within the same run with one EBOV survivor's sample (MP1702) diluted at 1/1,000 and repeated 10 times in the same assay (intra-assay variability). We also evaluated the reads of the same positive sample (MP1702) over 20 different runs (interassay variability) performed by 2 different persons (Fig. 1A and B).
FIG 1.
Interassay and intra-assay variabilities of EBOV-positive and -negative samples in the Luminex assay. An Ebola virus survivor's sample and a negative plasma sample were used to evaluate intra-assay and interassay variability of the novel Luminex assay. Plasma samples were tested at a 1/1,000 dilution in assay buffer and repeated 8 to 10 times within the same run (intra-assay variability [B and D]) or tested in 20 different runs over 3 weeks (interassay variability [A and C]). Panels A and B illustrate these variabilities for an EBOV-positive sample, while panels C and D illustrate the variabilities of an Ebola virus-negative sample.
The intra-assay MFIs varied from 5,188 to 6,177, 672 to 795, and 866 to 995 for NP, GP-Kissidougou/Makona, and GP-Mayinga, respectively. The coefficients of variation were thus 5.3%, 5.1%, and 5.1% for these proteins, in the same order. The interassay (sample MP1702) MFI varied from 5,919 to 8,046, 854 to 1,335, and 743 to 1,189 for NP, GP-Kissidougou, and GP-Mayinga, respectively. The interrun coefficients of variation were thus 8.6%, 13.6%, and 14.4% for the same antigens in the same order.
Similarly, intra-assay (n = 8) and interassay (n = 18 runs) variabilities were checked on an Ebola virus-negative sample diluted at 1/1,000 (Fig. 1C and D). The ranges of intra-assay MFIs were 124 to 145, 7 to 10, and 4 to 6 for NP, GP-Kissidougou/Makona, and GP-Mayinga, respectively (Fig. 1D). The ranges of interassay MFI variability were 119 to 219, 7 to 32, and 7 to 19 for NP, GP-Kissidougou/Makona, and GP-Mayinga, respectively (Fig. 1C). We concluded from these assessments that our novel assay is robust enough for further evaluations.
Cutoff calculation and compared performances of Luminex and ELISAs on EBOV recombinant proteins.
To set the cutoff for our Luminex assay, we tested 108 Ebola virus-negative samples and 94 samples from survivors of the 2014 EBOV outbreak in Guinea on the different EBOV proteins. The samples were tested at a 1/1,000 dilution, determined as described above, on NP, GP, and VP40 from the EBOV lineage. We used receiver operating characteristic (ROC) curve analysis to determine the cutoff values for the 4 antigens. The calculated cutoff values for MFI/100 beads for NP, GP-Mayinga, GP-Kissidougou/Makona, and VP40 were, respectively, 950, 381, 501, and 580 (see Fig. S1A to D in the supplemental material).
Next, we used these cutoff values to determine the number of positive samples among the Ebola virus-negative and EBOV-positive samples and to calculate sensitivity and specificity of the new assay for recombinant NP, GP, and VP40 individually and combined. Results from this analysis are summarized in Table 3. Fifteen EBV-negative samples reacted with NP (n = 6) and one or both of GP (n = 5) and VP40 (n = 4) in the Luminex assay, resulting in specificities of 94.4% for NP, 95.4% for GP, and 96.3% for VP40. Among the 94 samples from EBOV survivors, 90, 91, and 87 reacted with NP, GP, and VP40, resulting in sensitivities of 95.7%, 96.8%, and 92.5%, respectively. Overall, the accuracies of the Luminex assay were thus 95.0% for NP, 96.0% for GP, and 94.5% for VP40. We then compared these performances to the performance of a commercial ELISA. With the commercial ELISA, the specificities were 98.2% and 92.6% for recombinant NP and GP, respectively (Table 3). The sensitivities were 92.5% and 96.8% for NP and GP, respectively. The accuracies were 95.5% for NP and 94.5% for GP.
TABLE 3.
