Abstract
MicroRNAs (miRs) have been observed to be involved in the modulation of various physiopathological processes. However, the impacts of miRNAs on muscle atrophy have not been fully investigated. In the present study, the results demonstrated that miR-351 was differentially expressed in the tibialis anterior (TA) muscle at various times following sciatic nerve transection, and the time-dependent expression profile of miR-351 was inversely correlated with that of tumor necrosis factor receptor-associated factor 6 (TRAF6) at the mRNA and protein levels. The dual luciferase reporter assay indicated that miR-351 was able to significantly downregulate the expression levels of TRAF6 by directly targeting the 3′-untranslated region of TRAF6. Overexpression of miR-351 inhibited a significant decrease in the wet weight ratio or cross-sectional area of the TA muscle following sciatic nerve transection. Western blot analysis indicated that the protein expression levels of TRAF6, muscle ring-finger protein 1 (MuRF1) and muscle atrophy F-box (MAFBx) in denervated TA muscles were suppressed by overexpression of miR-351. These results demonstrate that miR-351 inhibits denervation-induced atrophy of TA muscles following sciatic nerve transection at least partially through negative regulation of TRAF6 as well as MuRF1 and MAFBx, the two downstream signaling molecules of TRAF6.
Keywords: microRNA-351, tumor necrosis factor receptor-associated factor 6, muscle atrophy, denervation, negative regulation
Introduction
Skeletal muscle atrophy occurs under various pathophysiological conditions, such as aging, starvation, disuse, severe injury, cancer and other cachectic diseases (1–3). Previous studies have been concerned with denervation-induced skeletal muscle atrophy following peripheral nerve injury (4,5). Therefore, isobaric tags for relative and absolute quantification coupled with two-dimensional liquid chromatography-tandem mass spectrometry were performed in order to identify the differentially expressed proteins in the tibialis anterior (TA) muscle 1 and 4 weeks following sciatic nerve transection in rats (4). The results demonstrated that tumor necrosis factor receptor-associated factor 6 (TRAF6) was upregulated in denervated TA muscle (4). Conversely, the results also demonstrated that knockdown of TRAF6 significantly attenuated glucocorticoid-induced myotube atrophy in vitro and in vivo (5).
As is well known, TRAF6 is a unique E3 ubiquitin ligase and adaptor protein. It is involved in activation of various signaling cascades including mitogen-activated protein kinase, nuclear factor-κB, and phosphoinositide 3-kinase/protein kinase B in response to various cytokines (6–8), and also important for the interaction with multiple components of the ubiquitin-proteasome system (UPS) in some cell types (9–11). Due to its biological traits, TRAF6 regulates skeletal muscle mass and UPS activation in denervation or starvation-induced muscle atrophy (6,12), and catalyzes lysine 63-linked polyubiquitination of several target proteins through its association with the dimeric ubiquitin-conjugating enzyme Ubc13/Uev1A (13).
MicroRNAs (miRNAs, miRs) are a novel class of small, non-coding RNAs of ~22 nucleotides in length. miRNAs negatively regulate the expression of target genes through interaction with the 3′-untranslated regions (3′-UTR) of the target mRNAs in many biological processes (14). There have been extensive studies of the functions of miRNAs in normal physiological phenomena and numerous diseases (15,16). For instance, previous studies have demonstrated that targeting miR-122 in patients with chronic HCV genotype 1 infection caused prolonged dose-dependent reductions in HCV RNA levels without evidence of viral resistance, which indicted that targeting miR-122 may be an effective and safe treatment strategy for HCV infection (17,18). However, the involvement of miRNAs in muscle-associated biological processes has not been fully investigated. To our knowledge, it has only been reported that miR-1 and miR-133 are able to promote differentiation and proliferation of the skeletal and cardiac muscle cells (19), and the expression of miR-351 in differentiated C2C12 cells is upregulated following treatment with dexamethasone (20). The underlying mechanisms, however, are still to be further explored. The present study aimed to examine whether certain miRNAs, such as miR-351, are able to regulate denervation-induced skeletal atrophy in a model of rat sciatic nerve injury and TA muscle denervation, providing a potential novel molecular target for the clinical treatment of denervation-induced muscle atrophy.
