Summary
Although the function of the mammalian pancreas hinges on complex interactions of distinct cell types, gene expression profiles have primarily been described with bulk mixtures. Here we implemented a droplet-based, single-cell RNA-seq method to determine the transcriptomes of over 12,000 individual pancreatic cells from four human donors and two mouse strains. Cells could be divided into 15 clusters that matched previously characterized cell types: all endocrine cell types, including rare epsilon-cells; exocrine cell types; vascular cells; Schwann cells; quiescent and activated stellate cells; and four types of immune cells. We detected subpopulations of ductal cells with distinct expression profiles and validated their existence with immuno-histochemistry stains. Moreover, among human beta- cells, we detected heterogeneity in the regulation of genes relating to functional maturation and levels of ER stress. Finally, we deconvolved bulk gene expression samples using the single-cell data to detect disease-associated differential expression. Our dataset provides a resource for the discovery of novel cell type-specific transcription factors, signaling receptors, and medically relevant genes.
Graphical abstract
Single-cell transcriptomics of over 12,000 cells from four human donors and two mouse strains was determined using inDrop. Cells were divided into 15 clusters that matched previously characterized cell types. Detailed analysis of each population separately revealed subpopulations within the ductal population, modes of activation of stellate cells, and heterogeneity in the stress among beta cells.
Introduction
The pancreas is a vertebrate-specific organ with a central role in energy homeostasis achieved by secreting digestive enzymes and metabolic hormones (Kimmel and Meyer, 2010). Most of the pancreas (95%) is comprised of two exocrine cell types: acinar and duct cells. Acinar cells produce digestive enzymes, including amylase, lipase, and peptidases (Whitcomb and Lowe, 2007), and duct cells secrete bicarbonate (Steward et al., 2005) and ferry the digestive enzymes to the gastrointestinal tract. Islets, about 5% of the pancreatic mass, are dispersed within the exocrine tissue and ducts and contain endocrine cells that secrete hormones for glucose homeostasis (Drucker, 2007). Islets contain five endocrine cell types: alpha cells (glucagon+), beta cells (insulin+), delta cells (somatostatin+), gamma cells (pancreatic polypeptide+), and epsilon cells (ghrelin+) (Mastracci and Sussel, 2012).
Dysfunction of the pancreas is clinically important, most notably in type 1 (T1D) and type 2 diabetes mellitus (T2D), pancreatitis, and cancer. Efforts to replace beta cells lost in type 1 diabetes have made the pancreas one of the focus organs of current regenerative biology. In particular, significant efforts have been made to produce insulin-secreting beta cells in vitro from pluripotent cells (Nostro et al., 2015; Pagliuca et al., 2014; Rezania et al., 2014; Russ et al., 2015; Xie et al., 2013). It may be argued that these efforts are limited by an incomplete understanding of the gene expression programs of adult beta cells. More generally, because the endocrine cell types all functionally interact, there is a clear need to systematically characterize all pancreatic cell types at the molecular level to understand their dysfunction in disease.
Significant advances in characterizing the transcriptomes of individual cells have been made using both in situ and RNA sequencing (RNA-seq) approaches. Although in situ methods are able to scale up to hundreds or even thousands of genes (Chen et al., 2015; Lee et al., 2014; Lubeck et al., 2014), RNA-seq has the unique advantage that it can detect transcripts comprehensively on a genomic scale. Single-cell RNA-seq has helped identify new cell types in the mouse brain (Zeisel et al., 2015) and mouse lung (Treutlein et al., 2014) and was recently used to characterize 70 individual human pancreatic cells (Li et al., 2016) and 341 cells from mouse islets (Xin et al., 2016). In the pancreas, fixed and sorted populations of alpha and beta cells have previously been compared using RNA sequencing (Blodgett et al., 2015; Dorrell et al., 2011; Morán et al., 2012; Nica et al., 2013). However, it has remained a challenge to analyze many human cells at a high-throughput scale (Blodgett et al., 2015). This stems both from the difficulty in obtaining tissues from human donors and developing a system that captures a sufficient number of cells. The inDrop method provides a systematic approach for capturing thousands of cells without pre-sorting (Klein et al., 2015). inDrop uses high-throughput droplet microfluidics that barcode the RNA from thousands of individual cells, implementing a sensitive linear amplification method for single-cell transcriptome library construction (cell expression by linear amplification and sequencing [CEL-seq]) (Hashimshony et al., 2012, 2016).
Comparing gene expression across species is an efficient method for inferring function from transcription data. The mouse model constitutes the most studied for pancreas and diabetes research; however, the findings are typically applied to humans, thereby highlighting the importance of inter-species similarities and differences. The size and organization of the endocrine pancreas have been characterized previously in humans and mice (Levetan and Pierce, 2013). In particular, mouse islets are organized as a core of beta cells surrounded by non-beta cells (Brissova et al., 2005), whereas, in humans, several patterns have been reported, such as clusters of beta cells within the islets (Orci and Unger, 1975), ribbon-like patterns (Grube and Bohn, 1983), or a diffuse unorganized form (Cabrera et al., 2006). These anatomical differences highlight the need to understand the molecular variants between these species as well as the conserved gene-regulatory complement across them.
In this work, we implemented the inDrop platform to study the human and mouse pancreas and to describe its constituent cell types at higher resolution than that provided by immuno-histochemical characterization and transcriptional analyses of bulk samples or cell type-enriched specimens. Analysis of four human pancreas donors and islets from two strains of mice, enriched for endocrine cells (islets), identifies all previously known cell types. Within these cell types, we find substructure in the ductal and beta cell populations. This approach provides a first detailed look at the gene expression program of the adult mammalian pancreas.
Results
Single-Cell RNA-Seq Identifies a Comprehensive List of Pancreatic Cell Types
Using inDrop, we isolated and sequenced ∼10,000 human and ∼2,000 mouse pancreatic cells from four cadaveric donors and two strains of mice, respectively, and processed them for transcriptomic analysis (Figure 1A). Our human donors spanned a range of ages and BMI health parameters (Table S1). From the human donors, we prepared and sequenced several separate batches, each of approximately ∼800 cells (on average, 100,000 reads were associated with each analyzed cell). Analysis of the sequence reads revealed on average ∼6,000 uniquely detected transcripts from ∼2,000 genes for each cell (Figures S1B and S1C).