Sensitivity, specificity, and accuracy of the Luminex assay compared to those of a commercial ELISA
| Protein(s) | Test | No. of EBV-negative samples (n = 108) testing positive | % specificity | 95% CI | No. of EBV-positive samples (n = 94) testing positive | % sensitivity | 95% CI | % accuracy | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| NP | ELISA | 2 | 98.20 | 93.4–99.5 | 87 | 92.50 | 85.4–96.4 | 95.50 | 91.7–97.6 |
| Luminex | 6 | 94.40 | 88.4–97.4 | 90 | 95.70 | 89.6–98.3 | 95.00 | 91.1–97.3 | |
| GP | ELISA | 8 | 92.60 | 86.1–96.2 | 91 | 96.80 | 91.3–98.9 | 94.50 | 90.5–96.9 |
| Luminex | 5 | 95.40 | 89.6–98.0 | 91 | 96.80 | 91.3–98.9 | 96.00 | 92.4–98.0 | |
| VP40 | Luminex | 4 | 96.30 | 90.9–98.6 | 87 | 92.50 | 85.4–96.4 | 94.50 | 90.5–96.9 |
| GP + NP | ELISA | 0 | 100 | 96.6–100.0 | 87 | 92.50 | 85.4–96.4 | 96.50 | 93.0–98.3 |
| GP + NP | Luminex | 1 | 99.10 | 94.9–99.8 | 90 | 95.70 | 89.6–98.3 | 97.50 | 94.3–98.9 |
| GP + NP + VP40 | Luminex | 1 | 99.10 | 94.9–99.8 | 87 | 92.50 | 85.4–96.4 | 96.00 | 92.4–98.0 |
Despite the fact that not all samples from EBOV survivors were identified in the antibody assays, we observed that when reactive, almost all samples (90/94) were reactive with NP and both GP proteins and 87/94 were reactive with all proteins (NP plus GP plus VP40). Two samples did not react with any of the proteins, and two samples were reactive with only a single antigen, GP or VP40. In contrast, among the EBV-negative samples, the majority reacted only with a single antigen, except one, which was reactive with all antigens, with MFIs/100 beads of 1,802, 1,549, 514, and 2,933 for NP, GP-Kissidougou/Makona, GP-Mayinga, and VP40, respectively.
In order to improve specificity, we defined an algorithm that requires positivity against more than one antigen to be considered positive for past EBV infection. Using the criterion of positivity with at least two antigens, the highest sensitivity (95.7%) was observed with the combination of NP and GPs, reaching a specificity of 99.1% and an accuracy of 97.5%. Using a criterion of positivity to three antigens decreased the sensitivity to 92.5% but maintained a similar specificity (see results for multiple proteins in Table 3) and an accuracy of 96.0%. Using the same strategy for the ELISA, the combination of NP and GP positivity had lower sensitivity (92.5%) than in the Luminex assay and a specificity of 100%, comparable to that in the Luminex assay. The ELISA accuracy is 96.5%. Thus, the two assays are comparable for specificity and accuracy, and the Luminex assay is more sensitive than the ELISA. Addition of VP40 antigen did not improve the Luminex assay performance.
Using the criterion of positivity with NP and GP proteins, four samples from 94 Ebola survivors were considered negative; one sample (MP1754) significantly reacted with Zaire VP40 only and one was negative with NP with a MFI of 650 (cutoff = 950) and positive with the two GPs. Finally, two samples did not react at all with any of the EBOV antigens. Moreover, these samples were also negative for IgM anti-EBOV NP and GP when tested at 1/1,000 and 1/200 dilutions. The four negative samples were collected from Ebola virus survivors at 268, 281, 358, and 450 days after discharge from the Ebola Treatment Centre. For the 90 reactive samples, this interval ranged between 31 and 532 days, with a median of 324 days. Interestingly, three of the four samples were also negative by the ELISA with both antigens and the fourth was negative with NP and positive with GP. Field investigations, including interviews with social workers and anthropologists, are ongoing to understand these serological results.
Of the 94 survivors included in the present study, 19 have been sampled twice and one patient was sampled three times. The median time between two sampling points was 31 days (range: 1 to 97 days). All the samples collected multiple times, but one gave concordant results with all the antigens tested by the Luminex assay. The discordant sample tested positive first (positive for NP, GP, and VP40) and then negative a month later (positive with GP and VP40 and marginally negative with NP, with signal MFI of 792/100 beads), most probably representing antibody decay with time.
Cross-reactions with other Ebola virus lineages.
One of the major advantages of the Luminex assay is the possibility of multiplex screening. We used this possibility to test for cross-reaction of samples from Zaire Ebola virus survivors against Sudan, Bunbibugyo, and Reston Ebola virus recombinant GP and VP40. We were not able to get recombinant Tai Forest Ebola virus recombinant protein. We thus tested the 94 survivors' samples against the three additional GP (SUDV, BDBV, and RESTV) and two VP40 (SUDV and BDBV) antigens. Results from this testing, and by extrapolating the same cutoff values as for GP-EBOV proteins, in the absence of appropriate controls for these lineages, showed that 77 (81.9%) samples cross-reacted with GP-SUDV, 48 (51.1%) with GP-BDBV, and 9 (9.6%) with GP-RESTV. For VP40, while 87 (92.5%) were positive with VP40-EBOV, 72 (76.6%) and 36 (38.3%) were positive with VP40-SUDV and BDBV, respectively. However, due to the lack of positive controls for SUDV, BDBV, and RESTV antigens, we cannot conclude with exact precision the magnitude of cross-reactions between these antigens and EBOV antigens, as the cutoff values might differ to some extent for homologous reactions. Among the 108 EBV-negative samples, 6 (5.6%) reacted with GP-SUDV, 4 (3.7%) with GP-BDBV, 1 (0.9%) with GP-RESTV, 58 (4.6%) with VP40-SUDV, and 3 (2.8%) with VP40-BDBV (Fig. 2C). Cross-reactions were also detected with the commercial ELISA for recombinant GP-SUDV; among the 94 EBOV survivors' samples tested, 78 (82.9%) were positive according to the assay criteria (Fig. 2B), comparable to what was observed with the Luminex assay. Of the 82 EBV-negative samples tested with this GP-SUDV-specific ELISA (Fig. 2D), seven (8.5%) were reactive, which is also comparable to observations in the Luminex assay.
