ABSTRACT
Extensively drug-resistant (XDR) tuberculosis (TB) cannot be easily or quickly diagnosed. We developed a rapid, automated assay for the detection of XDR-TB plus resistance to the drug isoniazid (INH) for point-of-care use. Using a simple filter-based cartridge with an integrated sample processing function, the assay identified a wide selection of wild-type and mutant sequences associated with XDR-TB directly from sputum. Four new large-Stokes-shift fluorophores were developed. When these four Stokes-shift fluorophores were combined with six conventional fluorophores, 10-color probe detection in a single PCR tube was enabled. A new three-phase, double-nested PCR approach allowed robust melting temperature analysis with enhanced limits of detection (LODs). Finally, newly designed sloppy molecular beacons identified many different mutations using a small number of probes. The assay correctly distinguished wild-type sequences from 32 commonly occurring mutant sequences tested in gyrA, gyrB, katG, and rrs genes and the promoters of inhA and eis genes responsible for resistance to INH, the fluoroquinolone (FQ) drugs, amikacin (AMK), and kanamycin (KAN). The LOD was 300 CFU of Mycobacterium tuberculosis in 1 ml sputum. The rate of detection of heteroresistance by the assay was equivalent to that by Sanger sequencing. In a blind study of 24 clinical sputum samples, resistance mutations were detected in all targets with 100% sensitivity, with the specificity being 93.7 to 100%. Compared to the results of phenotypic susceptibility testing, the sensitivity of the assay was 75% for FQs and 100% each for INH, AMK, and KAN and the specificity was 100% for INH and FQ and 94% for AMK and KAN. Our approach could enable testing for XDR-TB in point-of-care settings, potentially identifying highly drug-resistant TB more quickly and simply than currently available methods.
KEYWORDS: 10-color assay, three-phase PCR, XDR-TB, point-of-care test
INTRODUCTION
Drug-resistant tuberculosis (TB) is an increasing threat worldwide (1–5). The most worrisome cases of TB are caused by multidrug-resistant (MDR) Mycobacterium tuberculosis strains, which are already resistant to rifampin (RIF) and isoniazid (INH) and which acquire additional resistance to the fluoroquinolones (FQs) and any one of the newer injectable drugs, amikacin (AMK), kanamycin (KAN), or capreomycin (CAP). These highly resistant strains are termed extensively drug resistant (XDR) M. tuberculosis (6). XDR-TB is very difficult to treat and can lead to high rates of mortality (7–11), especially when an XDR-TB diagnosis is missed and appropriate treatment is delayed (10). Rapid diagnostic assays for the detection of XDR-TB are urgently needed to address these problems.
Culture and drug susceptibility testing of M. tuberculosis are expensive, time-consuming, and labor-intensive and present a serious biohazard to laboratory workers (12, 13), resulting in fewer accredited facilities in countries where M. tuberculosis is endemic. Even when available, culture-based susceptibility testing can take from weeks to months to complete. M. tuberculosis may also be tested for drug resistance using the alternative rapid, sensitive, and safer genotypic assays, which detect resistance by identifying mutations known to confer resistance to the first- and second-line drugs in a majority of clinical strains (14–23) and minimizes the biohazard. Genotypic testing approaches that can be reduced to a few manual steps are more amenable to use at the point of care, which can dramatically expand their availability to medically underserved populations.
The GeneXpert MTB/RIF (Xpert MTB/RIF) assay is an example of a genotypic assay that identifies M. tuberculosis while it simultaneously detects RIF resistance, if present (24). The Xpert MTB/RIF assay can be performed directly from clinical samples. It is entirely self-contained in a disposable test cartridge, and it can be operated by personnel with minimal training to provide on-demand results at the point of care in less than 2 h. Although the current assay is performed using a desktop GeneXpert MTB/RIF instrument that requires uninterrupted external power, an inexpensive, handheld, and battery-operated alternative platform for this assay recently announced by Cepheid should make it possible to perform Xpert MTB/RIF assays at the point of care in a clinical setting. The Xpert MTB/RIF assay has a clinical sensitivity of 96.8 to 99% for smear-positive sputum samples and 65.4 to 78.7% for smear-negative sputum samples (25, 26), and it identifies RIF resistance with approximately 94% sensitivity and 98% specificity (27). However, the drug resistance portion of the Xpert MTB/RIF assay provides information only on RIF resistance, and a rapid easy-to-use assay for the detection of other types of drug resistance in M. tuberculosis is still urgently needed.
An evolving body of work has indicated that most cases of resistance to the major TB drugs can be associated with approximately 25 mutations in six genes and promoter regions. Approximately 90% of cases of INH resistance can be detected by the presence of mutations in katG codon 315 and nucleotides −8 to −15 of the inhA promoter (28); approximately 90% of cases of FQ resistance can be detected by the presence of mutations in codons 88 to 94 of the gyrA gene, with mutations in codons 500 and 538 to 540 of the gyrB gene identifying resistance in an additional few percentage of cases (29–33). Mutations in positions 1401 and 1402 of rrs identify approximately 80% of cases of AMK resistance, and the same rrs mutations plus mutations at positions −8 to −37 of the eis promoter identify resistance in approximately 80% of cases of KAN resistance (34–36). The presence of a wild-type sequence (or of a few rare, previously identified polymorphisms that do not encode resistance) indicates susceptibility to the same drugs with close to 100% specificity when discordant results are carefully investigated.
Several molecular tests that detect most of the mutations responsible for XDR-TB have recently been developed (4, 14, 16–20, 23, 37–39). However, all of these tests require sophisticated laboratory facilities, making them unsuitable for near-patient TB diagnosis so that treatment may begin immediately (40, 41). Designing an Xpert MTB/RIF-like test to identify XDR mutations at the point of care has been challenging due to the difficulty in identifying all of the clinically relevant mutations associated with drug resistance in six separate genes or promoter regions plus an internal control in a single reaction tube. The need to detect resistance mutations directly from smear-negative clinical sputum samples that may contain small numbers of M. tuberculosis bacilli further challenges the assay design. Here, we describe the development of such a technology for use at the point of care which addresses and resolves all these challenges. This new assay is designed to complement the existing Xpert MTB/RIF assay, supplying additional information on the INH, FQ, KAN, and AMK resistance of isolates in clinical samples that are already positive for TB (with or without RIF resistance) using the Xpert MTB/RIF test. New large-Stokes-shift dyes, innovative three-segment nested thermal cycling, and melting temperature (Tm)-based mutation detection are used in combination to produce a 10-color, eightplex PCR which simultaneously amplifies seven genes and detects over 25 different mutations indicative of XDR-TB directly from sputum. The performance of this highly multiplexed assay in an analytical laboratory setting and its preliminary validation in a field setting are also described.
RESULTS
Large Stokes-shift fluorescent chemistries increase usable detection channels.