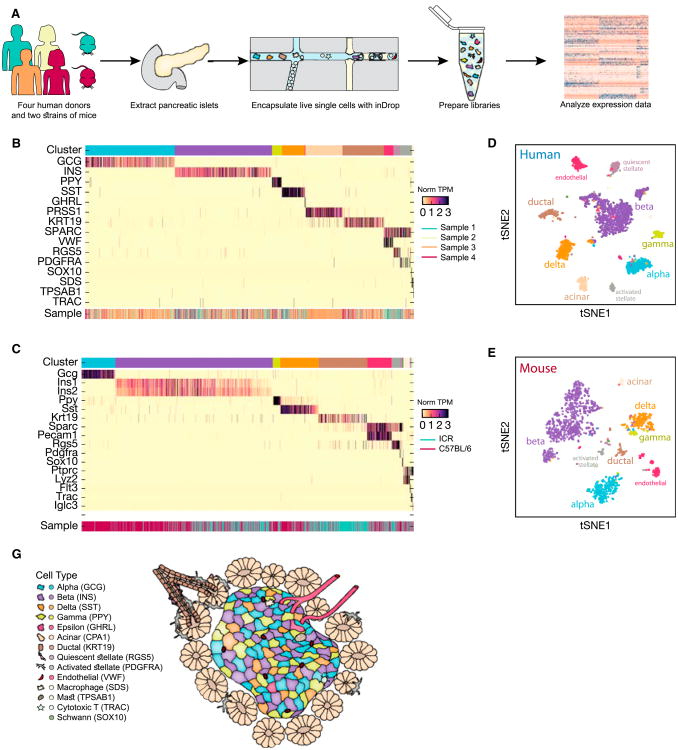
Figure 1. A Transcriptomic Map of the Human and Mouse Pancreas.
(A) Single-cell RNA-seq was carried out on human and mouse pancreatic islets using the inDrop microfluidics system to generate data that allow for quantification of transcript abundance across cells and genes.
(B and C) Heatmap of all cells clustered by recursive hierarchical clustering (STAR Methods), showing selected marker genes for every population of human (B) and mouse (C). The top bar indicates assigned cluster identity (legend shown in F). The bottom bar indicates the donor of origin.
(D and E) tSNE plot of cells from donor 1 based on the expression of highly variable genes for human (D) and mouse strain C57BL/6 (E). The detected clusters are indicated by different colors as shown in (F).
(F) Schematic of the pancreatic islet and the cellular neighborhood along with the identified cell types and their respective markers.
After filtering out cells with limited numbers of detected genes, our dataset contained 8,629 cells from the four donors and 1,886 mouse cells from both strains. When we visualized this dataset using t-distributed stochastic neighbor embedding (tSNE) (van der Maaten and Hinton, 2008), we found that they formed clear clusters, suggesting distinct cell types at the molecular level (Figures 1B–1E). Precisely delineating the clusters, however, was challenging because of the inherent hierarchical relationships among the cell types (Figures 2B and 2D). We thus devised an iterative hierarchical clustering method that restricts genes enriched in one cell type from being used to separate other cell types. Applying this method to the data, we identified 14 and 13 different cell populations across the human and mouse samples, respectively (Figures 1B–1E). We next sought to characterize the identities of these cell populations based upon expression of unique transcripts and with reference to the literature (Figures 1B and 1C; Table S2). For example, a particular cluster could be annotated as beta cells because it was enriched in the expression of insulin (INS) and several other markers (Figure S2). Similarly, we detected the cell types corresponding to alpha, gamma, delta, and epsilon cells within islets; acinar, ductal, quiescent and activated pancreatic stellate cells, and vascular cells; four immune cell types (tissue-resident macrophages, mast cells, B cells, and cytotoxic T cells); and Schwann cells (Figures 1C, 1E, and 1F).
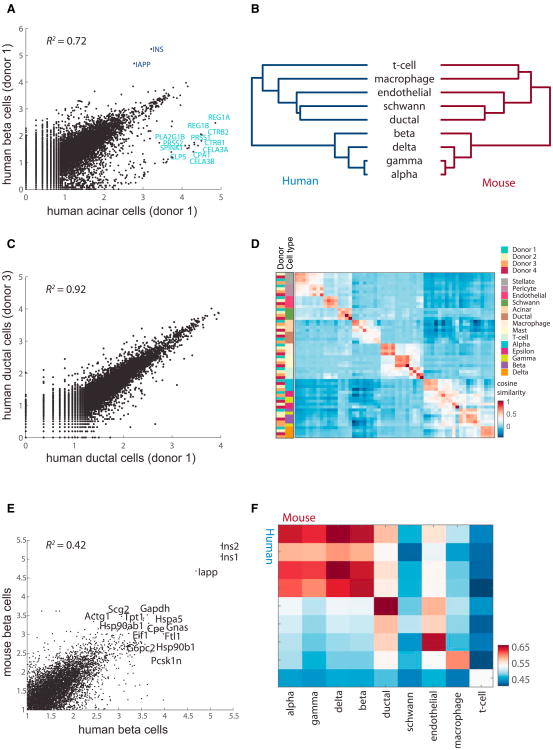
Figure 2. The Endocrine Transcriptome Is Readily Distinguished from the Other Cell Types and across Donors.
(A) Plot comparing the average expression (log10, transcripts per million [tpm]) of donor 1 beta and acinar cells. Genes with high differential expression are noted.
(B) Dendrogram showing relationships among the cell types in human (left) and mouse (right). The dendrogram was computed using hierarchical clustering with average linkage on the log10 tpm values of the highly variable genes.
(C) Same as (A) for the ductal cells of donors 1 and 3.
(D) Heatmap indicating correlations on the averaged profiles among donors for all cell types (STAR Methods).
(E) Same as (A) for human beta cells of donor 1 and mouse beta cells of mouse 1.
(F) Heatmap indicating Pearson's correlations on the averaged profiles among common cell types for human and mouse.
In addition to abundant cell types, our sequencing method and clustering approach also reliably detected rare cell types such as T cells and epsilon cells, which each constitute approximately 0.1% of a complex mixture of diverse cell types, allowing a detailed characterization of the cell types present in and near human islets. Although we identified cells from all cell types in each donor, the proportions of each cell type varied considerably (Figure S1D). Interestingly, donor 1 (17 years old) showed a particularly high number of Ghrelin+ epsilon cells, which are believed to be absent in adults (Mastracci and Sussel, 2012). Otherwise, the variation in the ratio of alpha cells to beta cells falls within the expected range (Fadista et al., 2014; Yoon et al., 2003). Because islet preparations deplete non-islet cell types, variation is expected in the purity and proportions of cell types. In addition, this variation is affected by differential responses to the culturing conditions of each cell type (STAR Methods). The observed correspondence of the clusters to previously characterized cell types is evidence for the quality of the dataset and its potential for new insights. Although we could not detect acinar cells in the mouse data, we did detect beta cells (Figure 1D). Our dataset provides a powerful resource for studying gene expression in a complex tissue across species. For example, Figure S2 shows the gene expression of the insulin gene in both humans and mice.
Relationships among Cell Types Reveal Consistent Cell Types across Donors and Species
Although inDrop captures less than 10% of the transcripts in a given cell by pooling together cells with correlated expression profiles, we derived a highly resolved transcriptome for each of the cell types. For example, comparing the transcriptome of two cell type clusters in one human pancreas revealed a large number of significantly differentially expressed genes (Figure 2A), with the three most highly expressed genes also among the most differentially expressed: INS, IAPP, and CPA1. Such differential expression provides strong evidence for the inference that these are beta (INS/IAPP+) and acinar (CPA1+) cells, respectively.