FIG 2.
Cross-reactions of EBOV antibody-positive plasma samples with recombinant proteins of different lineages of Ebola virus. Plasma samples from 94 EBOV survivors were tested by Luminex (A) or a commercial ELISA (B) against different lineages of Ebola virus recombinant proteins as indicated. Plasma samples from 108 EBOV-negative samples were also tested by Luminex (C) or a commercial ELISA (D) with the same antigens. The horizontal dotted red, blue, and magenta lines (A and C) indicate the cutoff values for NP, GP, and VP40. The cutoff value is the same for NP and GP for the commercial ELISA (determined by the manufacturer) and is indicated by the horizontal dotted green line (B and D). A high proportion of EBOV survivors' samples cross-react with recombinant glycoprotein from SUDV in the Luminex and ELISAs.
Cost effectiveness of the novel Luminex assay.
We calculated the cost to run one sample with the novel assay by including the cost of all the reagents and consumables. The cost to test one sample with one single antigen is $1.59. To test one sample with nine antigens as we did in this study, the cost is $4.09. Under the same conditions and by using the ELISA kits as we used in the present study, for one antigen and one sample, the cost is $8.36; for six antigens, the cost is thus $75.24. Thus, the Luminex assay is cost-effective. For an in-house ELISA using generic reagents with the same recombinant proteins as the Luminex assay, the cost to test one sample with one antigen is $6, and for nine antigens, it is thus $54, and the Luminex assay still remains cost-effective. However, this difference between Luminex and ELISA should be lower if the costs of ELISA and Luminex plate readers are also included in the calculation.
DISCUSSION
In this study, our aim was to develop a serological assay for the simultaneous detection of antibodies to all Ebola virus lineages in human samples. Our data showed that the assay is as specific as and more sensitive, more accurate, and more cost-effective than a commercial ELISA for Zaire Ebola virus (EBOV) antibody detection.
Several serological assays have been developed for epidemiological surveys on past Ebola virus outbreaks in humans and animals. These studies can be split into two major categories: those using whole viral lysates in ELISA or Western blotting and those using recombinant proteins, mainly the nucleoprotein (NP), the glycoprotein (GP), and the 40-kDa viral protein (VP40). The main advantage of using whole-viral-lysate extracts is their potential large spectrum of detection, thus allowing a high level of sensitivity. Using whole virus also potentially minimizes the effect of the kinetics of immune response toward different Ebola virus antigens. For example, it has been shown that IgG antibodies directed against NP were detected in all survivors during the symptomatic phase, while those directed against GP were never detected in this phase (21). A major drawback in using whole viral lysates in ELISA and similar methods is the potential number of false-positive samples, as observed in the early ages of HIV diagnosis, specifically in Africa, where extraordinarily high HIV prevalence rates, up to 40%, were reported in certain areas (22, 23). Subsequently, setting up of algorithms by combined methods and interpretation rules (ELISA and Western blotting and/or immunofluorescence) brought reported HIV seroprevalence rates closer to field realities (24). High rates of EBV antibodies have also been reported with ELISAs using whole-virus antigens in certain areas from Central Africa with documented EBV outbreaks but also in other areas, raising thus also the concern of false-positive results and the need to develop serological algorithms and/or interpretation rules as done in the past for serological diagnosis of HIV.
Based on the observations in our study on antibody reactivity against different antigens for 94 EBOV survivors, we considered a sample positive for EBV antibodies when it was positive with more than a single antigen, and we found that the ideal combination was reactivity against both recombinant NP and GP. With this combination, the specificity of the Luminex assay, which was 94.5% for individual recombinant NP and GP, increased to 99.1% (Table 3). Similarly, the specificity of the commercial ELISA we used for comparison (25), which was 98.2% for NP and 92.7% for GP, reached 100% when our algorithm was used, thus dramatically eliminating false-positive results. Since we used plasma samples from patients in France, it is very likely that the antibody reactivity to Ebola virus antigens is false positive.
We found, with this largest series of Zaire Ebola virus survivors tested by serology so far, that the novel Luminex assay presented a specificity and sensitivity similar to and higher than, respectively, those of the commercial ELISA. Of note, this performance was achieved with samples diluted 1/1,000 (compared to 1/200 for ELISA), thus sparing scarce biological material. It has been described that the Luminex approach, besides allowing the multiplexing of several antigens, is also in general more sensitive than ELISA using the same antigens (26).