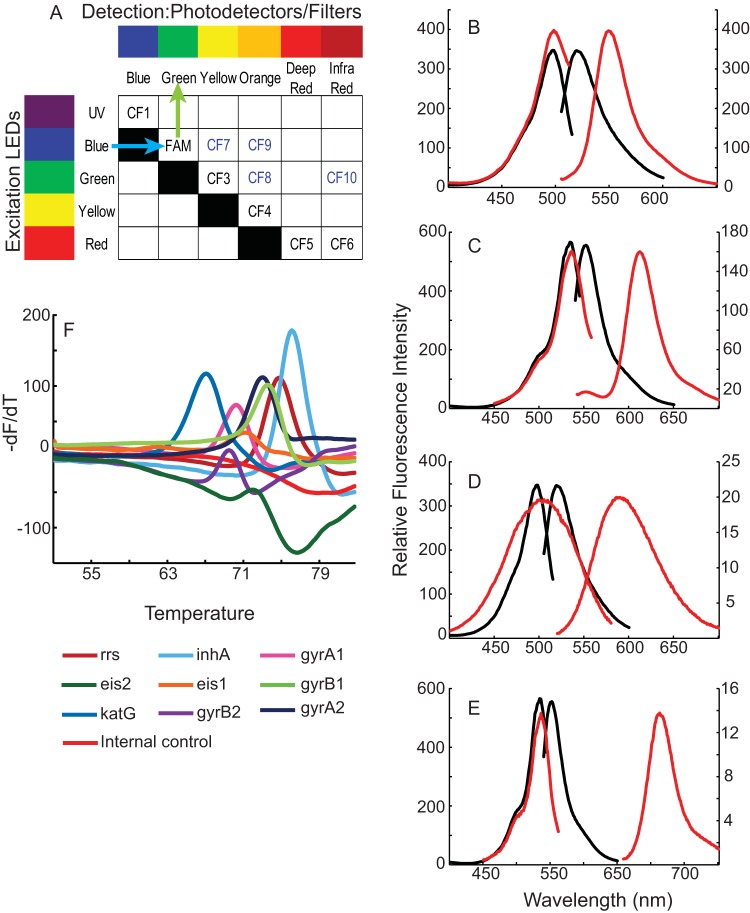
Our assay for the detection of XDR-TB (referred to here as the XDR assay) required nine different probes to identify mutations in seven M. tuberculosis-specific amplicons. An additional 10th probe was needed to confirm amplification of the assay's internal control (IC). The number of probes that can be distinguished in a single multiplexed homologous amplification assay is limited by the number of fluorophores that can be reliably resolved. Most homologous detection systems differentiate among fluorophores using an on-axis detection strategy, whereby each fluorophore has a distinct excitation and emission spectrum (Fig. 1A, CF1 to CF6). On-axis detection is usually limited to four fluorophores, given the narrow range of wavelengths available over the visible spectrum (Fig. 1A, 6-carboxyfluorescein [FAM]-CF5). However, we were able to resolve up to six different fluorophores, using an on-axis strategy, by adding one fluorophore with excitation in the near ultraviolet (UV) (Fig. 1A, CF1) and another fluorophore that emitted in the near infrared (IR) (Fig. 1A, CF6) (24). We sought to further increase multiplexing by developing what we have termed off-axis fluorophores that produce a larger Stokes shift (farther toward the red) than conventional fluorophores. These proprietary off-axis fluorophores have excitation spectra similar to those of on-axis fluorophores, but their unique combination of excitation and emission spectra enabled us to assign each one to a distinct channel that distinguished them from on-axis fluorophores. Large Stokes-shift fluorophores CF7 (which excites blue and emits yellow), CF8 (which excites green and emits orange), CF9 (which excites blue and emits orange), and CF10 (which excites green and emits IR) (Fig. 1A) were created to utilize four off-axis channels, thereby increasing the detection capacity of the GeneXpert MTB/RIF system to 10. Each of these large-Stokes-shift fluorophores produced strong signals when linked to the oligonucleotide probes in aqueous buffer as well as narrow excitation and emission spectra (Fig. 1B to E). We also developed a proprietary quenching molecule with sufficient spectral overlap to efficiently quench all of the new fluorophores, allowing us to incorporate the new off-axis, large-Stokes-shift fluorophores into molecular beacon probes.
FIG 1.
Highly multiplex mutation detection using a combination of on-axis and off-axis fluorophores. (A) Schematic showing the method for resolving up to 20 different fluorophores using only five excitation light-emitting diodes (LEDs) and six detection channels. In the current assay, 4 large off-axis Stokes-shift fluorophores (CF7 to CF10) are combined with 6 on-axis fluorophores (CF1 to CF6) to resolve 10 different fluorophores. UV, ultraviolet. (B to E) The excitation and emission spectra of the large-Stokes-shift fluorophores are shown (red curves) in comparison to the excitation and emission spectra of on-axis fluorophores (black curves) with similar excitation maxima, as shown by the overlapping of the excitation maxima of the different fluorophore types in each panel. The scale for the on-axis fluorophores is shown on the left-hand y axis; the scale for the off-axis fluorophores is shown on the right-hand y axis. (B) FAM and CF7; (C) CF3 and CF8; (D) FAM and CF9; (E) CF3 and CF10. (F) Melt curves of 10 different fluorophore-labeled sloppy molecular beacons specific to drug resistance targets in M. tuberculosis and the assay internal control target, simultaneously detected in a real-time PCR instrument normally designed to detect only six fluorophores. Note the absence of cross talk between channels. -dF/dT indicates the negative first derivative of the normalized fluorescent melt intensity of the probes with respect to temperature.
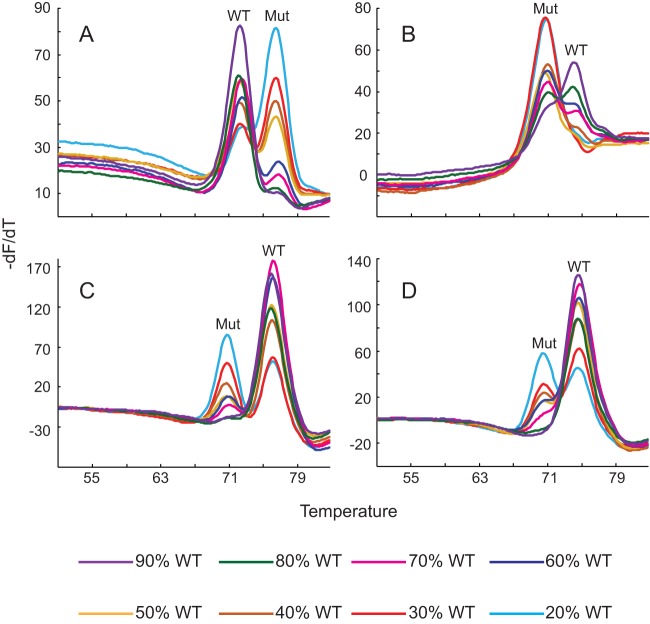
FIG 6.
Detection of heteroresistance. Melting temperature profiles of four different probes obtained from assays using mixtures containing various concentrations of wild-type (WT) and mutant (Mut) DNA, as shown. The wild-type and mutant Tm peaks are indicated in each panel. (A) gyrA1 probe profile with gyrA D94G and wild-type gyrA; (B) gyrA2 probe profile with gyrA D94A and wild-type gyrA; (C) inhA probe profile with inhA promoter C(−15)T and wild-type inhA promoter; (D) rrs probe profile with rrs A1401G and wild-type rrs.
Achieving highly multiplexed melting temperature analysis and PCR in an integrated sample processing cartridge.
We have previously described a series of real-time PCR assays that together detect most cases of resistance to FQs, AMK, and KAN in M. tuberculosis (34, 42). Resistance was detected by hybridizing sloppy molecular beacons (SMBs) to amplicons of drug resistance targets and then measuring the melting temperature (Tm) of the SMB-target hybrid. Drug-susceptible wild-type targets were identified by the presence of characteristic Tm values. Drug resistance mutations in these targets were identified by a shift in Tm values away from the wild-type Tm. However, the drug resistance assays were performed in multiple tubes, and our desire to add INH resistance testing as well as an internal control assay would have increased the number of required tubes even further. We therefore investigated whether we could make use of our 10 resolvable fluorophore-quencher pairs to combine all the different resistance detection assays into a single assay tube in the GeneXpert MTB/RIF system with an enhanced spectral resolution. We labeled 10 SMBs with the 10 fluorophores (and the universal quencher) designed to be detected in 1 of the 10 channels shown in Fig. 1A. Nine of the SMBs had a probe region that was strongly complementary (with no more than 3 mismatches) to short regions in the inhA promoter (one SMB) or the katG gene (one SMB) (for INH resistance detection), the gyrA (two SMBs) or gyrB (two SMBs) gene (for FQ resistance detection), and the rrs gene (one SMB) or eis promoter (two SMBs) (for KAN and AMK resistance detection). A 10th SMB was designed to be complementary to a region of the Bacillus globigii genome and was used as part of an IC for sample processing and PCR. All 10 SMBs as well as the primers (see Tables S1 and S2 in the supplemental material) and all necessary reagents needed to amplify the eight target amplicons in a two-step heminested PCR were integrated into a filter-based sample processing/real-time PCR detection cartridge identical to that used in the Xpert MTB/RIF assay, except for the differences in PCR reagents (24). The full assay was then tested on clinical sputum samples spiked with known numbers of CFU of drug-susceptible M. tuberculosis, using a GeneXpert MTB/RIF instrument designed to perform all the sample processing and PCR steps automatically. The first phase of the PCR was a conventional symmetrical PCR, while the second, nested phase was performed as an asymmetrical PCR (43) to allow the efficient generation of single-stranded amplicons, which are the best targets for Tm analysis. Our results confirmed that the 10-fluorophore set allowed us to differentiate among these 10 SMBs using the same excitation light-emitting diodes, filters, and detectors contained in a GeneXpert MTB/RIF instrument originally designed to differentiate only six fluorescent probes. The 10 individual first-derivative melt curve profiles were clearly visible without any interfering cross talk from one channel to the other (Fig. 1F).
Three-phase nested single-primer amplification dramatically enhances the LOD.