The same approach can be used to assign all clusters of cells to known cells types. Extending the analysis to all pairs of cell types detected in both humans and mice, we computed the dendrogram shown in Figure 2B. A deep branch separating four cell types from the others contained alpha, beta, gamma, and delta cells and was thus clearly identifiable as endocrine. Among these endocrine cell types, the alpha and gamma cells formed a group. Comparing this dendrogram with one based on the analogous mouse cell types, we found a nearly identical pattern of relationships (Figure 2B, right). The endocrine cells form a cluster to the exclusion of the other cell types. This confirms the expected separation and relationships of the endocrine cells as a distinct set of cell types.
We next considered the coherence of the cell types across individuals of the same species. Comparing ductal cells across samples (human donors 1 and 3), we found a correlation of R2 = 0.92 (Figure 2C), well above the differences between cell types within each donor (e.g., R2 = 0.72 in Figure 2A), suggesting that the average transcriptome of cell types is conserved. Overall, cell types generally grouped together despite originating from different individuals (Figure 2D). In particular, the endocrine cell types constituted a distinct cluster of cell types relative to the nine other cell types. Also, in mice, cell types were highly correlated across strains (Figure S3; R2 = 0.9). Comparing across species, we found that human and mouse beta cells are reasonably well correlated (Figure 2E). Comparing across all cell types common to both species (Figure 2F), we found that the four endocrine cell types (alpha, beta, gamma, and delta) are the most correlated.
Endocrine-Specific Gene Expression
Our dataset represents a treasure trove for the study of gene expression specific to pancreatic endocrine cells. We illustrate its relevance to drug discovery (FFAR4), functional maturation of beta cells (UCN3), genetic association studies (DLK1), and mouse models of human disease (LEPR). Agonists of FFAR4/GPR120, a G protein-coupled receptor, are currently being developed as therapeutics for diabetes because they exert incretin-like effects to lower blood glucose (Holliday et al., 2012). Our data provide evidence that FFAR4 is expressed in both beta and delta human cells (Figure 3A, left), opening up the question of whether a potential therapeutic effect of FFAR4 agonists associates with one or both of these cell types. To confirm the expression of FFAR4 in both human beta and delta cells, we performed immunohistochemistry stains on dissociated islet cells and detected FFAR4 protein in both cell types (Figure 3B). However, previous work in mice suggested a delta specificity of Ffar4 (Stone et al., 2014). Using our single-cell mouse data, we found an expression pattern that matches neither the existing literature nor human data: we detected Ffar4 in alpha, beta, gamma, and delta cells (Figure 3A, right). This highlights a gap in our knowledge about FFAR4/Ffar4 as well as a key difference between mice and humans that brings into question the relevance of mechanisms regarding FFAR4/Ffar4 inferred in mice.
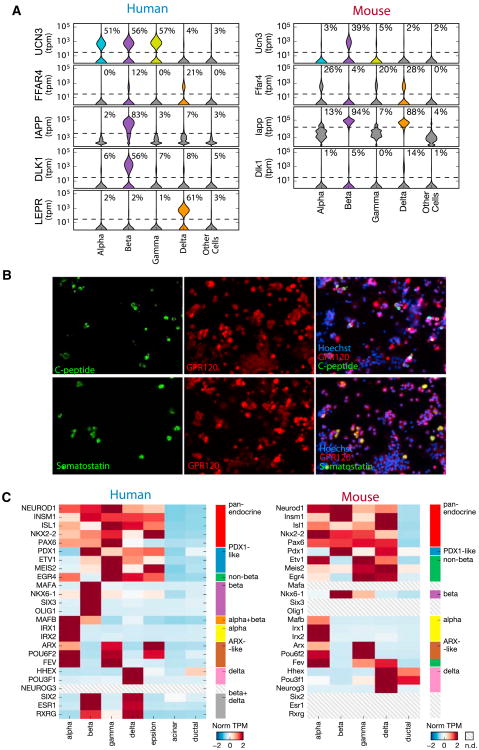
Figure 3. Endocrine Transcriptomes Reveal Novel Expression Patterns of Key Genes.
(A) Violin plots for expression of UCN3, FFAR4, LEPR, IAPP, and DLK1 across alpha, beta, gamma, and delta cell types for all human donors (left) and two mice (right). The percent number indicates the fraction of cells with detectable expression of the gene.
(B) FFAR4 (GPR120) is expressed in delta and beta cells of human islet cells. C-peptide and Somatostatin mark beta and delta cells, respectively. Note the co-positive yellow cells in both merged images showing FFAR4 expression in both beta and delta cells.
(C) Heatmap showing gene expression of transcription factors across the human (left) and mouse (right) endocrine cell types.
UCN3, initially discovered as a specific marker of beta cell maturation in mice (Blum et al., 2012), is known to be expressed in both alpha and beta cells in primates (van der Meulen et al., 2012). Our data confirm that Ucn3 is restricted to beta cells in mice (Figure 3A, right) and expand the list of human cell types that express UCN3 to include all human endocrine cells except delta cells (Figure 3A, left). This supports the recent finding that UCN3 signaling in human delta cells is part of a paracrine feedback mechanism (van der Meulen et al., 2015). The difference in expression pattern between mouse and human suggests an instance of gene expression evolution across mammalian pancreata.
Genome-wide association studies (GWASs) have implicated DLK1 as a non-immune-related gene involved in T1D (Wallace et al., 2010). Our study and others (Li et al., 2016) show that DLK1 is specifically expressed in human beta cells (Figure 3A, left), making it the only known T1D GWAS hit specific to beta cells and thus bolstering the case for its potential importance for T1D. By contrast, IAPP is believed to be specifically restricted to beta cells (Johnson et al., 1988), which our human expression data support; however, Iapp is more promiscuously expressed in mouse cells, with detectable expression in both beta and delta cells. Knockouts of leptin and the leptin receptors have been central models for obesity and T2D in mice (Drel et al., 2006). Several studies in mice have suggested a direct effect of leptin on leptin receptor-expressing beta cells (Morioka et al., 2007). Instead, our human data show that LEPR is a delta cell-specific gene in humans (Figure 3A, left), suggesting that leptin action in human pancreatic islets is mediated by delta cells. Together, these examples show the value of accurate cell type-specific mapping of genes implicated in islet function.
Although cell identity is often described by cell morphology, histology, surface markers, or physiological function, the gene expression programs that shape cell identity are orchestrated by transcription factors. Therefore, we chose to explore the cell type specificities of transcription factors detected in our dataset. Our analysis of human endocrine transcriptomes both confirmed the expected expression patterns of key transcription factors and revealed novel endocrine-specific transcription factor expression (Figure 3C, left; Table S3). We confirmed the known specificities of transcription factors in endocrine cells: MAFA, NKX6-1 (beta), IRX1, IRX2 (alpha), HHEX (delta and duct), MAFB (alpha and beta), ARX (alpha, gamma, and epsilon), PDX1 (all types except alpha), PAX6 (all types but lower in delta and epsilon), and NEUROD1, INSM1, and NKX2-2 (all types) (Benner et al., 2014; Bonal and Herrera, 2008; Murtaugh, 2007; Zaret and Grompe, 2008). Our data clearly recapitulate the known cell type specificities of pancreatic transcription factors.
