Abstract
Colorectal cancer is a prevalent malignancy worldwide, and investigations are required to elucidate the underlying carcinogenic mechanisms. Amongst these mechanisms, de novo carcinogenesis and the adenoma to carcinoma sequence, are the most understood. Metastasis of colorectal cancer to the liver often results in fatality, therefore, it is important for any associated risk factors to be identified. Regarding the treatment of the disease, it is important to manage not only the primary colorectal tumor, but also the liver metastases. Previously, through gene variation analysis, chromosomal loss has been indicated to serve an important role in liver metastasis. Such analysis may aid in the prediction of liver metastasis risk, alongside individual responses to treatment, thus improving the management of colorectal cancer. In the present study, we aimed to clarify a cause of the liver metastasis of colorectal cancer using comparative genomic hybridization analysis. A total of 116 frozen samples were analyzed from patients with advanced colorectal cancer that underwent surgery from 2004 to 2011. The present study analyzed mutations within tumor suppressor genes non-metastatic gene 23 (NM23), deleted in colorectal carcinoma (DCC) and deleted in pancreatic carcinoma, locus 4 (DPC4), which are located on chromosomes 17 and 18 and have all been reported to affect liver metastasis of colorectal cancer. The association between chromosomal abnormalities (duplication and deletion) and liver metastasis of colorectal cancer was evaluated using comparative genomic hybridization. Cluster analysis indicated that the group of patients lacking the long arm of chromosome 17 demonstrated the highest rate of liver metastasis. No significant association was observed between the frequency of liver metastases for synchronous and heterochronous colorectal cancer cases and gene variation (P=0.206). However, when these liver metastasis cases were divided into the synchronous and heterochronous types, the ratio of each was significantly different between gene variation groups, classified by the existence of the 17q deletion (P=0.023). These results indicate that the deletion of 17q may act as a predictive marker of liver metastasis in postoperative states.
Keywords: colorectal cancer, comparative genomic hybridization, liver metastasis, chromosome 17, cluster analysis
Introduction
Colorectal cancer is the second most common cause of cancer-associated mortality in Japan, and its incidence has been rapidly increasing in recent years (1,2) Colorectal cancer is a common form of cancer and investigations to elucidate the underlying oncogenic mechanisms, including the adenoma carcinoma sequence (3,4) and de novo carcinogenesis (5,6), are among the most advanced. However,. Regarding the treatment of the disease, it is important to target liver metastases, in addition to the primary lesion to maintain the best results. Liver metastasis is quite common in colorectal cancer, and the rate of liver metastasis is assumed to be between 20–30% in all cases of colorectal cancer (7). Radical cure excision is currently the preferred choice for the treatment of liver metastases and primary lesions (8). However, more recently, the use of neoadjuvant chemotherapy (NAC) or chemoradiotherapy (CRT) to reduce tumor size has become increasingly prevalent (8). Following NAC or CRT, excision of the primary lesion and distant metastasis is then performed (9). Early detection of the liver metastasis is important as long term survival related with size and a number of nodules of the liver metastasis (10).
Recently, targeted cancer therapies have been developed and approved for use in treating various types of cancer, and in certain therapies, treatment efficacy may be predicted by analyzing the gene variation of the primary lesion (11,12). If such gene variation analysis was employed to identify risk of liver metastasis, the management of the disease may be greatly improved. It was previously reported that abnormalities in gene and genomic copy numbers that contribute to lymph node metastases may be identified by comparative genomic hybridization (CGH) (13,14).
It is expected that identification of specific gene and genomic variations may be used in the clinic as tumor markers and prognostic predictive factors for metastasis (12,15); these prognostic markers may serve as important tools for colorectal cancer treatment.
Mutations in oncogenes, such as erb-B2 receptor tyrosine kinase 2 (ERBB2), or cancer-restraining genes, including tumor protein 53 (TP53), non-metastatic clone 23 (NM23), deleted in colorectal cancer (DCC) and deleted in pancreatic carcinoma, locus 4 (DPC4), are considered to have an effect on liver metastasis (16–24).
The present study aimed to clarify a cause of the liver metastasis of colorectal cancer using CGH analysis. In particular, we intended to demonstrate the association between liver metastasis and genome abnormality at chromosomes 17 and 18, on which important oncogenes and cancer-suppressing genes are located. The present study may lead to the identification of a liver metastasis marker in blood samples.
Materials and methods
Specimens
A total of 116 frozen tissue samples from surgical specimens of colorectal cancer were obtained from patients that had undergone surgery at the Department of Surgery, Juntendo University School of Medicine (Tokyo, Japan), under the institutional review board approved protocol (IRB2013145). The specimens were collected over a period of 8 years from October 2004 to December 2012. The perioperative breakdown was as follows: A total of 72 patients presented with no liver metastasis and 44 patients with liver metastasis. Recurrence of liver metastasis postoperative developed in 7 out of 72 patients in the no liver metastasis group, and 16 out of 44 patients in the liver metastasis group. In the present study, liver metastasis that was detected at the same time as the colorectal cancer surgery was classified as ‘synchronous’ metastasis, and those cases detected subsequent to the surgery were classified as ‘heterochronous’.