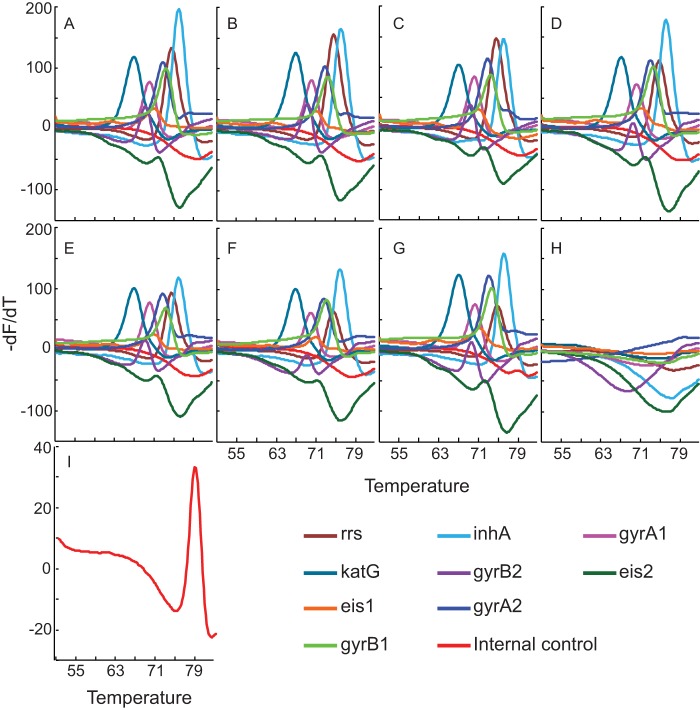
Multiplex PCR can adversely affect assay sensitivity, and we were unable to achieve a limit of detection (LOD) better than 1,639 CFU/ml sputum using a two-phase heminested and nested PCR (Fig. 2A), yet for the XDR assay to be used as a reflex test for positive Xpert MTB/RIF assays, its ideal LOD should be roughly equivalent to the Xpert MTB/RIF assay LOD of 132 CFU/ml (24). To this end, we devised a novel three-phase amplification scheme to further improve the assay LOD. The first amplification phase consisted of a symmetric PCR. This was followed by a second round of nested or heminested symmetric PCR. Finally, a third linear amplification was performed, but this time only one (nested) target primer was used to amplify each amplicon, generating single-stranded products to which the probes could specifically hybridize (Fig. 3). This three-phase amplification approach enabled us to fully optimize the first two PCR phases for sensitive symmetric amplification and to optimize the third amplification phase for the production of single-stranded targets that are ideal for probe-based Tm analysis. The total on-instrument assay time was approximately 110 minutes, permitting a time to the result that was approximately equal to that of the Xpert MTB/RIF assay. We found that three-phase amplification improved the LOD by almost 1 log unit to 300 CFU/ml (Fig. 2B). Even with counts below the established LOD, the assay showed 83%, 70%, and 20% success rates at 250 CFU/ml, 125 CFU/ml, and 62.5 CFU/ml, respectively. All Tm peaks remained strong and easily interpretable even in assays with samples that had counts below the LOD but that were positive (Fig. 4). Importantly, none of the 60 assays that were performed with samples containing M. tuberculosis at the sub-LOD target concentrations and that did not detect M. tuberculosis showed any Tm artifacts that could be erroneously interpreted as peaks corresponding to mutant sequences. Similarly, the >200 negative-control assays without added bacilli, performed over a period of 10 months, were negative for all the M. tuberculosis-specific probes. The IC assay remained positive for all the negative samples (controls or replicates with counts below the LOD), indicating a successful PCR (Fig. 4I).
FIG 2.
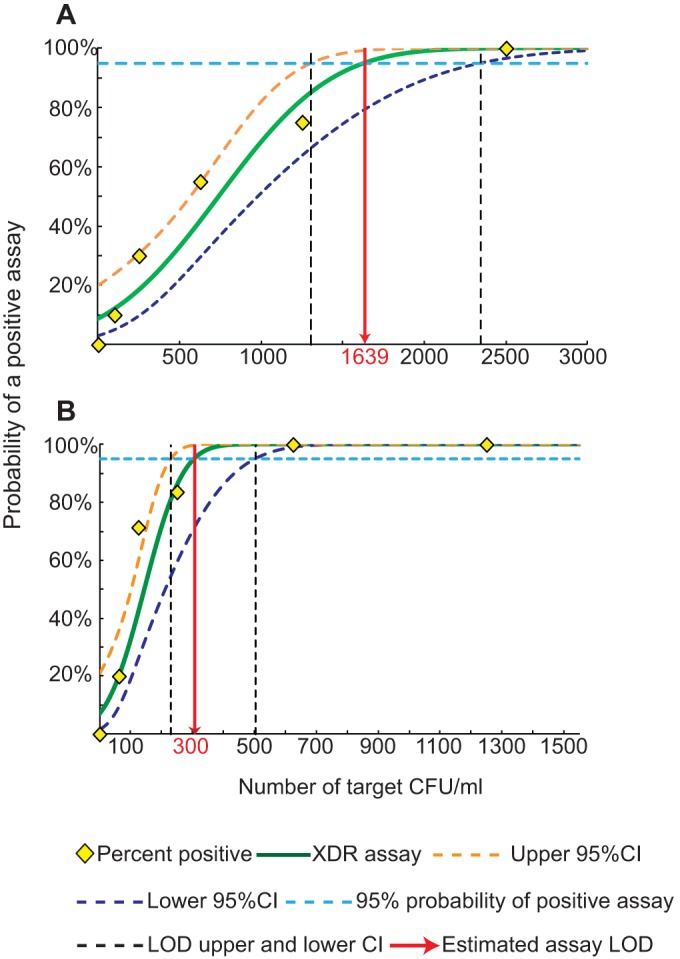
Assay limit of detection. One-milliliter sputum samples were spiked with the indicated numbers of M. tuberculosis CFU, and the success rate of the assay with each number of CFU was tested. The LOD was defined as the lowest number of CFU associated with a 95% probability of a successful assay. (A) Two-phase assay; (B) three-phase assay. CI, confidence interval.
FIG 3.
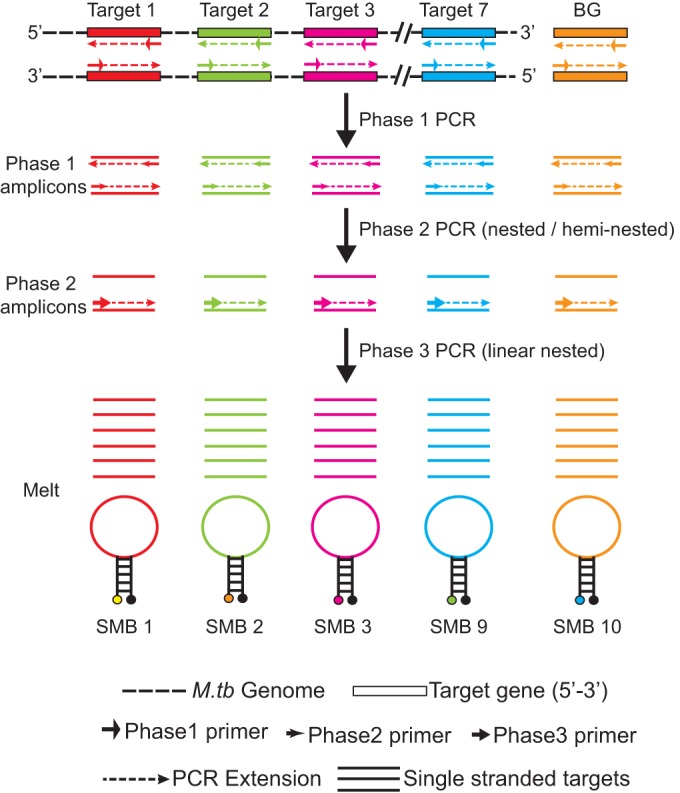
The three-phase, eightplex, 10-color PCR assay scheme. A sevenplex PCR amplifies the genes from M. tuberculosis (M.tb), and the eighth assay amplifies the Bacillus globigii (BG) internal control. Ten differently colored SMB probes target the eight amplicons generated.
FIG 4.
Assay dynamic range. One-milliliter sputum samples were spiked with 2.5 × 107 (A), 2.5 × 105 (B), 2.5 × 104 (C), 1,250 (D), 625 (E), 125 (F), and 62.5 (G) CFU/ml M. tuberculosis or were not spiked with M. tuberculosis (H and I). (A to H) Melt profiles of each of the nine M. tuberculosis detection probes; (I) melt profile of the internal control assay probe in the sputum sample into which M. tuberculosis was not spiked.
Accurate mutation detection.
The nine different M. tuberculosis-specific SMB probes in the Xpert MTB/RIF cartridge used for the XDR assay were designed to detect at least 32 different wild-type and mutant sequences in our test panel known to be associated with clinical INH, FQ, and aminoglycoside resistance (Table 1). We tested the performance of the assay with DNA from an extensive panel of MDR and XDR clinical isolates with a variety of common mutations in the target region, including double and triple mutations in the gyrA quinolone resistance-determining region (QRDR) (Table 1), as well as 50 different pan-susceptible clinical strains with wild-type sequences in all the target genes. Experiments were performed in eight different GeneXpert MTB/RIF system bays over a period of 3 months to confirm the reproducibility of each Tm value and the mutation detection capacity of the assay. The assay generated clear Tm peaks in the presence of each wild-type and mutant sequence (Fig. 5), with standard deviations (SDs) for each Tm being no more than ±0.3°C (Table 1). Mutant targets caused the Tm of the corresponding SMB to shift at least 2°C (equivalent to at least 6 standard deviations) above or below the Tm of the wild-type target. These substantial differences in Tm values between wild-type and mutant sequences (dTms) enabled us to generate probe-specific Tm windows (Table S6) unique to either wild-type or mutant sequences and to easily differentiate between drug-susceptible and drug-resistant isolates.
