ABSTRACT
New Delhi metallo-β-lactamase-1 (NDM-1)-producing Enterobacteriaceae has disseminated rapidly throughout the world and poses an urgent threat to public health. Previous studies confirmed that the blaNDM-1 gene is typically carried in plasmids but rarely in chromosome. We discovered a multidrug-resistant Escherichia coli strain Y5, originating from a urine sample and containing the blaNDM-1 gene, which did not transfer by either conjugation or electrotransformation. We confirmed the possibility of its chromosome location by S1-pulsed-field gel electrophoresis (PFGE) and XbaI-PFGE, followed by Southern blotting. To determine the genomic background of blaNDM-1, the genome of Y5 was completely sequenced and compared to other reference genomes. The results of our study revealed that this isolate consists of a 4.8-Mbp chromosome and three plasmids, it is an epidemic clone of sequence type (ST) 167, and it shows 99% identity with Escherichia coli 6409 (GenBank accession no. CP010371), which lacks the same blaNDM-1 gene-surrounding structure as Y5. The blaNDM-1 gene is embedded in the chromosome along with two tandem copies of an insertion sequence common region 1 (ISCR1) element (sul1-ARR-3-cat-blaNDM-1-bleo-ISCR1), which appears intact in the plasmid from Proteus mirabilis (GenBank accession no. KP662515). The genomic context indicates that the ISCR1 element mediated the blaNDM-1 transposition from a single source plasmid to the chromosome. Our study is the first report of an Enterobacteriaceae strain harboring a chromosomally integrated blaNDM-1, which directly reveals the vertical spreading pattern of the gene. Close surveillance is urgently needed to monitor the emergence and potential spread of ST167 strains that harbor blaNDM-1.
KEYWORDS: New Delhi metallo-β-lactamase-1, chromosome, ISCR1, Enterobacteriaceae
INTRODUCTION
The bacteria containing the New Delhi metallo-β-lactamase gene (blaNDM-1) have swiftly spread worldwide and are considered a serious public health concern (1, 2). Studies confirmed that mobile-resistance elements, such as plasmids, transposons (Tn) and insertion sequences (IS), are responsible for the distribution of this superresistant gene, and they are regarded as major mechanisms driving the dramatically increased prevalence of carbapenem-resistant Enterobacteriaceae isolates (3–5).
Plasmids constitute the main vehicles carrying the blaNDM-1 gene and are the primary media for its horizontal dissemination. Additional blaNDM-1 plasmids ranging from 40 kb to 400 kb in size have been fully sequenced and documented for their effective transferability within species (3–5). Recent surveys demonstrated the large similarities of the genetic structures surrounding the blaNDM-1 gene among different members of the Enterobacteriaceae family, which further highlights the potential for the extensive spread of this determinant of resistance. Such transposable elements, like IS26 and IS5, which are frequently disseminated among diverse species or strains, most likely constitute the other horizontal-transfer mechanisms and are directly involved in the spread of the blaNDM-1 gene cross-species (6).
Reports have cited the emergence of the blaNDM-1 gene in the chromosomes of nonfermenter strains but not in Enterobacteriaceae isolates(7). Notably, few reports have revealed vertical-spreading patterns associated with this gene. Therefore, neither evidence nor research on the vertical-spreading pattern of the blaNDM-1 gene has been discussed recently. In this study, we report the discovery of the first clinical strain of Escherichia coli sequence type 167 (ST167) with two tandem chromosomal copies of blaNDM-1 originating from China. We completely sequenced the genome to analyze the genomic background of antimicrobial-resistance determinants and reveal their dispersal mechanism.
RESULTS
Antimicrobial susceptibility analysis.
Antimicrobial susceptibility results showed that the E. coli strain Y5 was resistant to multiple antimicrobial agents, including cephalosporins, carbapenems, fluoroquinolones, and aminoglycosides, but it was susceptible to colistin and tigecycline. Additionally, the stain was positive for the metallo-β-lactamase (MBL) phenotype (Table 1).
TABLE 1.