Genomic DNA sample preparation
Cancer cells from the tumor specimens were collected for genomic DNA extraction. A total of 5–20 frozen sections (16 µm thick) were manually microdissected. Under the guidance of a pathologist, normal cells were removed leaving >95% tumor cells. Genomic DNA was extracted using the Quick Gene SP kit DNA tissue (Wako Pure Chemical Industries, Ltd., Osaka, Japan), and the quality was confirmed by standard agarose gel electrophoresis. Human Male Genomic DNA or Human Female Genomic DNA (Merck-Millipore, Billerica, MA, USA) was used as sex-matched normal reference DNA.
Preparation of whole genome DNA microarray
DNA microarray was created by Link Genomics Inc., (Tokyo, Japan) with complete coverage of the human genome, using 12,310 individually amplified bacterial artificial chromosome (BAC) clones. All BAC clones were cultured from a single colony and validated by polymerase chain reaction (PCR) amplification (using Blend Taq® DNA polymerase; Toyobo Co., Ltd., Osaka, Japan) using clone-specific primers (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). These primer sequences were designed by DNASIS Pro software (Hitachi Solutions, Tokyo, Japan), and confirmed only to amplify the target gene with UCSC in-Silico PCR (UCSC Genome Informatics Group, Santa Cruz, CA, USA; https://genome.ucsc.edu/cgi-bin/hgPcr). The extracted BAC DNA was BstYI-restricted and amplified by ligation-mediated PCR. The products were sent to be printed on glass slides (GE Healthcare Life Sciences, Uppsala, Sweden) with an ink-jet type spotter (NGK Insulators, Ltd., Nagoya, Japan).
Analysis of genome copy number by CGH array
AluI and RsaI-restricted genomic DNA was labeled by random priming with Alexa555-dCTP (test DNA) and Alexa647-dCTP (reference DNA) using the BioPrime® Plus Array-CGH Indirect Genomic Labeling system (Invitrogen; Thermo Fisher Scientific, Inc.). The labeled test and reference DNA were then ethanol precipitated in the presence of Cot-1 DNA (Thermo Fisher Scientific, Inc.), redissolved in a hybridization mix and subsequently denatured at 70°C for 10 min. Following incubation at 42°C for 5 min, the mixture was applied to a Whole Genome DNA Microarray (LinkGenomics, Tokyo, Japan) covered with an MAUI® Mixer Hybridization Chamber (BioMicro Systems, Inc., Salt Lake City, USA). Following incubation at 42°C for 48 h, the slides were washed with 2X saline sodium citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) buffer and 0.1X SSC/0.1%SDS buffer three times, respectively. Subsequent to rinsing with 0.01X SSC buffer and airdrying, the slides were scanned using the SureScan Microarray Scanner (Agilent Technologies, Inc., Santa Clara, USA), and analyzed with GenePix Pro 4.0 imaging software (Axon Instruments, Inc., Union City, CA, USA). Normalization was performed using global-normalization methods (25). The total amount of genomic DNA in one cell was regarded to be almost same, and the median values of each array sample was adjusted (log ratios for each array were adjusted to be zero). The fold change in the number of genome copies in the tumor tissue compared with the normal tissue was then calculated. Thresholds for gains and losses were set to 1.2 and 0.8, respectively.
Determination of target chromosomes
The present study focused on chromosomes 17 and 18, which contain regions/genes closely associated with cancer, including 17p13 containing TP53, 17q12 containing ERBB2, 17q21 containing NM23 and 18q21.1–3 containing DPC4 and DCC. NM23, DCC and DPC4 have been reported as tumor suppressor genes that reportedly affect the liver metastasis of colon cancer (15). Using CGH, the current study evaluated the presence of abnormalities, including duplications and deletions, in each chromosomal region presenting with liver metastasis of colorectal cancer. Data mining was performed using cluster analysis, and the data was tested for statistical significance.
To analyze the association between gene mutations of the primary lesions and liver metastasis, 18p21.3 was used as the locus for DCC, 18p21.1 as DPC4 and 17q21 as NM23. Cluster analysis lead to the dividing of patients into three groups, which included: Normal genome, genomic deletion and genomic duplication (Fig. 1).
Figure 1.