TABLE 1.
Tm profile and mutations tested by the XDR assay
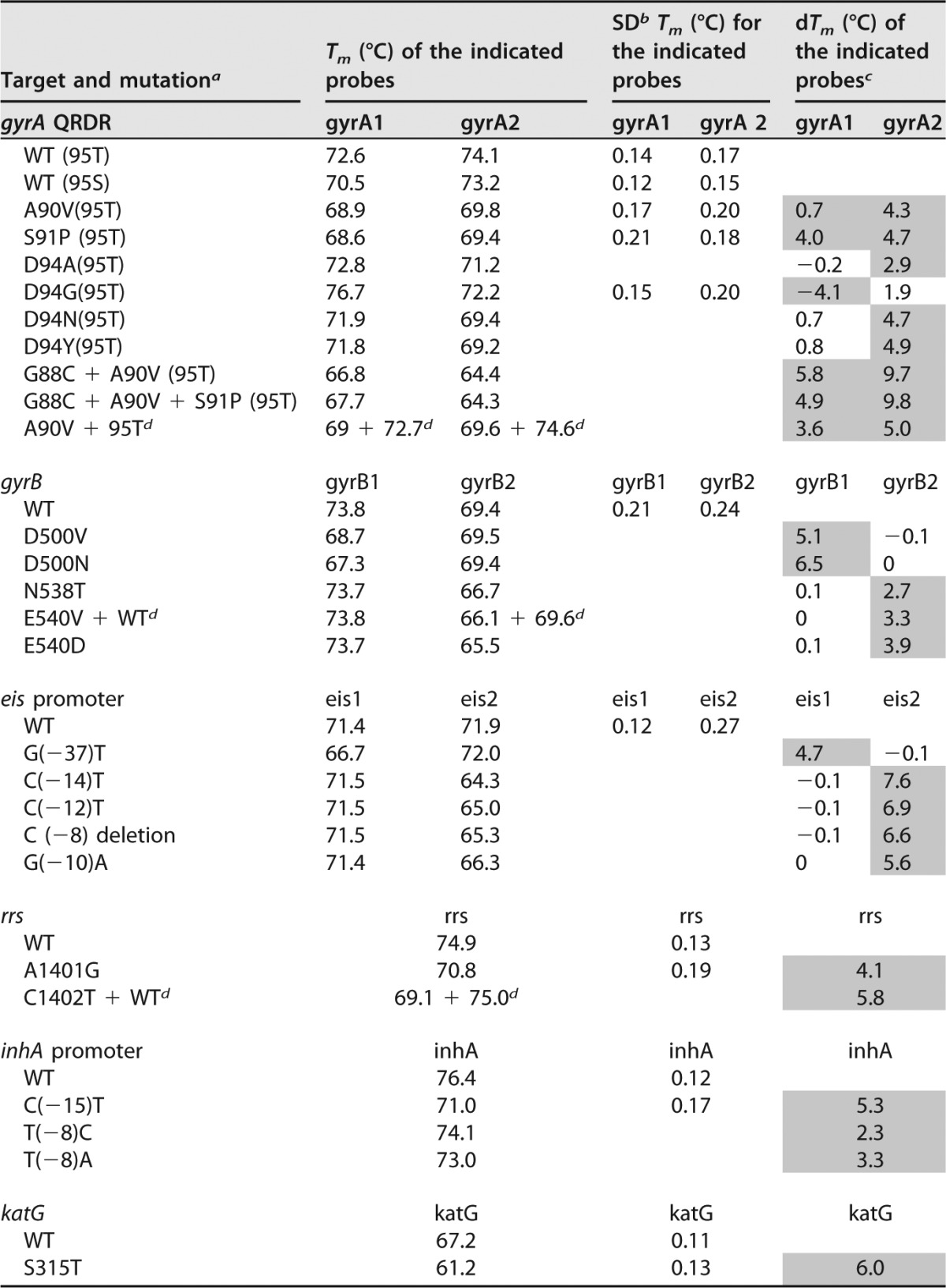
a WT, wild type; other letters represent amino acids except for the promoter region and rrs gene targets, where they represent nucleotides.
b The SD values are shown only for the Tm values obtained from at least 10 different clinical samples.
c The shaded cells represent the difference in Tm (dTm) values that enable the detection of the mutations. All the gyrA QRDR mutations in our study set contained the threonine polymorph at codon 95.
d Mixed DNA showing double Tms from both the mutant and the wild-type sequences in the mixture.
FIG 5.
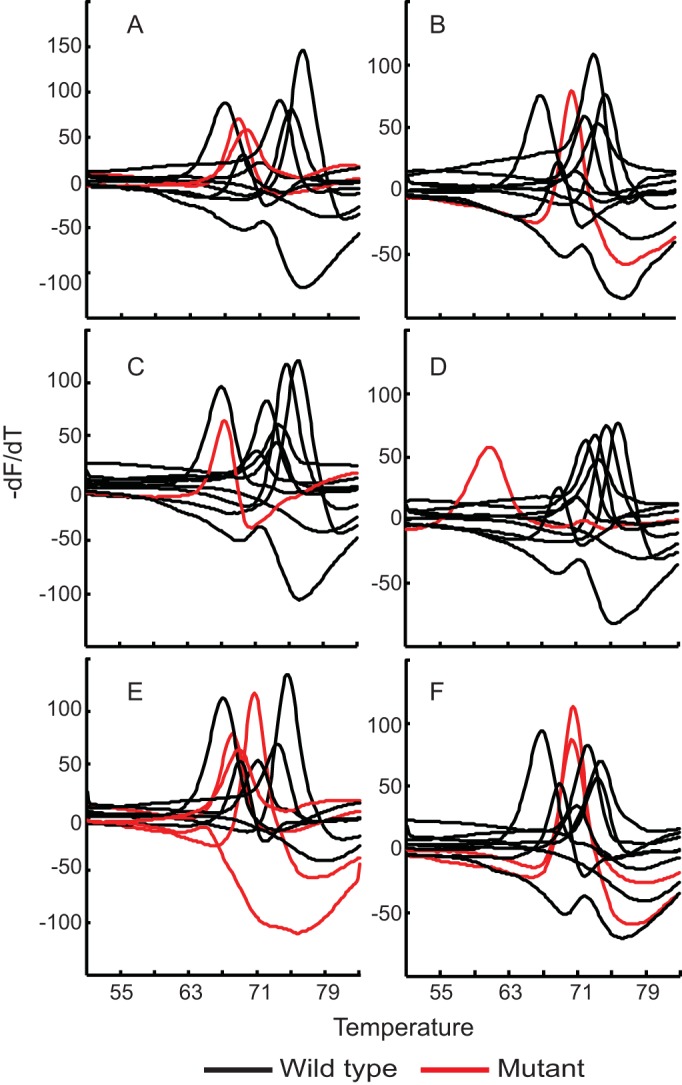
Resistance mutation detection in clinical M. tuberculosis isolates. Melt curves generated during tests of drug-resistant clinical M. tuberculosis isolates are shown for the gyrA A90V (A), inhA promoter C(−15)T (B), gyrB N538T (C), katG S315T (D), gyrA S91P, inhA promoter C(−15)T, and eis promoter C(−12)T (E), and rrs A1401G and inhA promoter C(−15)T (F) mutations. For the gyrA A90V and S91P mutants, both the gyrA probes showed distinct Tm peak shifts (red lines), as observed in panels A and E. For all other mutants, each individual mutation detection probe showed a Tm peak shift associated with the corresponding mutation (red lines in panels B to F). All other Tm peaks retain the wild-type profile (black lines).
There is increasing evidence that different resistance mutations may be associated with different MICs and may confer variable susceptibilities to different drugs within a given drug category (34, 35, 44–46). We investigated whether the Tm generated by each SMB could be used to specifically identify individual mutations (Table 1). Indeed, the two gyrA SMBs could distinguish between the S91P/A90V, D94A, D94G, and D94N/Y mutations and the two gyrB SMBs could distinguish between the D500V, D500N, and N538T/E540V/D mutations. Similarly, the rrs SMB could distinguish between nucleotide 1401 and 1402 mutations, the two different phylogenetic gyrA polymorphisms at position 95 could be specifically identified, and double and triple mutations in gyrA could also be identified as such. We were also able to distinguish among individual inhA and eis promoter mutations by virtue of their different Tm patterns (Table 1).