In addition to this advantage of applying an algorithm within a single assay, the Luminex approach also reveals potential cross-reaction to related antigens. In our current study, we could not specify with certainty the level of cross-reactions of samples from Zaire Ebola virus survivors toward the other three Ebola virus lineages using recombinant GP and/or VP40 because we lacked positive controls from these infections. However, if we interpreted these reactions in the same way as for EBOV glycoprotein (i.e., with the same cutoffs), the levels of cross-reactions were 81.9%, 51.1%, and 9.6% for SUDV, BDBV, and RESTV, respectively, while 76.6% and 38.3% cross-reacted with VP40 from SUDV and BDBV, respectively. The level of cross-reactions observed with recombinant GP-SUDV (81.9%) in our assay is very close to what we observed with the commercial ELISA for the same antigen (82.9%), thus indirectly validating our assumptions, especially on the cutoff. Several studies reported extensive serological cross-reactions between nucleoproteins of the different Ebola virus lineages. For example, a recent study by Natesan and coworkers (27) reported on reactions and cross-reactions of samples collected from 37 and 20 survivors of SUDV and BDBV infections, respectively. They found that 100% of samples from survivors of BDBV infection cross-reacted with NPs from SUDV, while this proportion was only 25% with GPs. SUDV survivors' samples cross-reacted with EBOV recombinant GP and NP at proportions of 40% and 50%, respectively, while BDBV survivors' samples cross-reacted with EBOV GP and NP at proportions of 10% and 50%, respectively.
Of the 94 Zaire Ebola virus survivors' samples tested by Luminex with nine antigens representing three of the four Ebola virus lineages, 2 were nonreactive with the nine antigens, 1 reacted strongly with VP40-EBOV and faintly with GP-RESTV, and 1 reacted with GP-EBOV from the Mayinga and Kissidougou/Makona strains. Three of the four samples were also negative by both NP and GP commercial EBOV ELISAs. The fourth sample was positive with GP and negative with NP by these two commercial ELISAs. Lack of detectable antibodies to EBOV NP and GP was not due to the difference in the time span since viral infection. Indeed, the delay for the group of 4 patients with no antibodies and the 90 with antibodies was not different statistically. In a follow-up study of survivors from the 1999-2000 Sudan Ebola virus outbreak, Sobarzo and colleagues (28) identified IgG antibodies to NP, VP40, and VP30 and to complete Sudan Ebola virus antigens in 120 survivors from samples collected 6 months (n = 40), 2 years (n = 48), and 10 years (n = 32) after viral infection. No antigen at any time point could interact with 100% of samples. The best rate was observed 10 years after infection with the nucleoprotein, where 26 of the 32 (81.2%) samples tested were positive. The authors detected IgG (and not IgM) antibodies only, but this fact alone or in addition to possible technical issues cannot explain why survivors of an Ebola virus infection lack humoral immune responses to viral antigens. This observation, together with our own results in the current report, suggest that some survivors of Ebola virus infection might lack detectable antibodies by currently available tools, or that the initial diagnosis of infection based on clinical symptoms could have included false-positively declared Ebola virus infections. A study in Gabon (17) showed also the maintenance of antibodies in survivors up to 9 to 12 years after infection, but it used an ELISA with whole-virus lysate, which could not differentiate among the different viral antigens. Nevertheless, a recent study showed presence of antibodies 40 years after infection in patients who survived the first outbreak from Yambuku, in 1976, suggesting thus a longstanding antibody response (29). Moreover, in the same study, neutralizing antibodies were still detected in a subset of patients. More studies are still needed to elucidate the humoral responses to different antigens during and after acute EBV infection.
In conclusion, we have developed a sensitive and specific assay for the serological screening of EBOV infection in humans. Moreover, we developed an algorithm to interpret the serological results to improve specificity of antibody-positive results. This assay can now be used to screen at a large scale samples from humans across Africa to identify eventual past outbreaks or asymptomatic infections among individuals who were in close contact with patients who died from or survived the disease. The assay can also be easily adapted to test animals in order to provide important information on where the virus circulates between zoonotic outbreaks in humans. The assay uses small volumes of plasma and can simultaneously detect antibodies against nine antigens or more in a cost-effective way.
MATERIALS AND METHODS
Human samples.