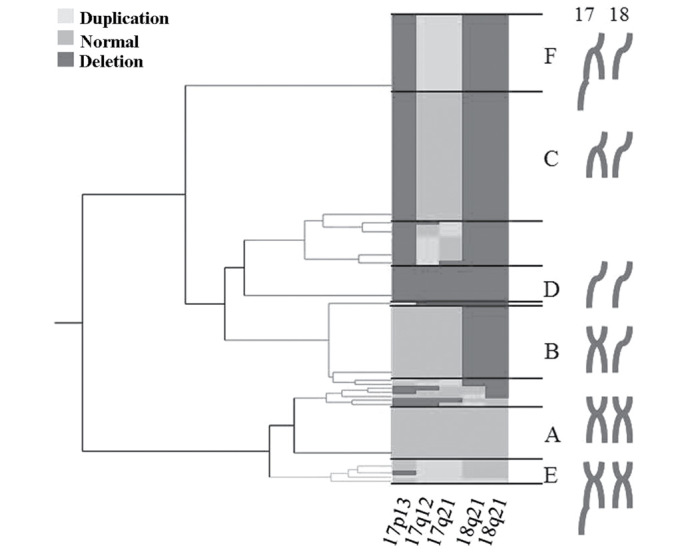
Classification of genomic variation as determined by cluster analysis. Cluster analysis was performed focusing on chromosome 17 and 18 and identified 2 duplication groups and 3 deletion groups.
Classification of genomic variation
The three patient groups were classified as follows: Group A, normal genome (n=13, 11.2%); group B, 18p and 18q deletion (n=21, 18.1%); group C, 18p, 18q and 17p deletion (n=43, 37.1%); group D, overall deletion of 17p, 17q, 18p and 18q, resulting in 1n (n=9, 7.8%); group E, 17q duplication alone (n=6, 5.2%); and group F, 17q duplication and 17p, 18p and 18q deletion (n=19, 16.4%). Based on these classifications, the affect of the varying genomic aberrations on liver metastasis was analyzed (Fig. 2).
Figure 2.
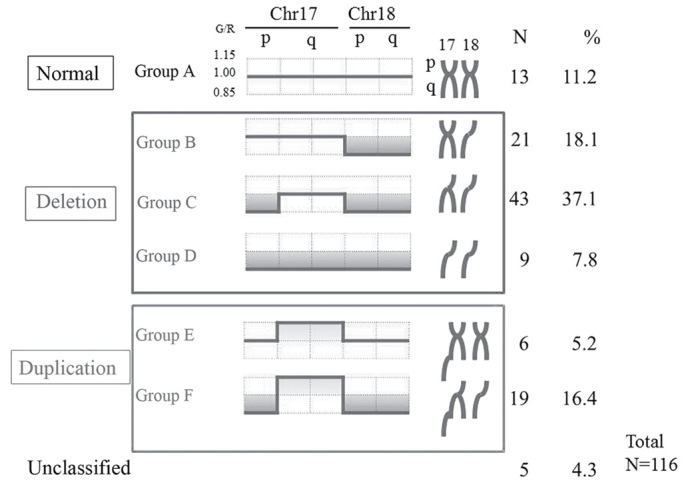
Classification of the genomic variation. Following cluster analysis, patients were classified into three groups: Normal, deletion and duplication. Chr, chromosome. G/R, green/red ratio;
Statistical analysis
Cluster analysis was used for data mining and Pearson's χ2 test was used to determine significant differences. JMP, version 9.0 (www.jmp.com), was used to perform the appropriate statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Genomic duplications
Six genomic variations were classified into three groups, normal (Group A), deletion (Group BCD) and duplication (Group EF). When these variations were reclassified into two groups, Group ABCD (without duplication) and Group EF (with the duplication), there was no difference in the metastasis categories between these two groups (P=0.26; 3×2 Pearson's χ2 test). The ratios of non-metastasis, heterochronous and synchronous metastasis cases were 64% (16/25), 8% (2/25) and 28% (7/25) in Group EF, respectively, and 50.6% (39/77), 3.9% (3/77) and 45.4% (35/77) in Group ABCD, respectively. In the present study, no effect of genomic duplication was detected on the liver metastasis; however, the authors plan to analyze additional cases, with focus to the duplication in future. The subsequent paragraph shows the results of the analyses on the normal (Group A) and deletion (Group BCD) groups, in order to focus on deletions of cancer suppressor genes.
Genomic deletions
The overall incidences of liver metastasis, including synchronous and heterochronous cases, in groups A-D were 30.8 (4/13), 33.3 (7/21), 51.2 (22/43) and 66.7% (6/9), respectively, with no significant differences observed (P=0.206; Fig. 3). The cases of liver metastasis were then divided into synchronous and heterochronous colorectal cancer types. The incidences of synchronous liver metastasis in groups A-D were 23.1 (3/13), 33.3 (7/21), 48.8 (21/43) and 44.4% (4/9), respectively (P=0.067; Fig. 4). The incidences of heterochronous liver metastasis in groups A-D were 7.7 (1/13), 0 (0/21), 2.3 (1/43) and 22.2% (2/9), respectively (P=0.067; Fig. 4). Therefore, although the differences between the synchronous and heterochronous cases were not significant, there was a tendency for metastasis to occur more prevalently in the synchronous type. Groups A-D were subsequently reclassified based on the presence or absence of the 17q deletion, with the incidence of liver metastasis compared between groups A + B + C (n=77) and D (n=9). In groups A + B + C and D, the incidence of the synchronous type was 40.3 (31/77) and 44.4% (4/9), respectively, and the incidence of the heterochronous type was 2.6 (2/77) and 22.2% (2/9), respectively. There was a significant difference in metastasis types between the two groups, Group ABC (no deletion on 17q) and Group D (with deletion on 17q) (P=0.023; by 2×3 Pearson's χ2 test; Fig. 5). Only in the heterochronous cases, metastasis ratios were significantly different between Group ABC and Group D (P=0.0087; <0.05/3; significant by Pearson's χ2 test with Bonferroni correction).