Materials and methods
Animal surgery and treatments
A total of 126 adult male Sprague-Dawley rats (weight, 200 g; age, 2 months) were supplied by the Experimental Animal Center of Nantong University (Nantong, China) with routine provision of food and water, and were maintained at 24°C with a 12:12 h light/dark cycle. All animal experiments were carried out in accordance with the institutional animal care guidelines of Nantong University and approved by the Administration Committee of Experimental Animals, Jiangsu Province.
A total of 72 rats underwent surgery for sciatic nerve transection as described previously (21). The TA muscle was harvested from a cohort of rats at 0, 3, 7 and 14 days post-surgery. Another cohort of 54 rats was randomly divided into two groups to receive a multi-point injection of miR-351 agomir and negative control agomir (both at 2.5 nmol in 50 µl vehicle; Guangzhou RiboBio Co., Ltd., Guangzhou, China) into the TA muscle. The injection was administered a total of 5 times at a regular interval of 2 days, the rats were sacrificed by cervical dislocation and the TA muscle was dissected.
Dual luciferase reporter assay
HEK-293T cells were cultured in growth medium consisting of Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml of streptomycin (Sigma-Aldrich, St. Louis, MO, USA), and 100 U/ml of penicillin (Sigma-Aldrich) in a 37°C, 5% CO2 incubator. The cells were co-transfected with 600 ng of the wild-type or mutant 3′-UTR luciferase reporter (Guangzhou RiboBio Co., Ltd., Guangzhou, China) and 20 µM of the miR-351 agomir or the negative control duplexes using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in 24-well plates. The sequences of these reporters were as follows: TRAF6 3′-UTR: 5′-ACGCTCATCAATTTGTCAGGGAA; TRAF6 3′-UTR mutant: 5′-ACGCTCATCAATTTGAGTCCCTA. Following transfection for 24 h, the cells were harvested by adding 100 µl of lysis buffer (Promega Corporation, Madison, WI, USA). The activity of firefly and Renilla luciferases was measured from the cell lysates using the dual luciferase reporter assay system (Promega Corporation). TargetScan prediction (www.targetscan.org/vert_71) was then used to deduce the biological targets of miRNAs.
Muscle morphological observation
At 14 days post surgery, the animals in the two groups that had been injected with miR-351 agomir and negative control agomir were anesthetized with 30 mg/kg pentobarbital (Merck Millipore, Darmstadt, Germany) and perfused through the left ventricle sequentially with saline and 4% paraformaldehyde. The TA muscle injected with either miR-351 agomir or negative control agomir was harvested and weighed, in order to calculate the wet weight ratio of the muscle.
For Masson's trichrome staining, the harvested TA muscle were post-fixed in 4% paraformaldehyde at 4°C for 24 h, dehydrated in a graded ethanol series, cleared in xylene, embedded in paraffin, and cut into 5 µm-thick transverse sections using a freezing microtome. Hematoxylin and eosin staining was applied to every section, and images were captured under a light microscope (magnification, ×40). The average cross-sectional area (CSA) of the TA muscle were then determined using a Leica Q5501 W image analysis system (Leica Mycrosystems GmbH, Wetzlar, Germany) as described previously (22).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the TA muscle (100 mg/group), harvested as described previously, using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). RT to cDNA was carried out with a SuperScript First-Strand Synthesis system (Invitrogen; Thermo Fisher Scientific, Inc.). All primers were purchased from Generay Biotech Co., Ltd. (Shanghai, China). The primers used in this study were as follows: TRAF6 forward, 5′-GCAGAGGAATCACTTGGCACG-3′ and reverse, 5′-CACGGACGCAAAGCAAGGTT-3′; miR-351 forward, 5′-ACACTCCAGCTGGGTCCCTGAGGAGCCCTTGG-3′ and reverse, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGGCTC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH forward, 5′-CAACGGGAAACCCATCACCA-3′ and reverse, 5′-ACGCCAGTAGACTCCACGACAT-3′. RT-qPCR was conducted in a StepOne™ Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using FastStart® SYBR Green qPCR Master Mix (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. The reaction mixture contained 1 µl cDNA from each sample that was mixed with 10 µl 2X Fast Start® SYBR Green qPCR Master Mix, 2 µl 10X ROX of the assays-on-demand kit (Biotium, Fremont, CA, USA), 1 µl primer, and 6 µl of RNase/DNase-free water (Biotium). A three-step fast cycle protocol was used, with cycling conditions as follows: Denaturation at 95°C, 10 sec; annealing at 60°C, 30 sec; extension at 70°C, 10 sec. The relative mRNA expression was measured through the 2−ΔΔCq method (23). U6 snRNA and GAPDH served as an internal control.