For Ebola virus antibody-negative samples (n = 108), plasma samples were used from leftover diagnostic samples of the virology laboratory of the University Hospital in Montpellier, France. Fifty microliters of each anonymized sample was aliquoted and kept frozen at −20°C until use. Presumed Ebola virus antibody-positive samples were from survivors of the 2014-2015 EBOV outbreak in Guinea and included in the PostEboGui study (30). Ebola virus infection diagnosis was based on clinical findings and on reverse transcription-PCR (RT-PCR) performed on blood specimens collected upon admission to the Ebola Treatment Centre. Samples from 94 patients were collected as dried blood spots (DBS). For 92 of them, the date between symptom onset and blood sampling was known and the median interval between these two dates was 350 days (interquartile range [IQR]: 293 to 421 days). For each patient, five spots of 50 μl of whole blood were prepared as described previously (31). Plasma was reconstituted from one DBS spot in 1 ml of incubation buffer, consisting of phosphate-buffered saline (PBS) containing 0.75 mol/liter of NaCl, 1% (wt/vol) bovine serum albumin (Sigma-Aldrich, St. Quentin Fallavier, France), 5% (vol/vol) fetal bovine serum (Gibco-Invitrogen, Cergy Pontoise, France), and 0.2% (vol/vol) Tween 20 (Sigma-Aldrich). By taking a hematocrit of 50%, reconstituted plasma from DBS under these conditions was at a dilution of 1/40 (25 μl of plasma in 1,000 μl of buffer). All crude and reconstituted plasma samples from DBS were heat inactivated at 56°C for 30 min at biosafety level 3 (BSL3) before further processing. After heat inactivation, plasma samples reconstituted from DBS were further incubated overnight at 37°C with continuous shaking for complete release of immunoglobulins.
Recombinant proteins and ELISA kits.
As antigens, we used different commercially available recombinant proteins derived from nucleoprotein (NP), viral protein 40 (VP40), and/or glycoprotein (GP) from different Ebola virus lineages. For Zaire Ebola virus nucleoprotein, we used partial NP (amino acids [aa] 488 to 739; MybioSource, San Diego, CA) derived from strain Mayinga 1976. Recombinant VP40 from the following viruses was used: EBOV (strain Kissidougou-Makona 2014, aa 31 to 326), SUDV (strain Gulu, aa 31 to 326), and BDBV (strain Uganda 2007, aa 31 to 326) from Sinobiologicals (Beijing, China). Recombinant glycoproteins from EBOV (Mayinga 1976, aa 1 to 650, and Kissidougou-Makona 2014, aa 1 to 650), SUDV (Uganda 2000, aa 1 to 637), and BDBV (Uganda 2007, aa 1 to 501) were also from Sinobiologicals. Recombinant RESTV glycoprotein (aa 1 to 650) was from IBT (Gaithersburg, MD).
Protein coupling to Luminex beads.
The protocol to couple primary amines bearing moieties (peptides and proteins) to beads has been described previously (20). Briefly, recombinant proteins (2 μg/1,25 × 106 beads) were covalently coupled on carboxyl functionalized fluorescent polystyrene beads (Luminex Corp., Austin, TX) with the BioPlex amine coupling kit (Bio-Rad Laboratories, Marnes-la-Coquette, France) according to the manufacturer's instructions. Unreacted sites were blocked with blocking buffer from the amine coupling kit (Bio-Rad Laboratories). Protein-coupled microsphere preparations were washed with PBS, counted with a hemocytometer, and stored in storage buffer (Bio-Rad) at 4°C in the dark until use.
Multiplex screening for anti-Ebola virus antibodies in human plasma with Luminex.
The buffer used for sample dilution and washing was the same as used to elute plasma DBS samples, described above, and is referenced throughout the text as assay buffer. Before use, recombinant protein-coupled beads were vortexed for 30 s and diluted to 4,000 beads/μl of assay buffer. Tests were performed in 96-well flat-bottom filter plates (Millipore, Tullagreen, Ireland). Plates were prewet with 100 μl of assay buffer, and 50 μl of bead mixture was subsequently added to each well. Liquid was aspirated with a vacuum manifold, and wells were then incubated with 100 μl of plasma or reconstituted DBS (final plasma dilution, 1:1,000) for 16 h at 4°C in the dark on a plate shaker at 300 rpm/min. After 5 washings with 100 μl of assay buffer, 50 μl of anti-human IgG-biotin-labeled (BD-Pharmingen, Le Pont De Claix, France) was added at a concentration of 4 μg/ml in each well and incubated for 30 min in the dark with shaking at 300 rpm. Plates were washed 5 times as described above, and 50 μl of streptavidin-R-phycoerythrin (Fisher Scientific/Life Technologies, Illkirch, France) at 4 μg/ml was added per well and incubated for 10 min with shaking at 300 rpm. Antigen-antibody reactions were then read on BioPlex-200 equipment (Bio-Rad, Marnes-la-Coquette. France). At least 100 events were read for each bead set, and the results were expressed as median fluorescence intensity (MFI) per 100 beads.
Detection of IgG to Ebola virus NP and GP by ELISA.
To validate our test, we compared the results of our xMAP assay to those obtained with a commercially available test. To that end, we used ELISA kits (Alpha Diagnostics, San Antonio, TX) to detect the presence of IgG antibodies to GP- and NP-EBOV and to GP-SUDV as separate tests on the same panel of samples. For ELISAs, samples were diluted at a 1/200 final plasma concentration in sample diluent buffer provided with the test, and ELISA was conducted per the manufacturer's instructions. At the end of the different incubations, optical densities (ODs) were read at 450 nm. We used the cutoff recommended in the user's guide provided in the ELISA kit.