Figure 3.
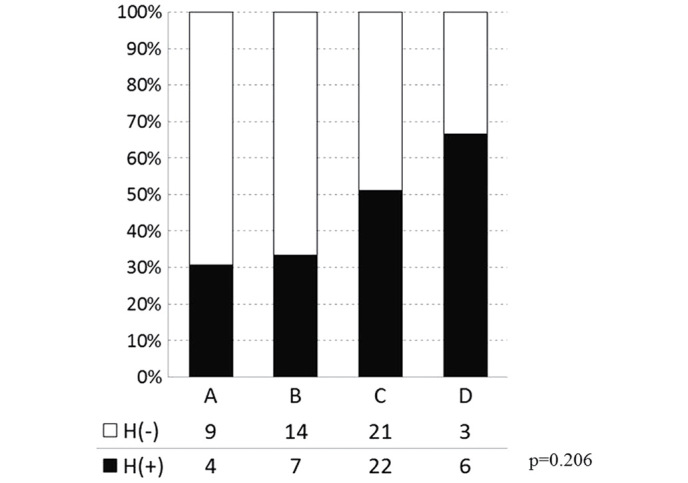
Liver metastasis and genomic variation in the deletion groups (A-D). No significant differences were observed between the genome deletion groups and the rate of liver metastasis. H(−), no liver metastasis; H(+), liver metastasis.
Figure 4.
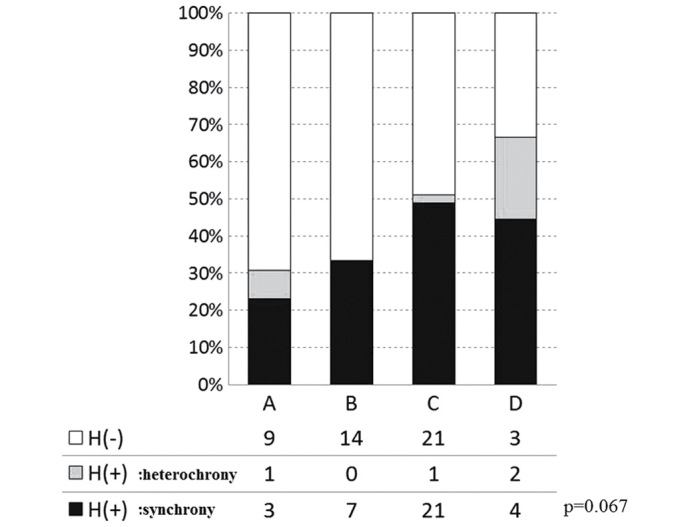
Liver metastasis and genomic variation in the synchronous and heterochronous cases. The ratio of liver metastasis incidence in four gene variations was marginal but not significant (P=0.067; 4×3 Pearson's χ2 test). Open bar H(−), no metastasis; gray bar H(+), heterochronous metastasis; closed bar H(+), synchronous metastasis.
Figure 5.
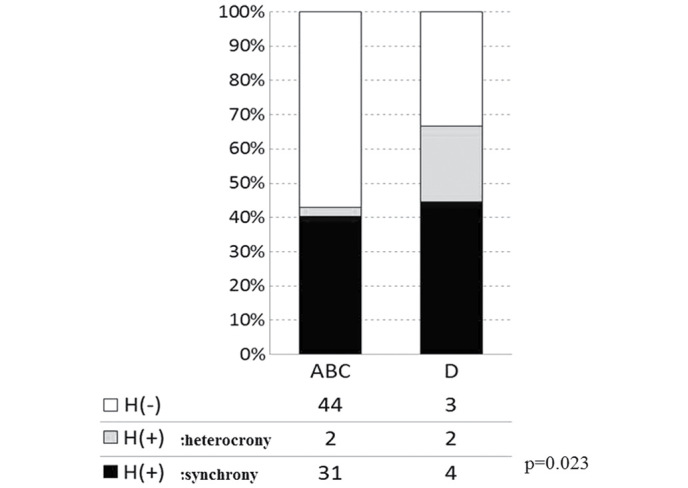
Liver metastasis and genomic variation (synchrony or heterochrony). Following reclassification of groups based on the presence or absence of 17q deletion, a significant difference was observed in the liver metastasis rate between the gene groups with (D) and without (ABC) 17q deletion (P=0.067; 2×3 Pearson's χ2 test).. Open bar H(−), no metastasis; gray bar H(+), heterochronous metastasis; closed bar H(+), synchronous metastasis.