Western blot analysis
The total protein of the TA muscles, harvested as previously described, was extracted using a T-PER tissue protein extraction reagent (Pierce Biotechnology; Thermo Fisher Scientific, Inc.) and quantified using a Coomassie Plus Bradford assay kit (Pierce Biotechnology; Thermo Fisher Scientific, Inc.). Samples containing 15 µg total protein were separated by 12% (w/v) SDS-PAGE and transferred to a polyvinylidene difluoride membrane (EMD Millipore, Bedford, MA, USA). Following incubation for 1 h with 5% (w/v) non-fat dry milk in Tris-buffered saline with Tween-20 [TBS-T; 0.05% (v/v) Tween-20], the membrane was probed with the following primary antibodies diluted in blocking buffer overnight at 4°C: Rabbit anti-TRAF6 polyclonal antibody (1:1,000; Abgent, Inc., San Diego, CA, USA), goat anti-muscle ring finger 1 (MuRF1) polyclonal antibody (1:1,000; R&D System, Minneapolis, MN, USA), rabbit anti-muscle atrophy F-box (MAFBx) polyclonal antibody (1:1,000; LifeSpan BioSciences, Inc., Seattle, WA, USA), and rabbit anti-β-tubulin polyclonal antibody (1:2,000; Abcam, Cambridge, MA, USA). Following washing with TBS-T, the membrane was incubated with secondary antibodies diluted in blocking buffer (LI-COR Biosciences, Lincoln, NE, USA) at room temperature for 1 h. These were anti-rabbit (dilution, 1:5,000; cat. no. 611-701-127; Rockland Immunochemicals, Inc., Pottstown, PA, USA) or anti-goat IgG (dilution, 1:6,000; cat. no. 605-706-125, Rockland Immunochemicals, Inc.). The images were scanned with the Odyssey infrared image system (LI-COR Biosciences), and the absorbance data were analyzed using PDQuest 7.2.0 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analyses
All data are presented as means ± standard error of the mean. Comparison between groups was assessed by unpaired Student's t test using SPSS 10.0 software (SPSS, Inc., Chicago, IL, USA). Unless otherwise specified, all assays were performed in triplicate. P<0.05 was considered to indicate a statistically significant result.
Results
Time-dependent expression profile of miR-351 and TRAF6 in TA muscle following sciatic nerve transection
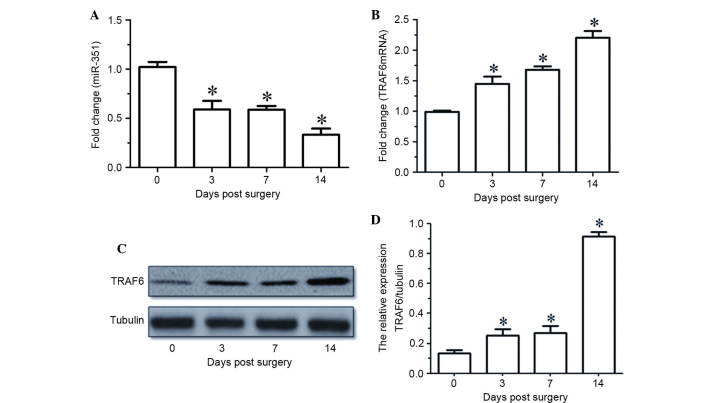
RT-qPCR indicated that the expression levels of miR-351 in the TA muscle were gradually decreased with time following sciatic nerve transection (Fig. 1A). The mRNA and protein expression levels of TRAF6 in the TA muscle were upregulated at various times following sciatic nerve transaction, as determined by RT-qPCR and western blot analysis (Fig. 1B-D). Notably, the temporal expression profile of miR-351 in the TA muscle was inversely correlated with that of TRAF6 during the period of post-sciatic nerve injury.
Figure 1.
(A and B) miR-351 and TRAF6 expression levels in TA muscles at various time points (0, 3, 7, 14 days) following sciatic nerve transection, as determined by reverse transcription-quantitative polymerase chain reaction. (C) Western blot analysis and (D) quantified results demonstrating the protein expression levels of TRAF6 in the TA muscles at various time points (0, 3, 7, 14 days) following sciatic nerve transection. Data are presented as means ± standard deviation, n=9 per animal group. *P<0.05, vs. 0 days post sciatic nerve transection. Tubulin was used as a loading control in the western blot analysis. miR, microRNA; TRAF6, tumor necrosis factor receptor-associated factor 6; TA, tibialis anterior.