Our data allowed us to discover novel endocrine-specific transcription factors. For instance, POU3F1 is a previously unreported factor specific to delta cells, as are SIX3 and OLIG1 to beta cells. We also identified new transcription factors that follow patterns of known, developmentally functional transcription factors. Two have profiles resembling PDX1: ETV1 and MEIS2 are restricted to non-alpha endocrine cells. POU6F2 and FEV resemble ARX: POU6F2 is detected in alpha, gamma, and epsilon cells, and although FEV is not detected in epsilon cells, this may be due to a relative under-sampling of the epsilon transcriptome. A similarly novel pattern involves transcription factors restricted to beta and delta cells, including SIX2, ESR1, and RXRG. Combinations of cell type-specific transcription factors have allowed reprogramming between pancreatic lineages (Li et al., 2014; Zhou et al., 2008), and the role of these transcriptional regulators in development and the maintenance of cell type identity merit further investigation.
Comparing across mice, we found an interesting pattern of conservation and divergence (Figure 3C). Pan-endocrine expression was conserved in mice (NEUROD1, INSM1, ISL1, NKX2-2, and PAX6). However, specific patterns of expression within the endocrine systems, such as ETV1 and MEIS2, which had a PDX1-like expression profile in the mouse, show “non-beta” expression in the mouse. This shift in expression specificities suggests that, although the overall functionality of the pancreas is conserved in evolutionary time, the specific expressions across cell types may actually be plastic.
Characterization of Pancreatic Stellate Activation and Schwann Cell Dedifferentiation
Pancreatic stellate cells have been implicated in ductal carcinoma angiogenesis and metastasis, pancreatic fibrosis, and unusual immune phenotypes associated with pancreatic cancers (Haber et al., 1999). Located primarily at the base of the acini and around vascular cells, quiescent stellate cells are a type of pericyte that plays a structural role in the pancreas and can be identified in microscopy by cytoplasmic lipid droplets that contain vitamin A (Figure 4A). By first examining the human data, we identified stellate cells in our dataset by their known marker gene PDGFRB along with specific gene expression of fibroblast growth factor (FGF), WNT, Activin A, transforming growth factor β (TGF-β), and numerous cytokines. We identified pancreatic stellate cells in each human donor, 457 in total (Figure 4B).
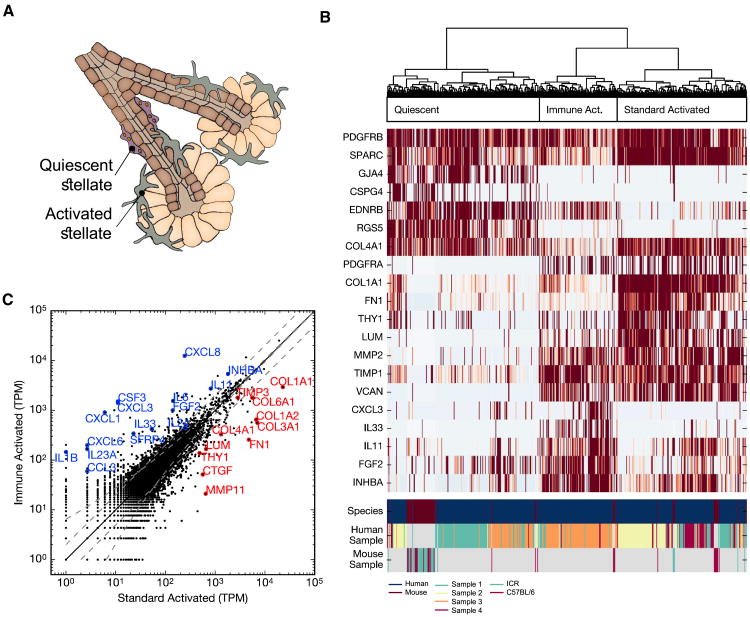
Figure 4. Multiple Modes of Pancreatic Stellate Cell Activation and Existence of Pancreatic Adult Neural Crest Stem Cells.
(A) Illustration of pancreatic stellate cells within the pancreas, indicating their typical periacinar localization and activation from a quiescent state.
(B) Hierarchical clustering of both human and mouse stellate cells on the basis of genes differentially expressed within human stellate cells and their mouse homologs. Genes highlighting the distinct clusters are displayed in the heatmap. Three groups of cells are indicated: quiescent, standard activated, and immune-activated. Most activated mouse stellate cells follow the pattern of standard activation. The three bottom rows indicate species and donor identities.
(C) Plot comparing the average expression (tpm) of the two distinct populations of human activated stellate cells reveals genes involved in immune signaling and secretion of the extracellular matrix, as indicated by annotated genes. Annotated genes are differentially expressed (fold change > 2 and above variation expected from Poisson sampling) and indicative of different biological functions.
The transcriptome of pancreatic stellate cells has not been characterized previously. When activated, these cells alter drastically, losing their lipid droplets, migrating, secreting ligands to several major signaling pathways such as WNT, TGFB, FGF, platelet-derived growth factor (PDGF), and Activin A, and producing large quantities of collagens, fibronectin, and other extracellular matrix components (Buchholz et al., 2005; Figure 4B). Consistently, we found a clear division between cells that expressed high levels of genes associated with stellate activation, such as collagen I and fibronectin, and those that did not. Although the proteins produced by activated stellate have received extensive attention (Buchholz et al., 2005), less focus has been placed on quiescent forms. In our data, among the genes enriched in these quiescent cells were several that may play functional roles, including ADIRF, associated with adipogenesis, and FABP4, associated with lipid transport in adipocytes.
We found a further division within activated cells (Figure 4B). One set of cells expressed genes related to the extracellular matrix more highly, whereas the other had highly specific expression of numerous cytokines, interleukins, and chemokines (Figure 4C). This latter phenotype is surprising and may explain the unusual interactions reported between stellates and immune cells. The cells of the immune-activated state are identified predominantly in two of our samples, whereas the standard activated state predominated in the other two. We also detect stellate cells in mouse islets. To determine which types of human stellates they resemble most closely, we identified genes that were enriched in the three types of human stellates and then used their expression to cluster all stellates. We found that most mouse stellates resembled the quiescent or standard activated stellates (Figure 4C). Although we cannot rule out that stellate cell activation is induced by post-isolation culture conditions rather than states present in the donor, our analysis nonetheless supports the existence of two modes of stellate cell activation.
The second intriguing population is of neural crest origin. We hypothesize that this population of 13 cells represents pancreatic Schwann cells responding to injury. These cells express known Schwann cell markers such as SOX10, S100B, CRYAB, NGFR, PLP1, and PMP22. However, components of the myelin sheath are lowly expressed or absent, and several genes shown previously to be upregulated in the Schwann cell response to nerve injury mark the population. These genes include SOX2, ID4, and FOXD3, which are transcription factors associated with Schwann cell dedifferentiation and repression of myelin sheath component expression, as well as GDNF, EDNRB, and NES (Jessen and Mirsky, 2008; Lang et al., 2015; Pereira et al., 2012). Based on this profile of expression of known Schwann cell and injury markers, we characterize this cell population as Schwann cells that dedifferentiated under extraction and culture conditions.