In addition, a significant difference was identified among the four variation groups in terms of the relapse rate of postoperative liver metastasis (including heterochronous cases and synchronous metastasis cases that showed re-recurrence on the liver that had clearly undergone tumor removal; Fig. 6). When Groups A-D were reclassified into Group AB (n=34) and Group CD (n=52), due to the positive or negative presence of deletions on 17p domains, a significant difference (P=0.009) was observed between these two groups (Fig. 7).
Figure 6.
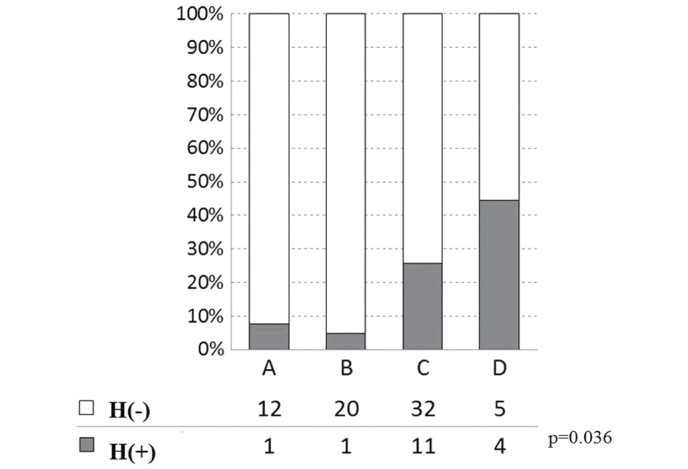
Postoperative liver metastasis and genetic variations. There was a difference in the relapse rate of postoperative metastasis [H(+)] among the four genomic variations, Groups A, B, C and D. Open bar H(−), no postoperative metastasis; gray bar H(+), postoperative metastasis, including heterochronous cases and synchronous cases with recurrence on the liver.
Figure 7.
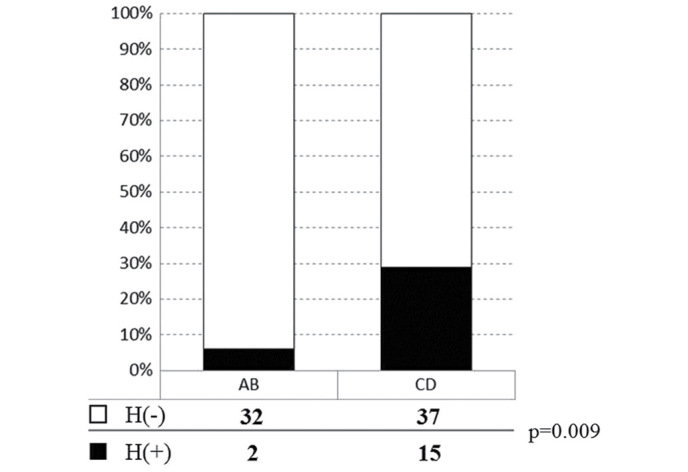
Postoperative liver metastasis and genomic variation. Following reclassification of groups based on the presence of absence of the 17p deletion, and comparisons between the A+B and C+D groups, a significant difference was demonstrated between the two groups. Open bar H(−), no postoperative metastasis; gray bar H(+), postoperative metastasis, including heterochronous cases and synchronous cases with recurrence on the liver.
Discussion
The present study revealed the impact of chromosome deletions on liver metastasis of colorectal cancer, and assumed that the impacts were primarily a result of lesion of cancer suppression genes on 17q. By contrast, deletions on chromosomal region 18p demonstrated no obvious effect on liver metastasis; however, the DCC and DPC4 genes are located in this region. In order to focus on deletion of cancer suppressor genes, we did not investigate the effects of duplications on chromosomes 17 and 18 in the present study.
The outcome tended to be favorable in the no liver metastasis group within the deletion groups. However, the number of patients presenting with duplications was particularly low, and their affect on the analytical results cannot be ruled out. Furthermore, a wide range of cases from stages II to III (26)were included in the no liver metastasis groups, therefore, a bias in the patients in the early stage of disease may have affected the outcome. Additional analysis with an increased number of patients followed by a prolonged observation period may be necessary.
Regarding the carcinogenesis and growth of primary colorectal cancer, de novo carcinogenesis and the adenoma to carcinoma sequence have been thoroughly investigated, and a number of genes, including fibroblast activation protein, Kirsten rat sarcoma viral oncogene homolog, mothers against decapentaplegic homolog 4 and p53, are considered to be involved in the development of the disease (26,27). Genes are universal elements, and in the absence of mutation, they may be reliably measured unlike clinicopathological factors, such as lifestyle and background. Furthermore, DNA has a higher physical stability compared with RNA, and its clinical application may be relatively straightforward (28).