Antimicrobial susceptibility testing of E. coli Y5 isolate
| Isolate | MIC (μg/ml)a |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FEP | CTX | SAM | TZP | CPS2/1 | IPM | MEM | IPM/IPM+EDTA | CST | AMK | TGC | MIN | CIP | ATM | |
| Y5 | >256 | >256 | >32 | >256 | >256 | >256 | >32 | >32/<1 | 0.5 | 32 | 1 | 24 | >32 | >256 |
FEP, cefepime; CTX, cefotaxime; SAM, ampicillin-sulbactam; TZP, piperacillin-tazobactam; CPS2/1, cefoperazone/sulbactam 2:1; IPM, imipenem; MEM, meropenem; CST, colistin; AMK, amikacin; TGC, tigecycline; MIN, minocycline; CIP, ciprofloxacin; ATM, aztreonam.
Location of the blaNDM-1 gene.
Southern blot results presented a positive signal band of ∼54 kb according to the XbaI-PFGE assay but not the S1-PFGE assay (see Fig. S1 in the supplemental material). Further plasmid transfer experiments were unsuccessful, which strongly suggested that the gene was most likely carried chromosomally.
Genomic sequence analysis.
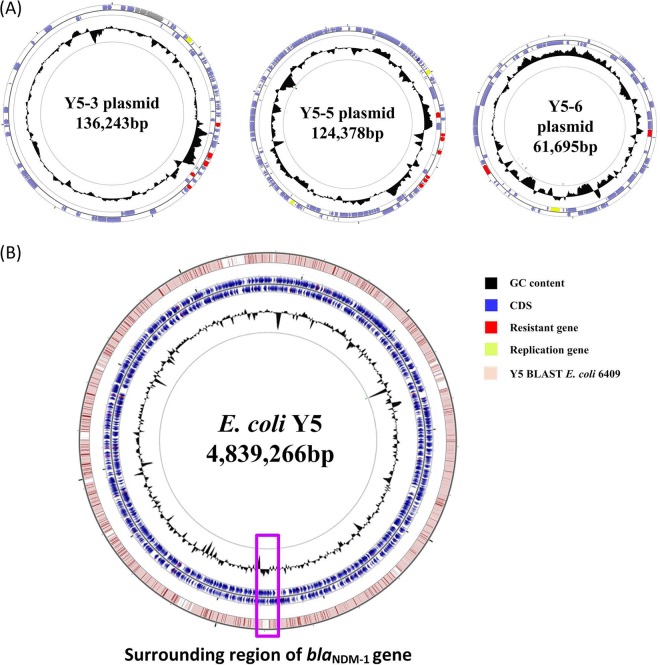
The assembly of whole-genome sequencing data generated a circular chromosome and three plasmids, with mean coverage and quality values of >82 and >43, respectively (Table 2). The size of the genome was 4,839,266 bp, with 50.81% GC content, 7 rRNA operons, 83 tRNAs, and 3,890 predicted protein-coding sequences (Table 2 and Fig. 1). Three plasmids measuring ∼61 kb to ∼136 kb in size and having between ∼50.29% and ∼52.76% GC content were grouped into identifiable replicon types (Table 2).
TABLE 2.
Genome and plasmids of E. coli Y5
| Genomic structure | Mean sequencing and alignment indices |
Size (kb) | GC content (%) | CDS no. | Accession no. | Resistance genes present | rep group | |
|---|---|---|---|---|---|---|---|---|
| Coverage | Quality | |||||||
| Y5 chromosome | 169.36 | 48.77 | 4,839 | 50.81 | 3,890 | CP013483 | mph(A), sul1, ARR-3, blaNDM-1, sul1, aadA16, aac(6′)Ib-cr, aac(3)-IId, bleo | — |
| Y5-3 plasmid | 123.79 | 47.49 | 136 | 51.95 | 90 | KT997783 | acc(3)-IId, aph(3¢)-Ia, strB, strA, sul2, tet(A) | repA |
| Y5-5 plasmid | 136.91 | 47.89 | 124 | 52.76 | 154 | KU043115 | aac(6′)Ib-cr, aadA5, blaOXA-1, blaCTX-M-15, sul1, tet(A) | repE |
| Y5-6 plasmid | 82.69 | 43.80 | 61 | 50.29 | 78 | KU043116 | blaCTX-M-14, blaCMY-42 | repE |
FIG 1.