Transcriptome Analysis of Ductal Cells Provides Evidence for Functional Subpopulations
Cell types are traditionally classified by function and marker gene expression. Single-cell transcriptome data enable us to further detect substructure within cell populations or the lack thereof. We developed a pipeline for systematically testing whether a given cell population contains structured variation in gene expression. Briefly, our approach proceeds by principal component analysis (PCA) of single cell types and then seeks to identify genes that have PC loadings greater than expected by chance, are not known as markers for other cell types, and do not trivially correlate with the total number of transcripts per cell or with expression of mitochondrial genes (a marker of cell stress). Applying our method to most cell type populations did not reveal biological differences among the cells (Figure S5A). For example, in alpha cells, searching for genes that contribute to PC1 more than expected (> , where N is the number of genes) revealed only genes correlating to total transcript abundance, suggesting that there is no detectable structure in this cell type (Figures S5B and S5C). This does not rule out the presence of cell-to-cell variability within these cell types, but it suggests that such variability may be post-transcriptional in nature or else that it is too weak to detect without very deep and sensitive sequencing of individual cells.
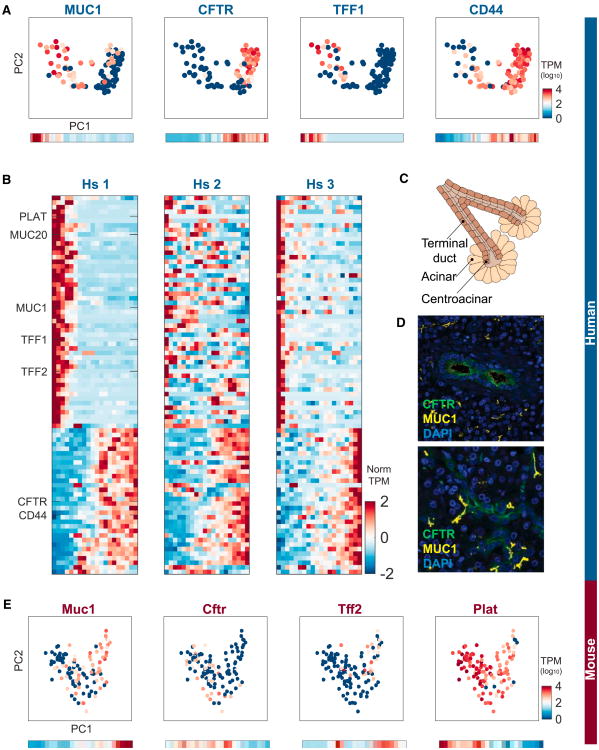
We did detect, however, substructure in the human ductal cell population. We found that the first PC (PC1) was significantly and strongly different from random, with cells continuously distributed along PC1, forming two lobes (Figure 5A). We ruled out an explanation of this spread according to the number of transcripts of each of these cells (Figure S5C). We found many genes whose expression is variable across PC1 (Figure S5D); for example MUC1 and CFTR function in mucosal restitution and repair (Hoffmann and Hauser, 1993) and secretion of HCO3, respectively (Ishiguro et al., 2009; Figure 5A). To characterize this variation further, we created a moving average profile for each gene across the range of PC1. A sharp change in expression is found in some genes, whereas others show a more gradual change across PC1. For example, gene CD44 shows a gradual mode of expression across PC1 (Figure 5A). Expanding this analysis to all genes with significant differences across PC1, we find a clear separation into two gene expression classes. The first is composed of genes enriched in low-PC1 cells. These genes are related to mucous secretion such as TFF1, TFF2, and MUC1 and protection of the ductal tissue from the digestive enzymes transported from acini. The second gene class shows highest expression in high-PC1 cells and then gradually diminishes across PC1. This class of genes is related to ion transport, including CFTR, consistent with the secretory function of ductal cells. Similar subpopulations were detected in human donors 2 and 3, providing further support for their existence (Figure 5B).
Figure 5. Subpopulations of Ductal Cells in the Human Pancreas.
(A) Differential expression of MUC1, CFTR, TFF1, and CD44 in PC space defined by the ductal cells. Bottom: moving averages for each gene computed based upon equidistant ranges across PC1.
(B) Heatmaps showing genes that are differentially expressed across PC1 for each of the three donors. Heatmaps including all genes names can be found in Figure S6.
(C) Schematic of the location of terminal and centroacinar ductal cells.
(D) Immunohistochemistry stains of human pancreata for CFTR and MUC1. Note their spatial isolation among the ductal cells.
(E) Differential expression of Muc1, Cftr, Tff2, and Plat in PC space defined by the mouse ductal cells. Bottom: moving averages for each gene computed based upon equidistant ranges across PC1.
Ductal cells have been known to exhibit two main morphologies: one forming the terminal duct and the other connecting to the acinus (Figure 5C;) (Rovira et al., 2010). However, the molecular properties distinguishing these cells have not been described. We hypothesized that the observed gene expression gradient across ductal cells is location-dependent. To test this, we applied immunohistochemistry stains to pancreatic tissues to study the expression of MUC1 and CFTR. We found a clear spatial separation between cells with higher levels of MUC1 and cells with higher levels of CFTR (Figure 5D).
If the ductal cell populations indeed relate to functional sub-cell types, it is reasonable to expect that their specialization would be conserved across species. We thus repeated the analysis in mice, and the PC analysis again revealed lobes (Figure 5E). Expression of Muc1 and Cftr showed inverse patterns, indicating that, among the mouse ductal cells, sub-cell populations also exist. Together, our results suggest that we can clearly distinguish two hitherto undescribed classes of ductal cells in molecular detail.
Heterogeneity of Beta Cells
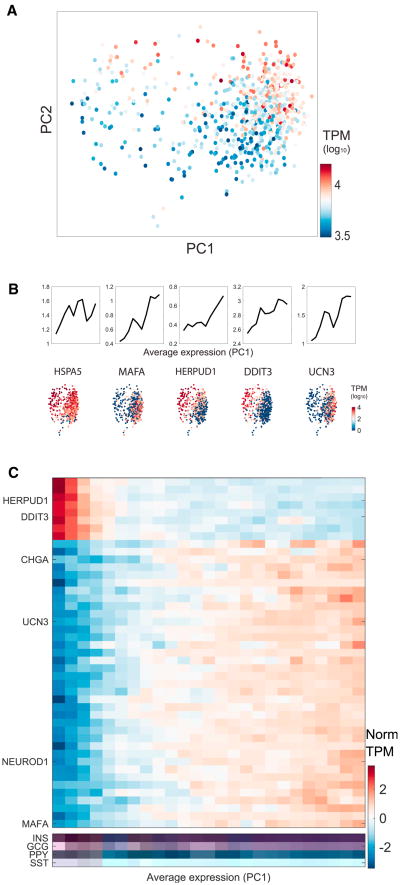
Applying our subpopulation method to beta cells, we also found evidence for significant heterogeneity among these cells. Initially, we observed two distinct subpopulations of beta cells in the donor 1 sample; however, we found that it stemmed completely from the number of detected transcripts per cell. We attributed the cluster of cells with low transcript counts to dead cells and excluded them from further analysis. Performing PCA on the remaining beta cells, we observed a distribution of cells that was not correlated with transcript counts (Figure 6A). Instead, the variation stemmed from variable expression of genes relevant to beta cell function, including UCN3 and the endoplasmic reticulum stress-inducible genes HERPUD1, HSPA5, and DDIT3 (Sharma et al., 2015; Figure 6B).