Detection of heteroresistance.
Patients infected with a mixture of susceptible and resistant clones (heteroresistant clones) have been increasingly described in both MDR-TB and XDR-TB cases (47–50). Detection of heteroresistance can be challenging for molecular assays; however, these mixtures of sequences may be detected as double peaks in Tm-based detection systems (42). We tested the ability of our XDR assay to detect heteroresistance in a series of DNA mixtures containing 0 to 100% mutant DNA against a background of wild-type DNA with a total of 10,000 genome equivalents (mutant plus wild type) per reaction. The DNA mixtures were distributed randomly and coded, and the assay was performed in a blind manner. The mutations tested were D94G and D94A in the gyrA QRDR, C(−15)T in the inhA promoter, and A1401G in the rrs gene. Mutations were detected either as a predominant mutant peak or as distinct double peaks corresponding to both wild-type and mutant Tm profiles. The assay was able to consistently detect as few as 20%, 20%, 30%, and 40% of the sequences with D94A, D94G, C(−15)T, and A1401G mutations, respectively (Fig. 6), whereas the results obtained by Sanger sequencing were 30%, 20%, 20%, and 30%, respectively, indicating the comparable performance of the two methods for the detection of heteroresistance.
Analytical specificity.
We tested the specificity of the assay for M. tuberculosis detection against a large panel of nontuberculous mycobacteria (NTM) (Table 2) as well as common Gram-positive and Gram-negative respiratory pathogens and colonizers (listed in the Materials and Methods section) using 108 CFU of each species. Several of our assay's primers and SMBs were highly specific for M. tuberculosis. We chose the inhA assay, from among our specific SMBs, to definitively identify M. tuberculosis in each test sample because it generated particularly robust Tm peaks with the highest Tm (Fig. 4). All of the NTM samples tested negative for M. tuberculosis on the basis of the results obtained with the inhA SMB. Next, we examined the cross-reactivity of all of the other M. tuberculosis-specific SMBs in the assay. No isolate in the bacterial panel produced a Tm peak for any of the M. tuberculosis SMBs except for the rrs SMB, which generated a wild-type Tm peak from several of the Gram-positive and Gram-negative bacteria tested. Similarly, no NTM produced a Tm peak in the wild-type window for any of the M. tuberculosis-specific SMBs except rrs, which did generate a wild-type Tm for most of the NTMs (Table 2). This was expected, as the rrs region targeted by our assay is highly conserved among most Mycobacterium species as well as some Gram-positive and Gram-negative bacteria. Several NTMs also produced Tm peaks in the assay's mutant Tm windows for the gyrA probes. The gyrA1 probe generated a Tm of 65.6°C with Mycobacterium xenopi and Tms of approximately 64°C with M. scrofulaceum and M. avium and 69.7°C with M. gordonae, and the gyrA2 probe generated Tms of 66.7°C, 66.4°C 66.5°C, and 70.3°C with M. scrofulaceum, M. malmoense, M. avium, and M. gordonae, respectively (Table 2). Cross-reactivity with these NTMs would not normally be a concern, because none would be mistaken for M. tuberculosis due to the lack of an inhA-specific Tm. However, there remained the possibility that NTMs could produce a false-positive resistance designation in rare cases where patients had dual infections with both M. tuberculosis and a cross-reacting NTM. Fortunately, our assay primers only weakly amplified NTMs. When we mixed 106 CFU of any of the five cross-reacting NTMs with 1,000 CFU of M. tuberculosis, only M. tuberculosis-specific wild-type Tm values were produced (Table S3), indicating that our assay would be able to correctly determine the drug resistance genotype of even small quantities of M. tuberculosis against a large NTM background.
TABLE 2.
Tm profiles from M. tuberculosis complex and NTM
| Species |
Tm (°C) for the following probe: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| gyrA1 | gyrA2 | gyrB1 | gyrB2 | inhA | katG | rrs | eis1 | eis2 | |
| M. tuberculosis | 70.4 | 73.2 | 73.6 | 69.4 | 76.4 | 67.3 | 74.9 | 71.3 | 72.0 |
| M. bovis BCG | 72.5 | 74.0 | 73.7 | 69.6 | 76.3 | 67.2 | 75.0 | 71.4 | 71.9 |
| M. scrofulaceum | 64.4 | 66.7 | 74.9 | ||||||
| M. flavescens | 75.0 | ||||||||
| M. vaccae | 74.8 | ||||||||
| M. triviale | 74.9 | ||||||||
| M. szulgai | 74.9 | ||||||||
| M. abscessus | 75.0 | ||||||||
| M. fortuitum | 74.8 | ||||||||
| M. smegmatis | 74.9 | ||||||||
| M. kansasii | 75.0 | ||||||||
| M. intracellulare | 75.0 | ||||||||
| M. simiae | 74.9 | ||||||||
| M. asiaticum | 75.0 | ||||||||
| M. marinum | 74.9 | ||||||||
| M. terrae | 75.0 | ||||||||
| M. celatum | 74.9 | ||||||||
| M. haemophilum | 75.0 | ||||||||
| M. malmoense | 66.4 | 74.8 | |||||||
| M. avium | 64.0 | 66.5 | 74.8 | ||||||
| M. xenopi | 65.6 | ||||||||
| M. gordonae | 69.7 | 70.3 | 75.0 | ||||||
Validation with clinical samples.
We performed a limited clinical evaluation of our assay's performance using a panel of 24 sputum samples that were selected from patients at a clinical site with high MDR-TB and XDR-TB prevalences and for which the investigators were blind to the sequencing or culture results. M. tuberculosis cultured from a separate sputum aliquot of the same patient was used to generate a reference genotype by Sanger sequencing and a reference drug resistance phenotype by use of the MGIT culture system. Assay performance comparisons were performed on all samples for which either sequencing or culture results, or both, were available. Compared to the results of Sanger sequencing, our assay detected 3/3 (100%) gyrA mutants, 5/5 (100%) katG mutants, 3/3 (100%) inhA mutants, and 2/2 (100%) rrs mutants as mutants (Table S4). All gyrA, katG, and inhA sequences which were identified to be wild type by Sanger sequencing were also identified to be wild type by our assay. None of the sputum samples in our study panel were shown to contain gyrB or eis promoter mutations by Sanger sequencing, and all of them were also identified to be wild type by our assay. The rrs assay detected 15/16 (93.8%) wild-type sequences as wild type. The single discordant sample for rrs was identified to be heteroresistant by our assay due to the presence of both the wild-type and mutant rrs peaks indicating the presence of mutant strains against a background of wild-type strains for that patient, which was not detected by Sanger sequencing. Compared to the results of phenotypic susceptibility testing, our assay identified 8/8 (100%) INH-resistant samples to be resistant, 2/2 (100%) AMK- and KAN-resistant samples to be resistant, and 3/4 (75%) FQ-resistant samples to be resistant (Table S5). The single discordant FQ-resistant sample did not show any mutations in either the gyrA or gyrB target upon Sanger sequencing, explaining why it was detected as susceptible by our assay. All phenotypically drug-susceptible samples were detected as wild type in their respective targets by our assay, except for the single sample that was detected as having a mixed sequence for the rrs gene target but that was phenotypically susceptible to AMK and KAN.
DISCUSSION
We have leveraged new large-Stokes-shift fluorophores, innovative microfluidics enabling three sequential PCR phases, and SMB probes that are able to query relatively large stretches of DNA targets to create a highly sensitive multiplex assay which detects all of the important mutations currently thought to be associated with drug-resistant M. tuberculosis. The two manual steps of the assay (adding a buffer to the sputum sample and then placing the sample into an assay cartridge) are identical to those of the Xpert MTB/RIF test, and our assay is performed on the same six-color GeneXpert MTB/RIF system platform. The operation of the Xpert MTB/RIF assay is so simple that it can be easily performed near patients in settings such as primary care clinics, where assay implementation can shorten the time to TB diagnosis and treatment (51).
The rapid mortality associated with untreated XDR-TB among HIV-positive patients highlights the need for a rapid and simple assay such as ours (8). Our assay should detect resistance in approximately 90% of INH-resistant isolates, 90% of fluoroquinolone-resistant isolates, and 80% of kanamycin- and amikacin-resistant isolates, according to previous studies of the mutations detected in the XDR assay (28, 29, 31, 33, 35). Thus, relatively few cases of drug resistance should be missed. Given the very high level of agreement between our XDR assay and Sanger sequencing, our assay should be highly specific. The only likely cause of a false resistance call would be a heteroresistant sample (which should arguably be treated as a resistant TB case) or the rare presence of a synonymous mutation in a probe binding site, as is occasionally seen with the Xpert MTB/RIF assay and rifampin resistance detection (52). The Xpert MTB/RIF assay and GeneXpert MTB/RIF instruments are already widely used in countries with a high burden of TB (26, 51), and our XDR assay can easily be added to sites that already detect TB using the GeneXpert MTB/RIF system. Thus, our new assay could be implemented with minimal additional costs, especially if it is used only as a reflex test on patients whose isolates are already identified to be RIF resistant by Xpert MTB/RIF assay.