Post-transcriptional downregulation of TRAF6 expression by miR-351
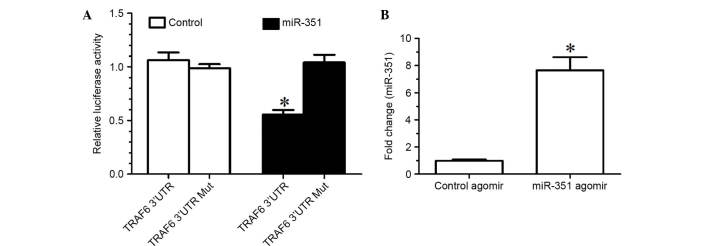
TargetScan prediction determined that the TRAF6 3′-UTR and miR-351 had a conservative matching area. To confirm that TRAF6 was negatively regulated by miR-351 through direct binding to the 3′-UTR of TRAF6, the wild-type 3′-UTR and mutant 3′-UTR of TRAF6 were constructed and inserted into the downstream region of the luciferase reporter gene, respectively. In the luciferase activity assay, miR-351 agomir or negative control agomir with p-Luc-UTR luciferase reporter were co-transfected into HEK 293T cells. Due to the presence of the wild-type 3′-UTR of TRAF6, miR-351 was able to significantly decrease the relative luciferase activity by >50%, whereas the relative luciferase activity of mutant 3′-UTR was not decreased by miR-353, suggesting that miR-353-induced decrease in the relative luciferase activity was sequence-specific (Fig. 2A). In other words, miR-351 is able to downregulate TRAF6 expression by directly targeting the 3′-UTR of TRAF6.
Figure 2.
(A) Relative luciferase activity of HEK-293T cells that were co-transfected with miR-351 agomir (miR-351) or negative control agomir (control) with p-Luc-UTR vectors. The Renilla luciferase vector was used as an internal control. (B) Expression levels of miR-351 in the TA muscle of rats treated with miR-351 agomir (2.5 nmol in 50 µl) was significantly increased compared with those of the rats treated with control agomir (2.5 nmol in 50 µl). *P<0.05, vs. the control. UTR, untranslated region; miR, microRNA; Mut, mutant.
Inhibition of denervation-induced TA muscle atrophy by overexpression of miR-351
The expression levels of miR-351 in the TA muscle of rats treated with miR-351 agomir were significantly increased compared with those treated with control agomir (Fig. 2B), suggesting that injection of miR-351 agomir into the TA muscle was an effective method for increasing the expression levels of miR-351 in the muscle.
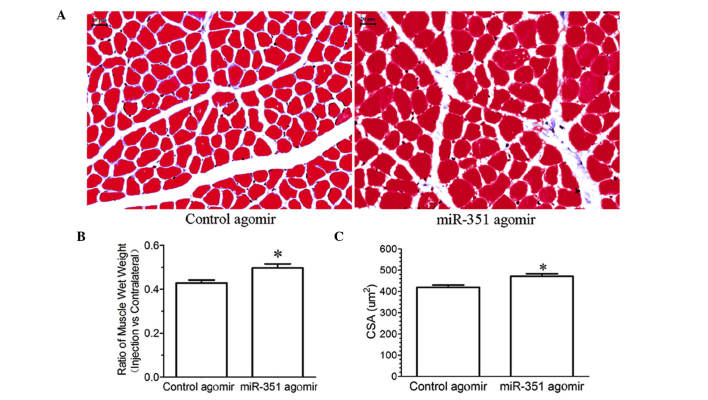
Morphological observation showed that treatment with miR-351 agomir inhibited a significant decrease in either the wet weight ratio or CSA of the TA muscle compared with control agomir (Fig. 3), suggesting that overexpression of miR-351 may inhibit denervation-induced muscle atrophy.
Figure 3.
(A) Masson's trichrome staining for determining the CSA of the TA muscle of rats injected with miR-351 agamir or control agamir. Scale bar = 20 µm; (B and C) Bar graphs showing the wet weight ratio and CSA of the TA muscle of rats injected with miR-351 agomir or control agomir. Data are presented as means ± standard deviation, n=8 per animal group. *P<0.05, vs. the control. CSA, cross-sectional area; TA, tibialis anterior.