Circular maps of the E. coli Y5 genome and its plasmids. (A) Circular graphs of three plasmids. (B) Circular graph of the Y5 genome sequence and genome alignment. Blue arrows denote coding sequences, red arrows denote resistance genes, and replication genes are denoted by green arrows. Genome alignment between Y5 and E. coli 6409 is the outer circle in pink, and GC content is the inner circle in black. The region surrounding the blaNDM-1 gene is highlighted with a purple frame.
The isolate belonged to an epidemic-resistant clone of ST167. Multiple resistance genes responsible for resistance to β-lactams, aminoglycosides, fluoroquinolones, macrolides, rifampin, sulfonamide, and tetracycline, were identified in the strain (Table 2).
Genomic background of the blaNDM-1 gene.
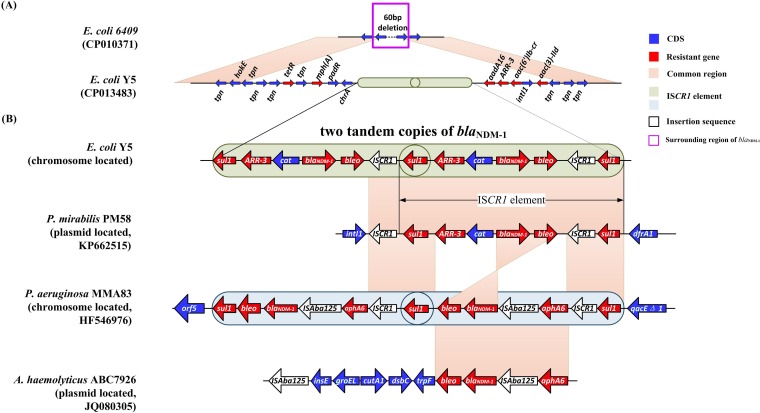
The blaNDM-1 gene was located upstream of an intI1 gene in the chromosome and was embedded within two tandem copies of a long-repeat fragment (Fig. 2). The fragment, which is an insertion sequence common region 1 (ISCR1) element, was flanked by two copies of sul1 and comprised a cluster of genes (sul1-ARR-3-cat-blaNDM-1-bleo-ISCR1).
FIG 2.
Alignment of sequences surrounding the region of blaNDM-1. Blue arrows are for the coding sequences and red arrows are for resistance genes. The pink boxes represent the common regions among different strains. ISCR1 elements are indicated in light green and gray. White arrows indicate insertion sequences. The region surrounding the blaNDM-1 gene is highlighted with a purple frame. mph(A), macrolide-2′-phosphotransferase I; padR, transcriptional regulator, PadR family; chrA, chromate transport protein A; sul1, sulfonamide resistance gene; ARR-3, rifampin resistance gene; catB3, chloramphenicol resistance gene; blaNDM-1, New Delhi metallo-β-lactamase gene; bleo, bleomycin resistance gene; IS, insertion sequence; aadA16, aminoglycoside-(3′) (9)-adenylyltransferase gene; arr3, rifampin ADP-ribosylation transferase; aac(6′)Ib-cr, aminoglycoside N(6′)-acetyltransferase gene; intl1, integron integrase gene; aac(3)-IId, aminoglycoside-(3)-N-acetyltransferase type II; dfrA1, dihydrofolate reductase; aphA6, aminoglycoside 3′-phosphotransferase type 6; qacEΔ1, QacEdelta1; tpn, transposer; hokE, HokE protein; tetR, transcriptional regulator, TetR family.
DISCUSSION
Enterobacteriaceae isolates positive for the blaNDM-1 gene have swept across the globe since its first report in 2009 (8). Many studies identified the gene location as resident within a plasmid and with horizontal transfer as the main spreading mechanism (6, 9–11). Our findings constitute the first report of its emergence in a chromosomal location in a clinical E. coli isolate revealing the vertical-spreading pattern for this gene.