The simple operation of our assay belies the actual complexity of its design. The assay can resolve 10 different fluorophores in a single PCR tube. The SMB probes that we used are products of multiple rounds of iterative design, which was needed to achieve reliable Tm separation between wild-type target sequences and all possible mutations. Finally, we were able to achieve a sensitivity equivalent to that of the duplex, heminested symmetric PCR assay used in the Xpert MTB/RIF test, even though our XDR assay amplified eight different amplicons and included an asymmetric amplification step. This was accomplished with a three-phase amplification scheme that took advantage of the exponential increase produced by the first two nested and heminested symmetrical amplification phases and the large amounts of single-stranded target produced by the final linear amplification phase. The linear amplification cycles in the final assay phase also eliminated any risk of exonuclease activity on the SMB probes, ensuring a low background and very robust Tm curves over the entire dynamic range of highly smear-positive to smear-negative samples. Our assay could theoretically accommodate the detection of additional mutations either by the development of fluorophores in another off-axis channel or by the use of two additional probes in the same channel that detect mutations over different temperature ranges.
The long probe length of the SMB probes and their mismatch tolerance capacity allowed us to design probes with programmable hybridization kinetics that could identify a wide range of mutations using a relatively small number of probes (34, 42, 53, 54). Our analytic studies demonstrated robust and reproducible Tm peaks that permitted error-free differentiation between wild-type and mutant target sequences, even at limiting concentrations of the target sequence. The mutation detection rate was 100% in all tests with clinical DNA samples, and the assay performed equally well in a small validation study performed on sputum. The explanation for the lack of detection of a single FQ-resistant isolate in a sputum sample by our assay may be the occurrence of a mutation somewhere outside the region with mutations that are known genetic causes of FQ resistance. Indeed, although the gyrA and gyrB targets we selected have been used to identify approximately 60 to 90% of cases of FQ resistance (42), some cases will remain undetected until the additional genetic causes of FQ resistance are identified and incorporated into future assays. It is unclear whether the result for one sputum sample that we misidentified as AMK and KAN heteroresistant is truly a false-positive result. It is possible that this sample truly contained both resistant and susceptible clones and that the susceptible clones in this mixed infection would eventually also become resistant to AMK and KAN in a patient undergoing treatment with these drugs, resulting in absolute resistance to aminoglycoside treatment.
In summary, our assay provides a potential new tool to identify XDR-TB so that treatment may begin immediately. If the results are confirmed by larger clinical trials, this new test should contribute to enhanced diagnosis, more rapid treatment, and lower rates of mortality and XDR-TB propagation.
MATERIALS AND METHODS
Human subjects approvals.
This study was approved by University of Medicine and Dentistry of New Jersey (now Rutgers University) Institutional Review Board protocol numbers 0120080314, 0120080291, and 0120090104, the Henan Provincial Chest Hospital (HPCH), and National Institute of Allergy and Infectious Diseases Institutional Review Board protocol NCT01071603. Subjects from HPCH provided written consent to participate in the study, including consent for the storage of sputum samples for future testing.
Sample selection.
Sputum samples were collected as part of a study of the natural history of suspected tuberculosis in patients presenting to a tertiary respiratory treatment hospital (Henan Provincial Chest Hospital) in Zhengzhou, China. This population was previously known to be enriched with individuals with highly drug-resistant TB. The enrolled patients were excluded if they had received more than 2 weeks of treatment for TB in the past 3 months. Sputum samples were collected during the study and banked while conventional drug susceptibility testing was performed (see the supplemental material for additional details about the susceptibility testing). Twenty-four sputum samples from individual patients with MDR-TB, XDR-TB, as well as drug-susceptible TB were identified from the stored samples and tested. The results of the XDR assay were analyzed by individuals who were blind to the conventional drug susceptibility testing results and were not used in any way to decide the treatment of the patients.
Cartridge configuration, assay composition, and testing procedure.
The assay used a filter-based sample processing/PCR cartridge (Cartridge A; Cepheid, Sunnyvale CA), which consisted of an integrated PCR tube, a multiposition fluidic valve, a filter to capture the bacteria, and 11 chambers that contained all the buffers needed for sample processing and PCR, as described previously (24). The complete details of the cartridge configuration and reagents are fully described in the supplemental material.
To perform a test, each sample (spiked sputum or cultured M. tuberculosis colonies) was first mixed at a 2:1 ratio with an NaOH- and isopropanol-containing sample reagent (SR) as described previously (24); the sample was then added to the sample loading chamber of the cartridge. The sample was then automatically processed as described fully in the supplemental material. The Tm values generated as output from the assay were then used to identify the presence of either wild-type or mutant targets through a look-up table of Tm windows (Table S6) for each wild-type sequence and each known mutant. A full description of the assay parameters is provided in the supplemental material.
Mutation panel challenge and reference Tm table.
DNA was isolated from clinical M. tuberculosis isolates, including isolates with a wide range of mutations in the target genes, as shown in Table 1. Fifty DNA samples obtained from different pan-susceptible isolates with the wild-type sequence in the target regions were also tested as controls and were originally isolated from clinical M. tuberculosis cultures at the International Tuberculosis Research Center, Masan, South Korea. The XDR assay cartridge was preloaded with the DNA of interest, the loaded cartridge was placed into a GeneXpert MTB/RIF instrument bay, and the assay was performed by selecting the automated 10-color XDR M. tuberculosis detection protocol specific for genomic DNA PCR from the instrument software (research use only; Cepheid, Sunnyvale, CA). The different wild-type and mutant DNA samples were run over a period of 3 months in eight different modules in the GeneXpert MTB/RIF instrument using different cartridge lots to check for the Tm reproducibility. The reference Tm table was generated with the Tm values obtained from these runs.
Sputum sample spiking and GeneXpert MTB/RIF assay.
The analytical sensitivity and limit of detection (LOD) of the XDR assay were determined by spiking an attenuated auxotrophic strain of M. tuberculosis H37Rv (mc26030) (55) into sputum and testing each sample according to our standard protocol. This strain was grown as described previously (55) and quantified by the plate dilution method, and 100-μl aliquots were frozen at −80°C until use. The assay LOD was defined as the number of bacteria that could be spiked into 1 ml of sputum such that all the nine M. tuberculosis-specific SMB probes generated distinct melt curves with unequivocal Tm values for ≥95% of samples tested. The 10th SMB, the IC, was also included in all tests, but its Tm was relevant only for M. tuberculosis-negative samples. Additional details for the spiking and testing protocol are available in the supplemental material.
Evaluation of the analytical specificity of the assay.
The analytical specificity of the assay was tested by using concentrated saturated bacterial liquid cultures (approximately 108 to 109 CFU/ml) or 108 genome equivalents of DNA (in the case of Gram-positive and Gram-negative bacteria for which cultures were not available). The Gram-positive and Gram-negative bacteria included the most common microflora found in sputum and in the upper respiratory tract and mouth, namely, Streptococcus agalactiae, Streptococcus pneumoniae, Streptococcus mitis, Streptococcus mutans, Streptococcus pyogenes, Staphylococcus aureus, Staphylococcus epidermidis, Haemophilus influenzae, Neisseria spp., Nocardia spp., Corynebacterium spp., Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Citrobacter freundii, Moraxella catarrhalis, and Enterobacter cloacae. The NTM species included a laboratory strain of Mycobacterium smegmatis and 19 different NTM isolates obtained from the ATCC repository (Manassas, VA), consisting of M. abscessus, M. scrofulaceum, M. celatum, M. haemophilum, M. asiaticum, M. kansasii, M. avium, M. flavescens, M. szulgai, M. terrae, M. fortuitum, M. intracellulare, M. marinum, M. xenopi, M. vaccae, M. simiae, M. triviale, M. malmoense, and M. gordonae. The Gram-positive and Gram-negative bacteria were not incubated in the sample reagent for more than 2 min to prevent any lysis before they were loaded in the sample chamber. The NTM were incubated for the usual 15 min, as was done for M. tuberculosis. The assay was performed using the cartridges specific for the XDR assay as described above.
Evaluation of the assay with clinical samples.