Suppression of the expression of TRAF6, MuRF1 and MAFBx in denervated TA muscle by miR-351
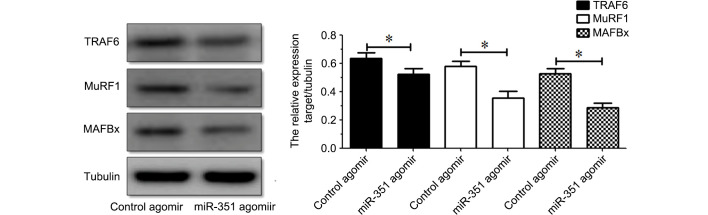
Western blot analysis indicated that the protein expression levels of TRAF6, MuRF1 or MAFBx in the TA muscle were inhibited following treatment with miR-351 agomir compared with those following treatment with control agomir (Fig. 4). The results indicated that overexpression of miR-351 is able to suppress the protein expression of TRAF6, MuRF1 or MAFBx in denervated TA muscles.
Figure 4.
(A) Western blot and (B) quantification showing the protein expression levels of TRAF6, MuRF1, and MAFBx in the TA muscle of rats injected with miR-351 agamir or control agamir. Data are presented as means ± standard deviation, n=9 per animal group. *P<0.05, vs. the control. Tubulin was used as a loading control in western blot analysis. miR, microRNA; TA, tibialis anterior; TRAF6, tumor necrosis factor receptor-associated factor 6.
Discussion
Recent evidence suggests that miRNAs are involved in modulation of atrophy, proliferation and differentiation of skeletal muscles (20,24,25). A previous study observed that miR-351 is transiently increased in early days of muscle regeneration (26). miR-351 inhibits the expression of E2f3, a key regulator of cell cycle progression and proliferation, promotes myogenic progenitor cell proliferation and protects early differentiating myogenic progenitor cell from apoptosis (26). These findings revealed that miR-351 is involved in muscle atrophy. The results of the present study identified miR-351 as a novel regulator of denervation-induced muscle atrophy. miR-351 was differentially expressed in the TA muscle at various times following sciatic nerve transection, and the temporal expression profile of miR-351 in the TA muscle was inversely correlated with that of TRAF6 at the mRNA and protein levels. The dual luciferase reporter assay indicated that miR-351 was able to significantly downregulate TRAF6 expression by directly targeting the 3′-UTR of TRAF6. The results further demonstrated that overexpression of miR-351 inhibited a significant decrease in the wet weight ratio or CSA of the TA muscle following sciatic nerve transection.
Studies have determined that TRAF6 expression is significantly upregulated in denervation- or starvation-induced muscle atrophy and TRAF6 affects denervation- or starvation-induced muscle atrophy through regulation of muscle-specific ubiquitin ligases MuRF1 and MAFBx (5,27). In addition, the E3 ubiquitin ligase activity of TRAF6 has proven essential for starvation-induced muscle atrophy (6,13). In this study, therefore, it was also determined whether MuRF1 and MAFBx, as two downstream signaling molecules of TRAF6, were regulated by miR-351. Western blot analysis indicated that the protein expression levels of MuRF1 and MAFBx as well as TRAF6 were all inhibited by overexpression of miR-351.
In conclusion, the results of the present study demonstrated that miR-351 has an inhibitory role in denervation-induced muscle atrophy through, at least in part, negative regulation of TRAF6 and MuRF1 and MAFBx (two muscle-specific ubiquitin ligases). Further research, however, is required in order to provide a good understanding of the detailed mechanism underlying miR-351 regulation in muscle atrophy.
Acknowledgements
This study was supported by the 863 Program (grant no. 2012AA020502), the 973 Program (grant nos. 2014CB542202 and 2014CB542203), the NSFC (grant nos. 81130080, 81171180 and 81301628), China Postdoctoral Science Foundation (grant no. 2016M591894), Fund of Doctoral Start-up of Nantong University (grant no. 15B18), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the Natural Science Foundation of Jiangsu Province (grant no. BK2012230), the Collegiate Natural Science Foundation of Jiangsu Province (grant nos. 13KJB180018 and 13KJ180005), the Natural Science Foundation of Nantong University (grant no. 142014). We thank Professor Jie Liu for assistance in the preparation of the manuscript.