Reports of carbapenem-resistant Enterobacteriaceae have dramatically increased recently and shown a geographical distribution not only in America and Europe but also in Asia (12). In China, many provinces, including Zhejiang, Hunan, and Henan, reported the emergence of Enterobacteriaceae positive for the blaNDM-1 gene (1, 3, 10, 13). The patient in this study was from Yuhuangding Hospital, located in Yantai, Shandong province, a coastal city in the Huadong region of China. The patient had never traveled abroad. Although there was no evidence to rule out a nosocomial infection, we preferred to consider that this Y5 strain might have originated from the patient herself, given that no further blaNDM-1-positive isolates were detected 1 month prior to or following the event in our hospital.
Enterobacteriaceae isolates producing NDM-1-type carbapenemase are often associated with lethal infectious diseases, especially when separated from blood or secretions (13). Bloodstream infections caused by such “super bugs” are always life-threatening due to their extreme drug resistance (13). However, our and previous studies proved that symptoms are mild and localized when urinary tract infections (UTIs) occur. Obstruction of the urinary tract is one of the most important direct risk factors involved in such infections; therefore, relief of the obstruction is as important as is the use of antibiotics (10). Approximately 80% of UTIs are caused by E. coli, a bacterium that is often found in the human gut, which escapes and translocates up the urinary tract to cause infection. Therefore, it is reasonable to postulate that this E. coli Y5 strain might have originated from and colonized in the gut of the patient, with subsequent infection occurring in an ascending manner.
The E. coli Y5 isolate reported here was a strain of ST167, an epidemic clone of significant public health concern that typically carries β-lactamase-resistant plasmids, including blaCMY, blaCTX-M, and blaSHV. blaNDM variants (blaNDM-5 and blaNDM-7) were recently detected in plasmids of E. coli ST167, further highlighting its potential as a threat to public health (9, 11, 14).
Sequence alignment of the genomes revealed high degrees of similarity (99% identity and 91% query coverage) between the Y5 strain and another ST167 strain (E. coli 6409 [GenBank accession no. CP010371]), except for an additional insertion around the blaNDM-1 gene. The insertion ranged 20 kb upstream and downstream of the site, exhibited >60% GC content, contained 15 drug-resistance genes, and substituted a 60-bp sequence of E. coli 6409 (Fig. 1 and 2). This genomic data provided direct evidence that blaNDM-1 was carried chromosomally and also proved that it was exogenous in origin. Further analysis of the insertion sequence surrounding blaNDM-1 revealed two tandem copies of an ISCR1 element. ISCR1, which was first reported by Wang et al., lacks terminal inverted repeats and is transposed by a mechanism called rolling-circle transposition. ISCR1 is often neighbored by a wide array of antibiotic-resistance genes and shows a potential for mobilizing adjacent antibiotic-resistance genes (15, 16).
Considering the important role of the ISCR1 element in blaNDM-1 transposition, sequence alignment was performed in our study. In 2013, Jovcic et al. reported that blaNDM-1 was carried in a similar ISCR1 element in Pseudomonas aeruginosa according to restriction enzyme-based cloning (7). Their ISCR1 element was embedded within a chromosome and also presented two tandem copies; however, the sequence within the element was significantly different (Fig. 2). They found a more composite Tn structure consisting of two insertion sequences (ISCR1 and ISAba125), a portion of which was highly homologous with the Tn125 in an Acinetobacter haemolyticus ABC7926 plasmid (GenBank accession no. JQ080305) (Fig. 2). Given that both ISCR1 and ISAba125 were capable of transfer, the determination of which is responsible for mobilizing the blaNDM-1 gene from the plasmid to chromosome is unlikely. Here, we revealed that the sequence of the ISCR1 element in the Y5 isolate showed 99% similarity to a plasmid, PM58, from P. mirabilis (GenBank accession no. KP662515.1) (Fig. 2) (13). Additionally, we observed an integrin-1 gene (intl1) upstream of two tandem copies of the ISCR1 element at a point where cassette integration usually occurs. This alignment information indicated that the ISCR1 element played an important role in the genetic transmission of blaNDM-1 and likely mediated its transposition from a single-source plasmid to the chromosome.