The evaluation of the assay with 24 clinical samples from patients with MDR-TB, XDR-TB, as well as pan-susceptible TB was performed at HPCH in China, and the results were analyzed in a blind manner at the New Jersey Medical School, Rutgers University. Two volumes of sample treatment reagent was added to 1 volume of sputum, the mixture was incubated for 15 min with occasional shaking, 2 ml was added to the investigational Xpert MTB/RIF cartridges for the XDR assay, and the assay was performed in the GeneXpert MTB/RIF instrument as described above. Details of the microbiological evaluation of the clinical samples are fully described in the supplemental material. For sequencing of the target genes, genomic DNA was isolated from the clinical isolates as described previously (34) and sequencing of the six target genes and promoter regions was performed as described previously (56, 57). The performance of the XDR assay was evaluated against that of the MGIT drug susceptibility testing assay and sequencing as the “gold standard.”
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by U.S. National Institutes of Health (NIH) grants AI080653 and AI111397. Partial funding for the study in South Korea and China was provided by the Intramural Research Program (IRP) of the National Institute of Allergy and Infectious Diseases, NIH, and through IRP's International Centers of Excellence in Research program's Sino-US International Research Center of Tuberculosis at the Henan Provincial Chest Hospital and by the Ministry of Science and Technology of China (2014DFA30340).
D.A. is one of a group of coinvestigators who invented the molecular beacon technology and who receive income from licensees, including a license to Cepheid for M. tuberculosis detection. To manage this conflict of interest, the income attributable to the GeneXpert MTB/RIF assay which he may receive has been irrevocably capped at $5,000 per year. D.A. also reports receiving two research contracts from Cepheid. D.A. and S.C. report the filing of patents for primers and probes for detecting drug resistance in M. tuberculosis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01771-16.
REFERENCES
- 1.Bonnet M, Sizaire V, Kebede Y, Janin A, Doshetov D, Mirzoian B, Arzumanian A, Muminov T, Iona E, Rigouts L, Rusch-Gerdes S, Varaine F. 2005. Does one size fit all? Drug resistance and standard treatments: results of six tuberculosis programmes in former Soviet countries. Int J Tuberc Lung Dis 9:1147–1154. [PubMed] [Google Scholar]
- 2.Lin J, Sattar AN, Puckree T. 2004. An alarming rate of drug-resistant tuberculosis at Ngwelezane Hospital in northern KwaZulu Natal, South Africa. Int J Tuberc Lung Dis 8:568–573. [PubMed] [Google Scholar]
- 3.Liu CE, Chen CH, Hsiao JH, Young TG, Tsay RW, Fung CP. 2004. Drug resistance of Mycobacterium tuberculosis complex in central Taiwan. J Microbiol Immunol Infect 37:295–300. [PubMed] [Google Scholar]
- 4.Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. 2003. Allele-specific rpoB PCR assays for detection of rifampin-resistant Mycobacterium tuberculosis in sputum smears. Antimicrob Agents Chemother 47:2231–2235. doi: 10.1128/AAC.47.7.2231-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Negi SS, Gupta S, Lal S. 2003. Drug resistance in tuberculosis in Delhi: a 2 year profile (2001-2002). J Commun Dis 35:74–81. [PubMed] [Google Scholar]
- 6.Raviglione MC, Smith IM. 2007. XDR tuberculosis—implications for global public health. N Engl J Med 356:656–659. doi: 10.1056/NEJMp068273. [DOI] [PubMed] [Google Scholar]
- 7.Faustini A, Hall AJ, Perucci CA. 2005. Tuberculosis treatment outcomes in Europe: a systematic review. Eur Respir J 26:503–510. doi: 10.1183/09031936.05.00103504. [DOI] [PubMed] [Google Scholar]
- 8.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 9.Goble M, Iseman MD, Madsen LA, Waite D, Ackerson L, Horsburgh CR Jr. 1993. Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N Engl J Med 328:527–532. doi: 10.1056/NEJM199302253280802. [DOI] [PubMed] [Google Scholar]
- 10.Iseman MD. 1993. Treatment of multidrug-resistant tuberculosis. N Engl J Med 329:784–791. doi: 10.1056/NEJM199309093291108. [DOI] [PubMed] [Google Scholar]
- 11.Iseman MD. 1999. Management of multidrug-resistant tuberculosis. Chemotherapy 45(Suppl 2):S3–S11. [DOI] [PubMed] [Google Scholar]
- 12.Kent PT, Kubica GP. 1985. Public health mycobacteriology: a guide for level III laboratory. Centers for Disease Control, U.S. Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 13.Heifets L, Linder T, Sanchez T, Spencer D, Brennan J. 2000. Two liquid medium systems, mycobacteria growth indicator tube and MB redox tube, for Mycobacterium tuberculosis isolation from sputum specimens. J Clin Microbiol 38:1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ajbani K, Shetty A, Mehta A, Rodrigues C. 2011. Rapid diagnosis of extensively drug-resistant tuberculosis by use of a reverse line blot hybridization assay. J Clin Microbiol 49:2546–2551. doi: 10.1128/JCM.02511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balasingham SV, Davidsen T, Szpinda I, Frye SA, Tonjum T. 2009. Molecular diagnostics in tuberculosis: basis and implications for therapy. Mol Diagn Ther 13:137–151. doi: 10.1007/BF03256322. [DOI] [PubMed] [Google Scholar]
- 16.Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, Hooks DP, Cowan LS, Plikaytis BB, Posey JE. 2011. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:2032–2041. doi: 10.1128/AAC.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dauendorffer JN, Guillemin I, Aubry A, Truffot-Pernot C, Sougakoff W, Jarlier V, Cambau E. 2003. Identification of mycobacterial species by PCR sequencing of quinolone resistance-determining regions of DNA gyrase genes. J Clin Microbiol 41:1311–1315. doi: 10.1128/JCM.41.3.1311-1315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, Liu S, Wang Q, Wang L, Tang S, Wang J, Lu W. 2013. Rapid diagnosis of drug resistance to fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol using genotype MTBDRsl assay: a meta-analysis. PLoS One 8:e55292. doi: 10.1371/journal.pone.0055292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feuerriegel S, Cox HS, Zarkua N, Karimovich HA, Braker K, Rusch-Gerdes S, Niemann S. 2009. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob Agents Chemother 53:3353–3356. doi: 10.1128/AAC.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillery N, Groessl EJ, Trollip A, Catanzaro D, Jackson L, Rodwell TC, Garfein RS, Lin SY, Eisenach K, Ganiats TG, Park D, Valafar F, Rodrigues C, Crudu V, Victor TC, Catanzaro A. 2014. The Global Consortium for Drug-Resistant Tuberculosis Diagnostics (GCDD): design of a multi-site, head-to-head study of three rapid tests to detect extensively drug-resistant tuberculosis. Trials 15:434. doi: 10.1186/1745-6215-15-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalokhe AS, Shafiq M, Lee JC, Ray SM, Wang YF, Metchock B, Anderson AM, Nguyen ML. 2013. Multidrug-resistant tuberculosis drug susceptibility and molecular diagnostic testing. Am J Med Sci 345:143–148. doi: 10.1097/MAJ.0b013e31825d32c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Grady J, Maeurer M, Mwaba P, Kapata N, Bates M, Hoelscher M, Zumla A. 2011. New and improved diagnostics for detection of drug-resistant pulmonary tuberculosis. Curr Opin Pulm Med 17:134–141. doi: 10.1097/MCP.0b013e3283452346. [DOI] [PubMed] [Google Scholar]
- 23.Rodwell TC, Valafar F, Douglas J, Qian L, Garfein RS, Chawla A, Torres J, Zadorozhny V, Kim MS, Hoshide M, Catanzaro D, Jackson L, Lin G, Desmond E, Rodrigues C, Eisenach K, Victor TC, Ismail N, Crudu V, Gler MT, Catanzaro A. 2014. Predicting extensively drug-resistant Mycobacterium tuberculosis phenotypes with genetic mutations. J Clin Microbiol 52:781–789. doi: 10.1128/JCM.02701-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-Lopez EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, Gler MT, Blakemore R, Worodria W, Gray C, Huang L, Caceres T, Mehdiyev R, Raymond L, Whitelaw A, Sagadevan K, Alexander H, Albert H, Cobelens F, Cox H, Alland D, Perkins MD. 2011. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 377:1495–1505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. 2014. Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev 1:CD009593. doi: 10.1002/14651858.CD009593.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazbon MH, Brimacombe M, Bobadilla del Valle M, Cavatore M, Guerrero MI, Varma-Basil M, Billman-Jacobe H, Lavender C, Fyfe J, Garcia-Garcia L, Leon CI, Bose M, Chaves F, Murray M, Eisenach KD, Sifuentes-Osornio J, Cave MD, Ponce de Leon A, Alland D. 2006. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 50:2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Z, Wang J, Lu J, Huang X, Hu Z. 2011. Association of mutation patterns in gyrA/B genes and ofloxacin resistance levels in Mycobacterium tuberculosis isolates from East China in 2009. BMC Infect Dis 11:78. doi: 10.1186/1471-2334-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi R, Zhang J, Li C, Kazumi Y, Sugawara I. 2006. Emergence of ofloxacin resistance in Mycobacterium tuberculosis clinical isolates from China as determined by gyrA mutation analysis using denaturing high-pressure liquid chromatography and DNA sequencing. J Clin Microbiol 44:4566–4568. doi: 10.1128/JCM.01916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh P, Jain A, Dixit P, Prakash S, Jaiswal I, Venkatesh V, Singh M. 2014. Prevalence of gyrA and B gene mutations in fluoroquinolone-resistant and -sensitive clinical isolates of Mycobacterium tuberculosis and their relationship with MIC of ofloxacin. J Antibiot (Tokyo) 68:63–66. doi: 10.1038/ja.2014.95. [DOI] [PubMed] [Google Scholar]