A number of studies have reported the possible benefits of cluster analysis in studies investigating gene variation in colorectal cancer (29–32). The present study performed data mining employing cluster analysis and observed a tendency toward differences in the incidences of synchronous and heterochronous liver metastases among groups A-D. Following reclassification of the patients based on the presence or absence of the 17q deletion, the incidences of synchronous and heterochronous liver metastases were compared. Significant differences were observed in the incidences of each type between groups A + B + C and D, suggesting that chromosomal region 17q contains genes that serve as a risk factor for heterochronous liver metastasis.
In addition, there is a significant difference in the relapse rate of the postoperative liver metastasis among groups A, B, C and D. A difference was also identified in the rate of the metastasis between the two groups (AB and CD), reclassified by the presence of the 17p domain including TP53. Such findings suggest that the lesion of TP53 gene, located in the 17p region, may act as a suppressor of postoperative liver metastasis.
Survival rates associated with 17q21 deletion and duplication were not examined in the present study due to the possibility of prolonged survival occurring as a result of postoperative oral and drip-infusion chemotherapy, which prevent the recurrence of liver metastasis. It is also possible that the outcome of the patient may be markedly affected by the timing of cancer diagnosis.
It cannot be ruled out that gene aberrations that are non-detectable by CGH (including translocations), unknown genes and regions not selected in the process of cluster analysis, may have affected the outcome and liver metastasis of the patients in the present study. It is also possible that the outcomes may have been modified by other factors, including disease stage; thus, additional analysis involving a larger sample size and a prolonged observation period may be necessary.
Korn et al (27) reported that similar gene mutations existed between metastatic and primary lesions of colorectal cancer, and Nakao et al (19) demonstrated that the abnormal chromosomal regions were physically close between synchronous and heterochronous liver metastases of colorectal cancer. However, certain studies have identified that a significantly higher incidence of gene alterations (including duplication and deletions) exist in metastatic lesions of the lymph nodes, the liver and the lungs than in the associated primary colorectal cancer lesions (33–39). Therefore, despite studies concerning the molecular mechanism of colorectal carcinogenesis being particularly advanced compared to those investigating other solid cancers, the overall picture has not yet been fully clarified. Additionally, it cannot be ruled out that the clinical classification of liver metastasis into synchronous and heterochronous types may have no true meaning.
Liver metastasis markedly affects the prognosis of patients with colorectal cancer. If gene tests can detect the risk of metastasis with high-sensitivity, their clinical application may be anticipated, with such tests possibly aiding the selection of treatment and adjustment of the course observation periods.
Future studies may aim to identify gene mutations that serve as true metastatic markers by comparing gene mutations between primary and metastatic lesions in the liver, lungs and lymph nodes within the same patients. Analyzing similarities and differences between the primary and metastatic lesions, in addition to comparing gene mutations with those in the primary lesions of non-metastatic patients, may aid the understanding of which particular variations are involved in colorectal cancer metastasis.
In conclusion, in the present study, it was demonstrated that the 17q chromosomal region contains genes that serve as risk factors for heterochronous liver metastasis. The possibility was also suggested that lesions on the 17p domain may become a risk factor for liver metastasis following surgery. Analysis of gene mutations and genomic aberrations in surgical specimens of primary lesions may aid the identification of metastatic markers for colorectal cancer.