Figure 6. Heterogeneity of Beta Cells Reveals the Unfolded Protein Response.
(A) PCA on beta cells colored by the transcript numbers. Beta cells were filtered for having at least 3,000 detected transcripts.
(B) Moving average of HSPA5, MAFA, HERPUD1, DDIT3, and UCN3 sorted by PC1 score. Bottom: PCA plots colored by the expression of the same genes.
(C) Heatmap showing expression of genes that contribute more to PC1 than expected. The profiles were computed as the moving average of 30 bins across PC1. Heatmaps including all genes names can be found in Figure S6.
Overall, we found that genes with low PC1 values were correlated with genes that function in endoplasmic reticulum (ER) stress and that high PC1 values were correlated with functional beta genes (Figure 6C). Recent work has linked the unfolded protein response in beta cells to proliferation (Sharma et al., 2015). Also known as ER stress, the unfolded protein response in beta cells is triggered by high levels of insulin synthesis. Beta cells are known to be engaged in ER stress because of the high demand for insulin secretion (Marchetti et al., 2007). Recent studies have also shown that, within human and mouse beta cell populations, cells exhibiting active ER stress are more likely to proliferate (Sharma et al., 2015).
Further work is required to characterize the beta cell heterogeneity detected here. We cannot rule out that a heterogeneous stress signature in beta cells reflects a variable response to pancreatic isolation, transport, and dissociation for inDrop analysis; however, it is notable that none of the other cell types nor samples showed a comparable heterogeneous ER stress response. The heterogeneity appears distinct from that of the ductal cells because we find a diverse gradient of molecular phenotypes as opposed to mainly two sub-types. Moreover, there was an absence of genes with clear “on/off” gene expression levels across the beta cell gradient that might be detectable by in situ analysis.
Deconvolving Bulk Gene Expression Samples Using Single-Cell RNA-Seq
Gene expression studies comparing complex tissues in bulk offer the possibility of studying gene-regulatory differences across a large number of normal and pathological samples. Often, however, these mask the biological signal because of the variability in cell type proportions. Although single-cell data remain prohibitively expensive to apply across hundreds of samples, the possibility exists to leverage them toward overcoming difficulties in studying bulk gene expression data. Specifically, statistical deconvolution methods have been used successfully for estimating cell type proportions and cell type-specific differential expression in silico from bulk tissue data (Shen-Orr and Gaujoux, 2013). The availability of single-cell gene expression datasets now offers improved means for these methods because, aggregated at the cell-type level, single-cell data provide characteristic profiles from a large number of minimally perturbed primary cells.
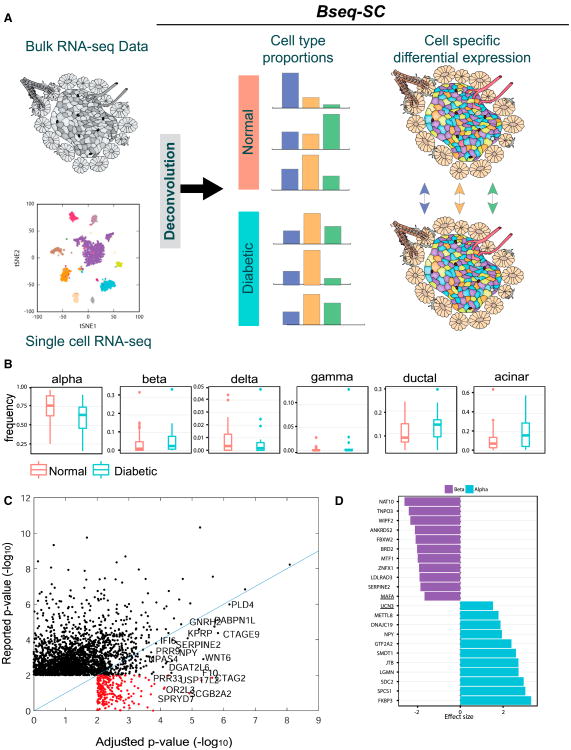
Building upon previous gene expression deconvolution methods, we developed a bulk sequence single-cell deconvolution analysis pipeline (Bseq-SC). Overall, the method integrates bulk data with single-cell gene expression to identify cell-type specific marker genes, estimate the proportion of each cell type in measured bulk tissue RNA-seq samples, adjust individual gene expression samples for variation in cell type proportion so that differential expression because of proportion differences is removed, and perform cell type-specific differential gene expression analysis among groups (Figure 7A). An important benefit of this approach is that, although single-cell analysis is expensive, it need only be applied once on a given tissue to deconvolve all bulk samples.
Figure 7. BSeq-SC Uses Single-Cell RNA-Seq to Deconvolve Bulk Heterogeneous Tissue Data and Decouples Disease-Associated Differential Expression from Cell Type Proportion-Associated Differences.
(A) Schematic of the BSeq-SC analysis. Single-cell RNA-seq information is used to deconvolve bulk pancreatic islet RNA-seq samples to estimate the cell type proportion of key cell types. Statistical deconvolution is used to leverage the variation in cell type frequencies between individuals to estimate average cell type-specific expression in diabetic versus healthy individuals and to compute cell type-specific differential expression.
(B) Proportion differences of pancreatic cell types between normal and diabetic samples.
(C) The majority of genes identified as differentially expressed between normal and diabetic in bulk samples are not significantly different following statistical adjustment to cell type proportions derived from the deconvolution. Genes that were not significant in the un-adjusted bulk analysis but were found to be significant after adjustment are shown in red.
(D) Cell type-specific effect size in alpha (blue) and beta cells (purple) for the top ten significant genes between hyper- and normoglycemic groups. The ER stress genes UCN3 and MAFA were upregulated and downregulated in alpha and beta cells, respectively (see the complete list in Figure S9D).
To test Bseq-SC's ability to detect a biological signal, we analyzed a previously published bulk RNA-seq dataset (Fadista et al., 2014) and asked whether applying our understanding of the structure of pancreatic cell type expression reveals insight into the regulatory differences between normal and diabetic pancreata. This dataset of bulk islet RNA-seq contained data for 82 samples, including 51 normoglycemic, 15 impaired glucose tolerance (IGT), and 12 T2D patients and 11 donors without HbA1c information (Fadista et al., 2014). Because of a sample size requirement, we merged the two hyperglycemic sub-groups (IGT and T2D) into one group and compared it with the normoglycemic group. We first identified a set of discriminative marker genes, derived from the single-cell data, which can best be used to estimate the proportion of each cell type in the bulk samples. We tested the capability of these to identify cell proportions using a simulation (STAR Methods). Next, we estimated the proportion of each of the six major cell types (alpha, beta, gamma, delta, acinar, and ductal cells) in all bulk samples (Figure 7B). No significant proportion differences were detected between normoglycemic and hyperglycemic samples (p < 0.05). However, this analysis revealed broad distributions of proportions among individuals for each cell type as well as markedly different proportion estimates of alpha, beta, acinar, and ductal cells among normal and hyperglycemic individuals, suggesting that much of the reported differences in bulk gene expression data may be affected by proportion variability.