- 32.Wang JY, Lee LN, Lai HC, Wang SK, Jan IS, Yu CJ, Hsueh PR, Yang PC. 2007. Fluoroquinolone resistance in Mycobacterium tuberculosis isolates: associated genetic mutations and relationship to antimicrobial exposure. J Antimicrob Chemother 59:860–865. doi: 10.1093/jac/dkm061. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, Liu Z, Li C, Galwey N, Deng J, Zhou Y, Zhu Y, Gao Y, Wang S, Huang Y, Wang M, Zhong Q, Zhou L, Chen T, Zhou J, Yang R, Zhu G, Hang H, Zhang J, Li F, Wan K, Wang J, Zhang XE, Bi L. 2013. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet 45:1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 34.Chakravorty S, Lee JS, Cho EJ, Roh SS, Smith LE, Lee J, Kim CT, Via LE, Cho SN, Barry CE III, Alland D. 2015. Genotypic susceptibility testing of Mycobacterium tuberculosis for amikacin and kanamycin resistance using a rapid sloppy molecular beacon-based assay identifies more cases of low-level drug resistance than phenotypic Lowenstein-Jensen testing. J Clin Microbiol 53:43–51. doi: 10.1128/JCM.02059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, Rodwell TC. 2012. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One 7:e33275. doi: 10.1371/journal.pone.0033275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maus CE, Plikaytis BB, Shinnick TM. 2005. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob Agents Chemother 49:3192–3197. doi: 10.1128/AAC.49.8.3192-3197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo T, Jiang L, Sun W, Fu G, Mei J, Gao Q. 2011. Multiplex real-time PCR melting curve assay to detect drug-resistant mutations of Mycobacterium tuberculosis. J Clin Microbiol 49:3132–3138. doi: 10.1128/JCM.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan M, Kalantri S, Flores L, Pai M. 2005. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect Dis 5:62. doi: 10.1186/1471-2334-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossau R, Traore H, De Beenhouwer H, Mijs W, Jannes G, De Rijk P, Portaels F. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob Agents Chemother 41:2093–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albert H, Bwanga F, Mukkada S, Nyesiga B, Ademun JP, Lukyamuzi G, Haile M, Hoffner S, Joloba M, O'Brien R. 2010. Rapid screening of MDR-TB using molecular line probe assay is feasible in Uganda. BMC Infect Dis 10:41. doi: 10.1186/1471-2334-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ling DI, Zwerling AA, Pai M. 2008. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J 32:1165–1174. doi: 10.1183/09031936.00061808. [DOI] [PubMed] [Google Scholar]
- 42.Chakravorty S, Aladegbami B, Thoms K, Lee JS, Lee EG, Rajan V, Cho EJ, Kim H, Kwak H, Kurepina N, Cho SN, Kreiswirth B, Via LE, Barry CE III, Alland D. 2011. Rapid detection of fluoroquinolone-resistant and heteroresistant Mycobacterium tuberculosis by use of sloppy molecular beacons and dual melting-temperature codes in a real-time PCR assay. J Clin Microbiol 49:932–940. doi: 10.1128/JCM.02271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyllensten UB, Erlich HA. 1988. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A 85:7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Chen Z, Li Y, Xia W, Chen X, Chen T, Zhou L, Xu B, Xu S. 2012. Characterization of gyrA and gyrB mutations and fluoroquinolone resistance in Mycobacterium tuberculosis clinical isolates from Hubei Province, China. Braz J Infect Dis 16:136–141. doi: 10.1016/S1413-8670(12)70294-5. [DOI] [PubMed] [Google Scholar]
- 45.Huyen MN, Cobelens FG, Buu TN, Lan NT, Dung NH, Kremer K, Tiemersma EW, van Soolingen D. 2013. Epidemiology of isoniazid resistance mutations and their effect on tuberculosis treatment outcomes. Antimicrob Agents Chemother 57:3620–3627. doi: 10.1128/AAC.00077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sirgel FA, Warren RM, Streicher EM, Victor TC, van Helden PD, Bottger EC. 2012. gyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother 67:1088–1093. doi: 10.1093/jac/dks033. [DOI] [PubMed] [Google Scholar]
- 47.Hofmann-Thiel S, van Ingen J, Feldmann K, Turaev L, Uzakova GT, Murmusaeva G, van Soolingen D, Hoffmann H. 2009. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur Respir J 33:368–374. doi: 10.1183/09031936.00089808. [DOI] [PubMed] [Google Scholar]
- 48.Mekonnen D, Admassu A, Mulu W, Amor A, Benito A, Gelaye W, Biadglegne F, Abera B. 2015. Multidrug-resistant and heteroresistant Mycobacterium tuberculosis and associated gene mutations in Ethiopia. Int J Infect Dis 39:34–38. doi: 10.1016/j.ijid.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 49.Pholwat S, Stroup S, Foongladda S, Houpt E. 2013. Digital PCR to detect and quantify heteroresistance in drug resistant Mycobacterium tuberculosis. PLoS One 8:e57238. doi: 10.1371/journal.pone.0057238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolani MP, D'Souza DT, Mistry NF. 2012. Drug resistance mutations and heteroresistance detected using the GenoType MTBDRplus assay and their implication for treatment outcomes in patients from Mumbai, India. BMC Infect Dis 12:9. doi: 10.1186/1471-2334-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cox HS, Mbhele S, Mohess N, Whitelaw A, Muller O, Zemanay W, Little F, Azevedo V, Simpson J, Boehme CC, Nicol MP. 2014. Impact of Xpert MTB/RIF for TB diagnosis in a primary care clinic with high TB and HIV prevalence in South Africa: a pragmatic randomised trial. PLoS Med 11:e1001760. doi: 10.1371/journal.pmed.1001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alonso M, Palacios JJ, Herranz M, Penedo A, Menendez A, Bouza E, Garcia de Viedma D. 2011. Isolation of Mycobacterium tuberculosis strains with a silent mutation in rpoB leading to potential misassignment of resistance category. J Clin Microbiol 49:2688–2690. doi: 10.1128/JCM.00659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakravorty S, Aladegbami B, Burday M, Levi M, Marras SA, Shah D, El-Hajj HH, Kramer FR, Alland D. 2010. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J Clin Microbiol 48:258–267. doi: 10.1128/JCM.01725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakravorty S, Kothari H, Aladegbami B, Cho EJ, Lee JS, Roh SS, Kim H, Kwak H, Lee EG, Hwang SH, Banada PP, Safi H, Via LE, Cho SN, Barry CE III, Alland D. 2012. Rapid, high-throughput detection of rifampin resistance and heteroresistance in Mycobacterium tuberculosis by use of sloppy molecular beacon melting temperature coding. J Clin Microbiol 50:2194–2202. doi: 10.1128/JCM.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL, Jacobs WR Jr. 2002. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med 8:1171–1174. doi: 10.1038/nm765. [DOI] [PubMed] [Google Scholar]
- 56.Lee J, Armstrong DT, Ssengooba W, Park JA, Yu Y, Mumbowa F, Namaganda C, Mboowa G, Nakayita G, Armakovitch S, Chien G, Cho SN, Via LE, Barry CE III, Ellner JJ, Alland D, Dorman SE, Joloba ML. 2014. Sensititre MYCOTB MIC plate for testing Mycobacterium tuberculosis susceptibility to first- and second-line drugs. Antimicrob Agents Chemother 58:11–18. doi: 10.1128/AAC.01209-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song T, Park Y, Shamputa IC, Seo S, Lee SY, Jeon HS, Choi H, Lee M, Glynne RJ, Barnes SW, Walker JR, Batalov S, Yusim K, Feng S, Tung CS, Theiler J, Via LE, Boshoff HI, Murakami KS, Korber B, Barry CE III, Cho SN. 2014. Fitness costs of rifampicin resistance in Mycobacterium tuberculosis are amplified under conditions of nutrient starvation and compensated by mutation in the β′ subunit of RNA polymerase. Mol Microbiol 91:1106–1119. doi: 10.1111/mmi.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.