References
- 1.Yoshida M, Kondo K, Tada T. The relation between the cancer screening rate and the cancer mortality rate in Japan. J Med Invest. 2010;57:251–259. doi: 10.2152/jmi.57.251. [DOI] [PubMed] [Google Scholar]
- 2.Ministry of Health, Labor and Welfare (MHLW), Government of Japan, corp-author. http://www.mhlw.go.jp/toukei/saikin/hw/jinkou/kakutei14/dl/11_h7.pdf. Vital statistics in Japan. 2014 Table 7: 3–1. Available at. Accessed Jan 30, 2016 (in Japanese) [Google Scholar]
- 3.Morson BC, Dawson IMP, editors. Gastrointestinal Pathology. Blackwell Scientific; Oxford, London, Edinburgh, Melbourne: 1972. [Google Scholar]
- 4.Muto T, Bussey HJ, Morson BC. The evolution of cancer of the colon and rectum. Cancer. 1975;36:2251–2270. doi: 10.1002/cncr.2820360944. [DOI] [PubMed] [Google Scholar]
- 5.Olschwang S, Hamelin R, Laurent-Puig P, Thuille B, De Rycke Y, Li YJ, Muzeau F, Girodet J, Salmon RJ, Thomas G. Alternative genetic pathways in colorectal carcinogenesis. Proc Natl Acad Sci USA. 1997;94:12122–12127. doi: 10.1073/pnas.94.22.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura Y. Genetic defects and pathogenesis of colorectal cancer. In: Nagasako K, editor. Early Colon Cancer: From Genesis to Diagnosis and Treatment. Igaku-Shoin, Ltd.; Tokyo: 1993. pp. 183–206. (In Japanese) [Google Scholar]
- 7.Sugihara K. Liver resection for colorectal metastases. Nihon Shokaki Geka Gakkai Zasshi. 2000;33:128–133. doi: 10.5833/jjgs.33.128. (In Japanese) [DOI] [Google Scholar]
- 8.McAuliffe JC, Qadan M, D'Angelica MI. Hepatic resection, hepatic arterial infusion pump therapy, and genetic biomarkers in the management of hepatic metastases from colorectal cancer. J Gastrointest Oncol. 2015;6:699–708. doi: 10.3978/j.issn.2078-6891.2015.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L, Li TJ, Zhang JW, Liu S, Fu BS, Liu W. Neoadjuvant chemotherapy followed by surgery versus surgery alone for colorectal cancer: Meta-analysis of randomized controlled trials. Medicine (Baltimore) 2014;93:e231. doi: 10.1097/MD.0000000000000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordlinger B, Guiguet M, Vaillant JC, Balladur P, Boudjema K, Bachellier P, Jaeck D. Association Française de Chirurgie: Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Cancer. 1996;77:1254–1262. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1254::AID-CNCR5>3.3.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Diep CB, Kleivi K, Ribeiro FR, Teixeira MR, Lindgjaerde OC, Lothe RA. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer. 2006;45:31–41. doi: 10.1002/gcc.20261. [DOI] [PubMed] [Google Scholar]
- 12.Kodeda K, Asting AG, Lönnroth C, Derwinger K, Wettergren Y, Nordgren S, Gustavsson B, Lundholm K. Genomic CGH-assessed structural DNA alterations in rectal carcinoma as related to local recurrence following primary operation for cure. Int J Oncol. 2012;41:1397–1404. doi: 10.3892/ijo.2012.1562. [DOI] [PubMed] [Google Scholar]
- 13.Ishido Y, Unotoro J, Kamiyama H, Yaginuma Y, Kasamaki S, Sakamoto K, Shindou T, Niwa S, Kamano T. The comparative genomic hybridization analysis of chromosomal aberrations related to lymph node metastasis of colorectal cancer. Juntendo Med J. 2005;51:534–539. doi: 10.14789/pjmj.51.534. [DOI] [Google Scholar]
- 14.Kamiyama H, Unotoro J, Ishido Y, Yaginuma Y, Kasamaki S, Sakamoto K, Shindo T, Niwa S, Kamano T. Analysis of DNA copy-number aberrations related with organ metastases of colorectal cancer by comparative genomic hybridization. J Jpn Coll Surg. 2005;30:831–835. [Google Scholar]
- 15.Brosens RPM, Belt EJTH, Haan JC, Buffart TE, Carvalho B, Grabsch H, Quirke P, Cuesta MA, Engel AF, Ylstra B, Meijer GA. Deletion of chromosome 4q predicts outcome in stage II colon cancer patients. Cell Oncol (Dordr) 2011;34:215–223. doi: 10.1007/s13402-011-0042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 17.Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Döhner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: Biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. doi: 10.1002/(SICI)1098-2264(199712)20:4<399::AID-GCC12>3.3.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 19.Nakao K, Hiratsuka K, Kamiyama G, Tsunoda A, Nishino S, Shibusawa M, Kusano M, Waldman FM. The genetic analysis of the liver metastasis due to colon cancer by Comparative Genomic Hybridization-Array (CGH-array) Cytometry Res. 2002;12:13–18. (In Japanese) [Google Scholar]
- 20.Nakao K, Tsunoda A, Suzuki N, Morohara K, Yamazaki K, Nishino N, Kusano M, Waldman FM. The analysis of genetic aberration in colorectal cancer with genomic array comparative hybridization (CGH) Cytometry Res. 2004;14:69–73. [Google Scholar]