To address this, we repeated the statistical analysis of the bulk tissue samples comparing the gene expression profiles of normal and hyperglycemic individuals, first according to the original publication (Fadista et al., 2014) and then including estimated cell type proportions as additional covariates into the model, hence accounting for their confounding variability. To maintain enough statistical power, we did not include all cell types in the model but, rather, estimated the expression profile of the four most variable cell types: alpha, beta, acinar, and ductal (STAR Methods). Comparing the differential expression observed in the bulk tissue with that found in the adjusted proportions (Figure 7C), we found that, of the 2,369 genes originally identified in the bulk data to be associated with high versus low HB1AC levels (p ≤ 0.01), 1,867 (73%) did not exhibit statistical significance following cell type proportion adjustment. The majority of the remaining genes (75%) showed a marked reduction in statistical significance. Conversely, we identified a set of 360 genes for which the statistical differential expression between groups increased following cell type proportion adjustment. These were made up of 127 genes that were already significant prior to adjustment and 233 newly significant genes. We propose that a select, smaller set of genes is differentially expressed in hyperglycemic relative to healthy pancreatic islets when the variability is controlled for cell type proportions.
Finally, we sought to identify the specific cell types that show differential expression using a regression-based method for statistical deconvolution (Shen-Orr et al., 2010). This method leverages the inherently large variability in cell type proportions among samples to infer average cell-type specific expression profiles to assess cell type-specific differential expression between groups. Again we limited the statistical deconvolution model to alpha, beta, acinar, and ductal cells. Our analysis identified a signal of cell type-specific differences in alpha cells (45 genes upregulated at a false discovery rate [FDR] of 0.05) and beta cells (24 genes downregulated at an FDR of 0.1) (Figure 7D). Notably, we detected cell type-specific differential expression across the genes characterizing ER stress in beta cells (Figure 6) and found that all six of them showed cell type-specific differences in alpha or beta cells (significant at a looser FDR of 0.15). UCN3, NEUROD1, and HERPUD1 were upregulated, and DDIT3 was downregulated in alpha cells, whereas MAFA, HERPUD1, and HSPA5 were downregulated in beta cells. (Figure 7D, underlined). This suggests that the potential beta cell heterogeneity identified in the single-cell data may be smaller in a diabetic relative to a normal state or, alternatively, represents a regulated cell state important for insulin glycemic control, which may be impaired under a pathological condition.
Discussion
We have shown that we can determine the transcriptomes of thousands of pancreatic cells, map them to known cell type identities, and point to their functional roles. We found that cells form distinct clusters when studied with tSNE, the common dimensionality reduction algorithm, as well as with an iterative hierarchical clustering method (Figure 1). The similarity among the summary transcriptomes of these clusters clearly distinguishes sets of cells. These sets are conserved across individuals (Figure 2), and each cell type has dominant genes specifically expressed in that cell type. This systematic and unbiased characterization method of analyzing pancreatic endocrine cells is consistent with previous analyses, suggesting a satisfying correspondence between classical studies and systematic transcriptome analysis. However, this approach extends the analysis beyond a few highly expressed marker genes by providing an atlas detailing gene expression across thousands of cells from four individuals. This approach also sheds light on pancreatic cell types that were previously understudied, such as pancreatic stellate cells (Figure 4).
By characterizing both human and mouse pancreatic cell types, we describe the ways in which corresponding cell types are similar or different. At the largest scale, the cell types themselves are conserved between mouse and human (Figure 1), and the transcriptomes of these cell types are very similar (Figure 2). Because this pattern of similarity matches our expectation, discrepancies between the two species are rare and, thus, potentially interesting. We identify several key differences in specific genes relevant to pancreatic biology as well as in the patterns of expression exhibited by transcription factors (Figure 3).
Beyond identifying the major cell types, we were also able to identify subpopulations within two of the observed cell types. In ductal cells, we detected a distinct subpopulation that may correspond to cells in contact with acinar cells and another that likely corresponds to terminal duct cells (Figure 5). More relevant to diabetes research, we detected classes of beta cells. Beta cells from one donor also showed a distinct population structure, with the primary axis characterized by a gradient of ER stress (Figure 6). Confirming the presence of this heterogeneity in vivo and understanding its significance will require further study.
Finally, we developed a method to deconvolve bulk RNA-seq data based on our single-cell data and were able to compare gene expression profiles among healthy and diabetic donors. This analysis revealed that a large number of genes identified in the bulk data as differentially expressed among diabetic and healthy donors were likely variable solely because of cell type proportion differences (false positives), whereas the differential expression of other genes between these two group was masked (false negatives) by the same phenomena. After cell proportion adjustments, we were able to detect differentially expressed genes in alpha and beta cells, including two genes also identified by our single-cell analysis as markers of a beta cell subpopulation marked by a potential unfolded protein stress response (Figures 6 and 7).
One limitation of our analysis is the lack of spatial resolution. However, the specific genes identified in our analyses of population substructure are clear candidates for follow-up experiments to couple the observed gene expression differences to spatial information. For example, it will be important to determine whether stressed beta cells are spatially concentrated in islets or uniformly dispersed. Furthermore, if diabetes ensues, then the pattern may change. Such a three-dimensional map will be important for an understanding of pancreatic function and potential therapeutics to the diseased state.
Star★Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Itai Yanai (itai.yanai@nyumc.org).
Experimental Model and Subject Details
Mouse and Human Sample Preparation
Institutional review board approval for research use of human tissue was obtained from the Harvard University Faculty of Arts and Sciences. Human islets were obtained from NDRI (The National Disease Research Interchange) and were recovered in CMRLS for 24 or 48h (Our human donors span a range of ages and BMI health parameters (Table S1)). Donor anonymity was preserved, and the human tissue was collected under applicable regulations and guidelines regarding consent, protection of human subjects and donor confidentiality. Animal studies were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the animals were handled according to approved institutional animal care and use committee (IACUC) protocol number [93-15]. The protocol was approved by the Committee on the Use of Animals in Research and Teaching of Harvard University Faculty of Arts & Sciences (HU/FAS). The HU/FAS animal care and use program is AAALAC International accredited, has a PHS Assurance (A3593-01) on file with NIH's Office of Laboratory Animal Welfare, and is registered with the USDA (14-R-0128). Animals were euthanized in accordance with AVMA Guidelines for the Euthanasia of Animals. Both ICR and C57BL/6 mice strain used were obtained from Jackson Laboratories. Mice were maintained on regular chow (Prolab Isopro RMH 3000, LabDiet) and on a 12 hr light/dark cycle with continuous access to food and water. Mouse islets were isolated (and pooled) from five C57BL/6 and ICR mice by perfusion of the common bile duct with 0.8 mM Collagenase P (Roche), digestion of the pancreata with 0.8 mM Collagenase P (Roche) and purification of the islets by Histopaque gradient (Sigma) centrifugation. Single cell dispersion was carried out as follows: islets were settled by centrifugation (250 rpm, 3 min), washed with 2 ml of PBS. Islets were centrifuged (250 rpm, 3 min) again and PBS was replaced with 1 ml of TrypLE Express. Samples were incubated at 37°C for 18-20 min. Following incubation, TrypLe was removed and 2ml of CMRLS media was added. Cells were gently dispersed mechanically using a P1000 pipette. Cells were then spun down (500 rpm, 3 min) and washed with 1 ml PBS prior to a final resuspension in PBS. Cells were then stored on ice for < 30 min awaiting encapsulation into droplets and immediate lysis. Immediately before running cells through the inDrop device, Optiprep was added to the cell suspension to a final concentration of 16%v/v, and the cell concentration was adjusted to 75,000 cells/mL. The cells were encapsulated at a rate of 12,000/hour, and approximately 80% of encapsulated cells were successfully barcoded.