References
- 1.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 2.Kandarian S. The molecular basis of skeletal muscle atrophy-parallels with osteoporotic signaling. J Musculoskelet Neuronal Interact. 2008;8:340–341. [PubMed] [Google Scholar]
- 3.Piccirillo R, Demontis F, Perrimon N, Goldberg AL. Mechanisms of muscle growth and atrophy in mammals and Drosophila. Dev Dyn. 2014;243:201–215. doi: 10.1002/dvdy.24036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H, Qiu J, Chen Y, Yu M, Ding F, Gu X. Proteomic and bioinformatic analysis of differentially expressed proteins in denervated skeletal muscle. Int J Mol Med. 2014;33:1586–1596. doi: 10.3892/ijmm.2014.1737. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Gong Y, Qiu J, Chen Y, Ding F, Zhao Q. TRAF6 inhibition rescues dexamethasone-induced muscle atrophy. Int J Mol Sci. 2014;15:11126–11141. doi: 10.3390/ijms150611126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul PK, Bhatnagar S, Mishra V, Srivastava S, Darnay BG, Choi Y, Kumar A. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol. 2012;32:1248–1259. doi: 10.1128/MCB.06351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays. 2003;25:1096–1105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Zhang J, Zhang L, van Dam H, ten Dijke P. UBE2O negatively regulates TRAF6-mediated NF-κB activation by inhibiting TRAF6 polyubiquitination. Cell Res. 2013;23:366–377. doi: 10.1038/cr.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J Biol Chem. 2010;285:2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamothe B, Campos AD, Webster WK, Gopinathan A, Hur L, Darnay BG. The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide and RANKL. J Biol Chem. 2008;283:24871–24880. doi: 10.1074/jbc.M802749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul PK, Kumar A. TRAF6 coordinates the activation of autophagy and ubiquitin-proteasome systems in atrophying skeletal muscle. Autophagy. 2011;7:555–556. doi: 10.4161/auto.7.5.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, Kumar A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol. 2010;191:1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang YB, Xu L, Yue S, Liu S, Giffard RG. Neuroprotection by astrocytes in brain ischemia: Importance of microRNAs. Neurosci Lett. 2014;565:53–58. doi: 10.1016/j.neulet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soares RJ, Cagnin S, Chemello F, Silvestrin M, Musaro A, De Pitta C, Lanfranchi G, Sandri M. Involvement of microRNAs in the regulation of muscle wasting during catabolic conditions. J Biol Chem. 2014;289:21909–25. doi: 10.1074/jbc.M114.561845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirby TJ, Chaillou T, McCarthy JJ. The role of microRNAs in skeletal muscle health and disease. Front Biosci (Landmark Ed) 2015;20:37–77. doi: 10.2741/4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 18.van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, Zeuzem S, Lawitz EJ, Rodriguez-Torres M, Kupcova V, Wiercinska-Drapalo A, et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014;111:53–59. doi: 10.1016/j.antiviral.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen H, Liu T, Fu L, Zhao S, Fan B, Cao J, Li X. Identification of microRNAs involved in dexamethasone-induced muscle atrophy. Molecular and cellular biochemistry. 2013;381:105–113. doi: 10.1007/s11010-013-1692-9. [DOI] [PubMed] [Google Scholar]
- 21.Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D, Liu M, Ding F, Gu X. Expression of myostatin RNA transcript and protein in gastrocnemius muscle of rats after sciatic nerve resection. J Muscle Res Cell Motil. 2006;27:37–44. doi: 10.1007/s10974-005-9050-5. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Chen JF, Callis TE, Wang DZ. microRNAs and muscle disorders. J Cell Sci. 2009;122:13–20. doi: 10.1242/jcs.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenberg I, Alexander MS, Kunkel LM. miRNAS in normal and diseased skeletal muscle. J Cell Mol Med. 2009;13:2–11. doi: 10.1111/j.1582-4934.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Melton DW, Gelfond JA, McManus LM, Shireman PK. MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation. Physiol Genomics. 2012;44:1042–1051. doi: 10.1152/physiolgenomics.00052.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A, Bhatnagar S, Paul PK. TWEAK and TRAF6 regulate skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care. 2012;15:233–9. doi: 10.1097/MCO.0b013e328351c3fc. [DOI] [PMC free article] [PubMed] [Google Scholar]