In summary, this is the first report of an E. coli isolate with chromosomally carried blaNDM-1, although plasmid-borne blaNDM-1 is already widespread. By aligning the genomic sequences surrounding this blaNDM-1 isolate, we discovered that the ISCR1 element was critical to blaNDM-1 mobility between chromosome and plasmid. Therefore, blaNDM-1 appears to be transmissible via multiple mechanisms and should be monitored with greater vigilance.
MATERIALS AND METHODS
Strain information and case history.
A female patient, aged 32 years-old, came to Yantai Yuhuangding Hospital reporting serious pain in the right portion of her back accompanied by fever (the highest point reaching 38.6°C) for 10 days in September, 2013. She was diagnosed with ureteral and kidney stones, hydronephrosis, and a urinary tract infection (UTI) according to computed-tomography scan results and was admitted to our hospital for a ureterolithotomy after her menstrual period. Her temperature and systemic inflammatory index were normal and the white blood cell (WBC) count was 8.11 × 109 cells/liter, with 78.2% neutrophils and 14.3% lymphocytes in the blood. However, the WBC count in a urine sample was 67 cells/μl and the red blood cell count was 65 cells/μl, indicating a local UTI. She received nonstandard antibiotic treatment, including cefepime (2 g, every 8 h [q8h]) for 1 day and piperacillin-tazobactam (3.375 g, q8h) for 3 days. E. coli strain Y5, which was extremely resistant to multiple antimicrobial agents, including piperacillin-tazobactam, cephalosporin, and carbapenem, was isolated from urine cultures (at >105 CFU/ml) using the Vitek 2 system (bioMérieux, France). Soon, the patient was transferred to Shandong Provincial Hospital (a higher-level hospital) for further treatment. A follow-up survey showed that she recovered shortly after surgery for removal of renal and ureteral calculi, and bacteria were no longer isolated.
Antimicrobial-susceptibility testing and blaNDM-1 location.
Antimicrobial susceptibility was measured with Etest strips (bioMérieux, Sweden) according to standard operating procedures. Briefly, direct colony suspension was adjusted to that of a 0.5 McFarland standard in saline (corresponds to ∼1.5 × 108 CFU/ml), and then the suspension was swabbed to a Müller-Hinton agar (MHA) plate. The strip was applied to the MHA surface and the plate was inoculated in ambient air at 35°C for 16 to 18 h (http://www.biomerieux-diagnostics.com/Etest). The MBL phenotype and MICs for 13 antibiotics were determined and interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (17). The presence of the blaNDM-1 gene was established by PCR and confirmed by sequencing.
Because previous reports identified the blaNDM-1 gene as being located in plasmids, we performed S1-pulsed-field gel electrophoresis (PFGE) and XbaI-PFGE, followed by Southern blotting with a blaNDM-1 probe (6). In accordance with published methods, conjugation mating was tested between E. coli Y5 and E. coli J53, and electrotransformation was tested using competent E. coli DH5α cells (6).
Whole-genome sequencing and annotation.
E. coli strain Y5 was cultured to mid logarithmic phase in 50 ml of LB medium at 37°C. DNA for sequencing was extracted via a QIAamp DNA minikit (Qiagen, Valencia, CA) and further purified using a PowerClean DNA cleanup kit (Mo Bio Laboratories, Carlsbad, CA) according to the protocols of the manufacturers. The quality of DNA was determined by gel electrophoresis and by a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). Single-molecule real-time (SMRT) sequencing reads were generated using a PacBio RS II platform (Pacific Biosciences, Menlo Park, CA). Sequencing reads were de novo assembled with the PacBio Hierarchical Genome Assembly Process 3 (HGAP3.0)/Quiver software package. The resulting assembly was confirmed with optical maps generated with 30-fold coverage on the Argus mapping station according to the OpGen protocol (OpGen, Inc., Gaithersburg, MD).