- 21.Heide I, Thiede C, Poppe K, de Kant E, Huhn D, Rochlitz C. Expression and mutational analysis of Nm23-H1 in liver metastases of colorectal cancer. Br J Cancer. 1994;70:1267–1271. doi: 10.1038/bjc.1994.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng ZS, Hsu S, Zhang ZF, Cohen AM, Enker WE, Turnbull AA, Guillem JG. High level of Nm23-H1 gene expression is associated with local colorectal cancer progression not with metastases. Br J Cancer. 1994;70:1025–1030. doi: 10.1038/bjc.1994.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cawkwell L, Lewis FA, Quirke P. Frequency of allele loss of DCC, p53, RBI, WT1, NF1, NM23 and APC/MCC in colorectal cancer assayed by fluorescent multiplex polymerase chain reaction. Br J Cancer. 1994;70:813–818. doi: 10.1038/bjc.1994.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi A. Genetic changes in liver metastasis of colorectal cancer and their clinical application. Jpn J Surg. 2001;102:370–5. (In Japanese) [PubMed] [Google Scholar]
- 25.Stoeckert CJ, Jr, Causton HC, Ball CA. Microarray databases: Standards and ontologies. Nat Genet. 2002;32:469–473. doi: 10.1038/ng1028. (Suppl) [DOI] [PubMed] [Google Scholar]
- 26.Japanese Society for the Cancer of Colon and Rectum (JSCCR), corp-author Japanese Classification of Colorectal Carcinoma. 6th. Kanehara & Co. Ltd.; Tokyo: 1998. (In Japanese) [Google Scholar]
- 27.Korn WM, Yasutake T, Kuo WL, Warren RS, Collins C, Tomita M, Gray J, Waldman FM. Chromosome arm 20q gains and other genomic alterations in colorectal cancer metastatic to liver, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Genes Chromosomes Cancer. 1999;25:82–90. doi: 10.1002/(SICI)1098-2264(199906)25:2<82::AID-GCC2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 28.Rooney PH, Boonsong A, McKay JA, Marsh S, Stevenson DA, Murray GI, Curran S, Haites NE, Cassidy J, McLeod HL. Colorectal cancer genomics: Evidence for multiple genotypes which influence survival. Br J Cancer. 2001;85:1492–1498. doi: 10.1054/bjoc.2001.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma W, Yu C, Zhang W, Wu S, Feng Y. The emergence of DNA in the RNA world: An in silico simulation study of genetic takeover. BMC Evol Biol. 2015;15:272. doi: 10.1186/s12862-015-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koehler A, Bataille F, Schmid C, Ruemmele P, Waldeck A, Blaszyk H, Hartmann A, Hofstaedter F, Dietmaier W. Gene expression profiling of colorectal cancer and metastases divides tumours according to their clinicopathological stage. J Pathol. 2004;204:65–74. doi: 10.1002/path.1606. [DOI] [PubMed] [Google Scholar]
- 31.Kawamura T, Mutoh H, Tomita Y, Kato R, Honda H. Cancer DNA microarray analysis considering multi-subclass with graph-based clustering method. J Biosci Bioeng. 2008;106:442–448. doi: 10.1263/jbb.106.442. [DOI] [PubMed] [Google Scholar]
- 32.Yamasaki M, Takemasa I, Komori T, Watanabe S, Sekimoto M, Doki Y, Matsubara K, Monden M. The gene expression profile represents the molecular nature of liver metastasis in colorectal cancer. Int J Oncol. 2007;30:129–138. [PubMed] [Google Scholar]
- 33.Fujita F, Shirane M, Mori K. The profiling of the human tumor strain using the onset of cyclopedic gene analysis. Gan To Kagaku Ryoho. 2010;37:1525–1529. (In Japanese) [PubMed] [Google Scholar]
- 34.Al-Mulla F, Keith WN, Pickford IR, Going JJ, Birnie GD. Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer. 1999;24:306–314. doi: 10.1002/(SICI)1098-2264(199904)24:4<306::AID-GCC3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 35.Aragane H, Sakakura C, Nakanishi M, Yasuoka R, Fujita Y, Taniguchi H, Hagiwara A, Yamaguchi T, Abe T, Inazawa J, Yamagishi H. Chromosomal aberrations in colorectal cancers and liver metastases analyzed by comparative genomic hybridization. Int J Cancer. 2001;94:623–629. doi: 10.1002/ijc.1522. [DOI] [PubMed] [Google Scholar]
- 36.Diep CB, Parada LA, Teixeira MR, Eknaes M, Nesland JM, Johansson B, Lothe RA. Genetic profiling of colorectal cancer liver metastases by combined comparative genomic hybridization and G-banding analysis. Genes Chromosomes Cancer. 2003;36:189–197. doi: 10.1002/gcc.10162. [DOI] [PubMed] [Google Scholar]
- 37.Knösel T, Schlüns K, Stein U, Schwabe H, Schlag PM, Dietel M, Petersen I. Chromosomal alterations during lymphatic and liver metastasis formation of colorectal cancer. Neoplasia. 2004;6:23–28. doi: 10.1016/S1476-5586(04)80050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang JK, Chen YJ, Lin CH, Yu IT, Lin JK. Genetic changes and clonality relationship between primary colorectal cancers and their pulmonary metastases - an analysis by comparative genomic hybridization. Genes Chromosomes Cancer. 2005;43:25–36. doi: 10.1002/gcc.20167. [DOI] [PubMed] [Google Scholar]
- 39.Knösel T, Schlüns K, Dietel M, Petersen I. Chromosomal alterations in lung metastases of colorectal carcinomas: Associations with tissue specific tumor dissemination. Clin Exp Metastasis. 2005;22:533–538. doi: 10.1007/s10585-005-5239-7. [DOI] [PubMed] [Google Scholar]