Calculation of cutoff, sensitivity, specificity, and accuracy.
We used receiver operating characteristic (ROC) curve analysis to determine the cutoff values for each antigen used in the Luminex assay. The ROC curve analysis is a graphic representation of the relationship between both the sensitivity and specificity of a diagnostic test, on samples with known disease status. The cutoff is at the optimum where sensitivity and specificity curves intersect. The sensitivity was then defined as the ratio of number of samples found to be positive with the assay to the number of true positives, and the specificity was defined as the ratio of number of samples found to be negative with the assay to the number of true negatives. The accuracy of the assay was defined as the number of correct assessments with the assay divided by the number of all assessments. The accuracy can also be deduced from ROC curve analysis by the value of the area under the curve (AUC). The ROC curve analysis was performed with the Life module of XLSTAT (Addinsoft, Paris, France) implemented in Micosoft Excel. The Wilson method (32) was used to calculate online (http://ww3.ac-poitiers.fr/math/prof/resso/cali/ic_phrek.html) the 95% confidence intervals (CI) around the proportions.
Ethical considerations.
All the participants in the PostEboGui cohort gave their written and informed consent. The study was approved by the Comité National d'éthique pour la recherche en santé of the Republic of Guinea (approval number 074/CNERS/015).
Supplementary Material
ACKNOWLEDGMENTS
We thank the study participants who consented to provide specimens.
This work was supported by the French Ebola Task Force Interministérielle (J.-F. Delfraissy and Y. Yazdanpanah), the Institut National de la Santé et de la Recherche Médicale (Y. Lévy), and the Institut de Recherche pour le Développement.
We report no conflicts of interest.
All the authors reviewed the manuscript. A.A. and M.P. designed the study and wrote the manuscript. A.T., M.S.S., A.K.K., V.F., and E.D. included the patients and collected samples. A.A., C.B., and F.B. performed the experiments.
Members of the PostEboGui Study Group include Ahidjo Ayouba, Sylvain Baize, Kaba Bangoura, Alimou Barry, Moumié Barry, Mohammed Cissé, Eric Delaporte, Jean-François Delfraissy, Christelle Delmas, Alice Desclaux, Saliou Bella Diallo, Mamadou Safiatou Diallo, Mariama Sadio Diallo, Jean François Étard, Cécile Etienne, Ousmane Faye, Ibrahima Fofana, Bruno Granouillac, Esther Hereth Hébert, Suzanne Izard, Djenaba Kassé, Alpha Kabinet Keita, Lamine Koivogui, Cécé Kpamou, Christine Lacarabaratz, Sandrine Leroy, Claire Levy Marchal, Yves Lévy, Vincent Mendiboure, N′Fally Magassouba, Laura March, Philippe Msellati, Harissatou Niane, Martine Peeters, Yves-Marie Pers, Hervé Raoul, Sidi Lamine Sacko, Ibrahima Savané, Mamadou Saliou Sow, Bernard Taverne, Abdoulaye Touré, Fodé Amara Traoré, and Yazdan Yazdanpanah.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01979-16.
REFERENCES
- 1.Pigott DM, Golding N. 2014. Mapping the zoonotic niche of Ebola virus disease in Africa. Elife 3:e04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keita S, De Clerck H, Tiffany A, Dominguez G, Loua M, Traore A, Kolie M, Malano ER, Heleze E, Bocquin A, Mely S, Raoul H, Caro V, Cadar D, Gabriel M, Pahlmann M, Tappe D, Schmidt-Chanasit J, Impouma B, Diallo AK, Formenty P, Van Herp M, Gunther S. 2014. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 3.WHO. 2016. Ebola situation reports. WHO, Geneva, Switzerland: http://apps.who.int/ebola/ebola-situation-reports Accessed 23 September 2016. [Google Scholar]
- 4.Baize S. 2015. Ebola virus in West Africa: new conquered territories and new risks—or how I learned to stop worrying and (not) love Ebola virus. Curr Opin Virol 10:70–76. doi: 10.1016/j.coviro.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Pigott DM, Millear AI, Earl L, Morozoff C. 2016. Updates to the zoonotic niche map of Ebola virus disease in Africa. Elife 5:e16412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa H, Miyamoto H, Nakayama E, Yoshida R, Nakamura I, Sawa H, Ishii A, Thomas Y, Nakagawa E, Matsuno K, Kajihara M, Maruyama J, Nao N, Muramatsu M, Kuroda M, Simulundu E, Changula K, Hang'ombe B, Namangala B, Nambota A, Katampi J, Igarashi M, Ito K, Feldmann H, Sugimoto C, Moonga L, Mweene A, Takada A. 2015. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. J Infect Dis 212(Suppl 2):S101–S108. [DOI] [PubMed] [Google Scholar]