The assembled genome was annotated using GeneMark and Rapid Annotation by using Subsystems Technology (http://rast.nmpdr.org/rast.cgi) (18, 19). Coding DNA sequences (CDS) and protein similarities were determined by BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi), whereas protein domains were identified by InterProScan (20). ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) was used to detect antimicrobial resistance genes at 98% identity (21). The BacWGSTdb (http://bacdb.org/BacWGSTdb/) server was used for in silico multilocus sequence typing (MLST) analysis (22). Whole genomes were compared using Artemis Comparison Tool release 11.1.1, and the CGView server (http://stothard.afns.ualberta.ca/cgview_server/) was used to generate graphical maps, sequence features, base composition plots, analysis results, and sequence similarity plots (23).
Accession number(s).
The chromosome and three plasmids of E. coli strain Y5 were deposited in GenBank under accession numbers CP013483, KT997783, KU043115, and KU043116 (Table 2).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant no. NSFC81471986 and NSFC 31170130) and by the Science and Technology Development Projects of Shandong Province, China (project no. 2013YD18041).
The authors declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01581-16.
REFERENCES
- 1.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng J, Qiu Y, Yin Z, Chen W, Yang H, Yang W, Wang J, Gao Y, Zhou D. 2015. Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J Antimicrob Chemother 70:2987–2991. doi: 10.1093/jac/dkv232. [DOI] [PubMed] [Google Scholar]
- 4.Alm RA, Johnstone MR, Lahiri SD. 2015. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: role of a novel insertion in PBP3. J Antimicrob Chemother 70:1420–1428. doi: 10.1093/jac/dku568. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Du X, Ji J, Chen Y, Jiang Y, Yu Y. 2012. Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J Antimicrob Chemother 67:2114–2122. doi: 10.1093/jac/dks192. [DOI] [PubMed] [Google Scholar]
- 7.Jovcić B, Lepsanovic Z, Begovic J, Rakonjac B, Perovanovic J, Topisirovic L, Kojic M. 2013. The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob Agents Chemother 57:3405–3407. doi: 10.1128/AAC.02312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang P, Xie Y, Feng P, Zong Z. 2014. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother 58:7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du XX, Wang JF, Fu Y, Zhao F, Chen Y, Wang HP, Yu YS. 2013. Genetic characteristics of blaNDM-1-positive plasmid in Citrobacter freundii isolate separated from a clinical infectious patient. J Med Microbiol 62:1332–1337. doi: 10.1099/jmm.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 11.Cuzon G, Bonnin RA, Nordmann P. 2013. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One 8:e61322. doi: 10.1371/journal.pone.0061322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Struelens MJ, Monnet DL, Magiorakos AP, Santos O'Connor F, Giesecke J, European NDM-1 Survey Participants . 2010. New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill 15:pii=19716 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19716. [DOI] [PubMed] [Google Scholar]
- 13.Qin S, Qi H, Zhang Q, Zhao D, Liu ZZ, Tian H, Xu L, Xu H, Zhou M, Feng X, Liu HM. 2015. Emergence of extensively drug-resistant Proteus mirabilis harboring a conjugative NDM-1 plasmid and a novel Salmonella genomic island 1 variant, SGI1-Z. Antimicrob Agents Chemother 59:6601–6604. doi: 10.1128/AAC.00292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Lou D, Xu Y, Shang Y, Li D, Huang X, Li Y, Hu L, Wang L, Yu F. 2013. First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol 13:282. doi: 10.1186/1471-2180-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Xie L, Zhang F, Ni Y, Sun J. 2015. Molecular characterization of ISCR1-mediated blaPER-1 in a non-O1, non-O139 Vibrio cholerae strain from China. Antimicrob Agents Chemother 59:4293–4295. doi: 10.1128/AAC.00166-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Wu K, Sun J, Wang Q, Chen Q, Yu S, Rui Y. 2012. Novel ISCR1-linked resistance genes found in multidrug-resistant Gram-negative bacteria in southern China. Int J Antimicrob Agents 40:404–408. doi: 10.1016/j.ijantimicag.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobail susceptibility testing; 20th informational supplement. CLSI M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206-214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulder N, Apweiler R. 2007. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol 396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 21.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruan Z, Feng Y. 2016. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res 44:D682-687. doi: 10.1093/nar/gkv1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181-184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.