- 7.Bermejo M, Rodriguez-Teijeiro JD, Illera G, Barroso A, Vila C, Walsh PD. 2006. Ebola outbreak killed 5000 gorillas. Science 314:1564. doi: 10.1126/science.1133105. [DOI] [PubMed] [Google Scholar]
- 8.Leroy EM, Rouquet P, Formenty P, Souquiere S, Kilbourne A, Froment JM, Bermejo M, Smit S, Karesh W, Swanepoel R, Zaki SR, Rollin PE. 2004. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science 303:387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- 9.Colebunders R, Van den Ende J. 2015. Filovirus epidemic in 1956 in Bili, DRC. Lancet Infect Dis 15:379. doi: 10.1016/S1473-3099(15)70092-7. [DOI] [PubMed] [Google Scholar]
- 10.Mulangu S, Borchert M, Paweska J, Tshomba A, Afounde A, Kulidri A, Swanepoel R, Muyembe-Tamfum JJ, Van der Stuyft P. 2016. High prevalence of IgG antibodies to Ebola virus in the Efe pygmy population in the Watsa region, Democratic Republic of the Congo. BMC Infect Dis 16:263. doi: 10.1186/s12879-016-1607-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broadhurst MJ, Brooks TJ, Pollock NR. 2016. Diagnosis of Ebola virus disease: past, present, and future. Clin Microbiol Rev 29:773–793. doi: 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ksiazek TG, West CP, Rollin PE, Jahrling PB, Peters CJ. 1999. ELISA for the detection of antibodies to Ebola viruses. J Infect Dis 179(Suppl 1):S192–S198. [DOI] [PubMed] [Google Scholar]
- 13.Krähling V, Becker D, Rohde C, Eickmann M, Eroglu Y, Herwig A, Kerber R, Kowalski K, Vergara-Alert J, Becker S. 2016. Development of an antibody capture ELISA using inactivated Ebola Zaire Makona virus. Med Microbiol Immunol 205:173–183. doi: 10.1007/s00430-015-0438-6. [DOI] [PubMed] [Google Scholar]
- 14.Becquart P, Mahlakoiv T, Nkoghe D, Leroy EM. 2014. Identification of continuous human B-cell epitopes in the VP35, VP40, nucleoprotein and glycoprotein of Ebola virus. PLoS One 9:e96360. doi: 10.1371/journal.pone.0096360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saijo M, Niikura M, Morikawa S, Ksiazek TG, Meyer RF, Peters CJ, Kurane I. 2001. Enzyme-linked immunosorbent assays for detection of antibodies to Ebola and Marburg viruses using recombinant nucleoproteins. J Clin Microbiol 39:1–7. doi: 10.1128/JCM.39.1.1-7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Zhu Y, Yang M, Zhang Z, Song D, Yuan Z. 2014. Nucleoprotein-based indirect enzyme-linked immunosorbent assay (indirect ELISA) for detecting antibodies specific to Ebola virus and Marbug virus. Virol Sin 29:372–380. doi: 10.1007/s12250-014-3512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becquart P, Wauquier N, Mahlakoiv T, Nkoghe D, Padilla C, Souris M, Ollomo B, Gonzalez JP, De Lamballerie X, Kazanji M, Leroy EM. 2010. High prevalence of both humoral and cellular immunity to Zaire ebolavirus among rural populations in Gabon. PLoS One 5:e9126. doi: 10.1371/journal.pone.0009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moyen N, Thirion L, Emmerich P, Dzia-Lepfoundzou A, Richet H, Boehmann Y, Dimi Y, Gallian P, Gould EA, Gunther S, de Lamballerie X. 2015. Risk factors associated with Ebola and Marburg viruses seroprevalence in blood donors in the Republic of Congo. PLoS Negl Trop Dis 9:e0003833. doi: 10.1371/journal.pntd.0003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernhard OK, Mathias RA, Barnes TW, Simpson RJ. 2011. A fluorescent microsphere-based method for assay of multiple analytes in plasma. Methods Mol Biol 728:195–206. doi: 10.1007/978-1-61779-068-3_12. [DOI] [PubMed] [Google Scholar]
- 20.Ahuka-Mundeke S, Ayouba A, Mbala-Kingebeni P, Liegeois F, Esteban A, Lunguya-Metila O, Demba D, Bilulu G, Mbenzo-Abokome V, Inogwabini BI, Muyembe-Tamfum JJ, Delaporte E, Peeters M. 2011. Novel multiplexed HIV/simian immunodeficiency virus antibody detection assay. Emerg Infect Dis 17:2277–2286. doi: 10.3201/eid1712.110783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baize S, Leroy EM, Georges-Courbot MC, Capron M, Lansoud-Soukate J, Debre P, Fisher-Hoch SP, McCormick JB, Georges AJ. 1999. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med 5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 22.Biggar RJ, Melbye M, Kestens L, de Feyter M, Saxinger C, Bodner AJ, Paluko L, Blattner WA, Gigase PL. 1985. Seroepidemiology of HTLV-III antibodies in a remote population of eastern Zaire. Br Med J (Clin Res Ed) 290:808–810. doi: 10.1136/bmj.290.6471.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de The G, Gessain A, Gazzolo L, Robert-Guroff M, Najberg G, Calender A, Peti M, Brubaker G, Bensliman A, Fabry F, Strobel M, Robin Y, Fortune R. 1985. Comparative seroepidemiology of HTLV-I and HTLV-III in the French West Indies and some African countries. Cancer Res 45:4633s–4636s. [PubMed] [Google Scholar]
- 24.Wendler I, Schneider J, Gras B, Fleming AF, Hunsmann G, Schmitz H. 1986. Seroepidemiology of human immunodeficiency virus in Africa. Br Med J (Clin Res Ed) 293:782–785. doi: 10.1136/bmj.293.6550.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewer K, Rampling T, Venkatraman N, Bowyer G, Wright D, Lambe T, Imoukhuede EB, Payne R, Fehling SK, Strecker T, Biedenkopf N, Krahling V, Tully CM, Edwards NJ, Bentley EM, Samuel D, Labbe G, Jin J, Gibani M, Minhinnick A, Wilkie M, Poulton I, Lella N, Roberts R, Hartnell F, Bliss C, Sierra-Davidson K, Powlson J, Berrie E, Tedder R, Roman F, De Ryck I, Nicosia A, Sullivan NJ, Stanley DA, Mbaya OT, Ledgerwood JE, Schwartz RM, Siani L, Colloca S, Folgori A, Di Msarco S, Cortese R, Wright E, Becker S, Graham BS, Koup RA, Levine MM, Volkmann A, Chaplin P, Pollard AJ, Draper SJ, Ballou WR, Lawrie A, Gilbert SC, Hill AV. 2016. A monovalent chimpanzee adenovirus Ebola vaccine boosted with MVA. N Engl J Med 374:1635–1646. doi: 10.1056/NEJMoa1411627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu N, Shichijo S, Nakagawa M, Itoh K. 2004. New multiplexed flow cytometric assay to measure anti-peptide antibody: a novel tool for monitoring immune responses to peptides used for immunization. Scand J Clin Lab Invest 64:535–545. doi: 10.1080/00365510410007008. [DOI] [PubMed] [Google Scholar]
- 27.Natesan M, Jensen SM, Keasey SL, Kamata T, Kuehne AI, Stonier SW, Lutwama JJ, Lobel L, Dye JM, Ulrich RG. 22 June 2016. Human survivors of disease outbreaks caused by Ebola or Marburg viruses exhibit cross-reactive and long-lived antibody responses. Clin Vaccine Immunol doi: 10.1128/cvi.00107-16. [DOI] [PMC free article] [PubMed]
- 28.Sobarzo A, Ochayon DE, Lutwama JJ, Balinandi S, Guttman O, Marks RS, Kuehne AI, Dye JM, Yavelsky V, Lewis EC, Lobel L. 2013. Persistent immune responses after Ebola virus infection. N Engl J Med 369:492–493. doi: 10.1056/NEJMc1300266. [DOI] [PubMed] [Google Scholar]
- 29.Rimoin A, Mukadi WP, Doshi R, Hoff N, Bramble M, Alfonso V, Mwanza A, Mukadi D, Nicholson B, Kebela B, Okitolonda E, Lu K, Steffen I, Olinger G, Hensley L, Simmons G, Muyembe-Tamfum JJ. 2016. Persistent immune response in Ebola survivors from Yambuku outbreak 40 years after infection, abstr 5. 8th International Symposium on Filoviruses, Antwerp, Belgium, 12 to 15 September 2016. [Google Scholar]
- 30.Sow MS, Etard JF, Baize S, Magassouba N, Faye O, Msellati P, Toure AI, Savane I, Barry M, Delaporte E. 3 May 2016. New evidence of long-lasting persistence of Ebola virus genetic material in semen of survivors. J Infect Dis doi: 10.1093/infdis/jiw078. [DOI] [PubMed] [Google Scholar]
- 31.Monleau M, Aghokeng AF, Eymard-Duvernay S, Dagnra A, Kania D, Ngo-Giang-Huong N, Toure-Kane C, Truong LX, Chaix ML, Delaporte E, Ayouba A, Peeters M. 2014. Field evaluation of dried blood spots for routine HIV-1 viral load and drug resistance monitoring in patients receiving antiretroviral therapy in Africa and Asia. J Clin Microbiol 52:578–586. doi: 10.1128/JCM.02860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson EB. 1927. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc 22:209–212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.