Method Details
Single Cell Sequencing Bioinformatics Pipeline
inDrop encapsulation of the cells and reverse transcription (RT) reaction was carried out as previously reported (Klein et al., 2015). Library preparation was carried out according to this protocol with the following minor changes: random hexamers were used during the second reverse transcription step (after linear amplification), eliminating the need for primer ligation. The number of PCR cycles required for final library enrichment ranged from 9-12 cycles. Paired end sequencing was performed on Illumina Hiseq 2500 machine. Reads lacking any of the expected sequences in read 1 (known cell barcode, adaptor sequence (W1), or beginning of the poly-T tail) were removed. Reads were trimmed using Trimmomatic (Bolger et al., 2014) (version 0.32) and split into individual cells based on their match against both cell barcode sequences (BC1 and BC2, each with 384 unique possible sequences), with errors of up to two nucleotides mismatch corrected. Read 2 was then mapped to the reference transcriptome using bowtie (Langmead and Salzberg, 2012) (version 1.1.1) and expression was quantified as previously described (Klein et al., 2015) with the added modification of ignoring reads containing a high fraction of single-base repeats after quality trimming.
Clustering
Clustering was carried out recursively: for every subset of cells, only highly expressed genes (fraction of total TPM above 0.5) and highly variant (Fano factor above mean-dependent threshold) were used. Then, hierarchical clustering was performed with Ward's criterion over correlations computed from the expression of the selected genes (log, TPM). This is carried out recursively, until a subset of cells is 3 (or less), has only 3 high-variance genes (or less) or has less than 2 genes enriched for that subset (75% of the TPMs). Finally, we examine the clustering and search for cases which were over-split (sample-specific effects, outliers of a few cells in a cluster) and assign them to merged final clusters.
Clustering Samples across Donors
For each donor, we identified the most variable 2000 genes with at least three counts in at least three cells. Variability was measured using the n-score noise statistic (Klein et al., 2015), which is closely related to the Fano factor and measures above-Poisson noise. From these genes, we selected those with the highest loading coefficients for each non-random principal component. For each pair of cell types, we computed the cosine similarity among every cell in the two groups to all other cells. These were summarized by an average score for each cell. The similarity between the cell types is then estimated as the average of these scores. Hierarchical clustering was used to order the similarity matrix.
Immunohistochemistry of FFAR4/GPR120
Human islets were obtained through the Integrated Islet Distribution Program and the National Disease Research Interchange (NDRI) and cultured (Walpita et al., 2012). For dissociation, islets were pelleted, washed in PBS, and centrifuged at 1000 rpm for 5 min at room temperature. Pelleted islets were incubated at 5000 IEQ/mL in Accutase (Life Technologies) at 37°C for 20 min. The pellet was resuspended in CMRL complete media, and an aliquot was removed for cell counting using Countess Automated Cell Counter (Thermo Fisher Scientific). ∼30,000 cells were seeded per well in 96-well plates (donor information can be provided if necessary). Cultures were then fixed for 20 min at room temperature using 3% paraformaldehyde in PBS and washed twice with PBS. Cells were permeabilized for 20 min at room temperature using 0.2% Triton X-100 in PBS and blocked for 2 hr at room temperature with 2% bovine serum albumin (BSA) in PBS. The C-peptide antibody GN-ID4 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development (NICHD) and maintained by the University of Iowa, Department of Biology (Iowa City, IA), the FFAR4/GPR120 antibody was from Novus Biologicals, AlexaFluor488 goat anti-rat and AlexaFluor594 goat anti-rabbit were from Life Technologies. Primary antibodies were diluted in 1% BSA in PBS and incubated overnight at 4°C, followed by three washes with 1% BSA in PBS. Cultures were then incubated with secondary antibodies diluted in 1% BSA in PBS for 1 hr at room temperature, followed by five washes with PBS. Plates were stored foil-sealed at 4°C with 100 μL/well PBS. Plates were imaged with an ImageXpress Micro automated microscope (Molecular Devices).
Immunohistochemistry of MUC1 and CFTR
Formalin fixed paraffin embedded tissue sample from resection specimens of pancreas were de-paraffinization using W-cap reagent (Bio-Optica), with epitope retrieval Citrate buffer for 20 min (95°C) and then transferred to warm DDW (65°C) container for additional 20 min. Sections were then transferred to DDW for 15 min in RT followed by 3 washes in DDW and moved to PBS. Sections were permeabilized for 2 hr at RT using 5% Goat serum, 0.2% triton in PBS. Conjugated MUC1 antibody (Alexa Fluor 647) and CFTR anti mouse primary and secondary antibody (Alexa Fluor 594) were obtained from Abcam. Conjugated and primary antibodies were diluted in blocking solution at required concentration and incubated overnight at 4°C in a moist light chamber. After 3 washes in PBS, secondary antibody was incubated for 1 hr in RT. Slides were washed 8 times in PBS and covered with DAPI containing mounting solution. Slides were imaged using laser scanning confocal microscope.
Deconvolution of Bulk Gene Expression Data
The Bseq-SC methodology was implemented in R using Bioconductor packages (Gentleman et al., 2004). Code will be made available as an R package at http://github.com/shenorrlab/bseq-sc.
Bulk-RNA-Seq Pre-processing
To evaluate cell-type specific differences between diabetic and healthy individuals we retrieved the Fadista et al. RNA-Seq dataset (GEO accession GSE50244) (Fadista et al., 2014). This dataset contains gene expression profiles of 82 donors, 21 of which were identified as diabetic and/or having a high level of hba1c and 47 of which were healthy controls. We downloaded the raw reads data from GEO dataset GSE50244 and TPM matrices were generated using Kallisto (Hensman et al., 2015). Reads were then aligned using ENSEMBL transcripts. This resulted in a gene expression matrix (tpm) of 34824 genes and 82 samples. We removed 2 samples (GSM1216808, GSM1216834) that appeared as outliers with respect to BMI/HBA1C values and PCA respectively.
Marker Selection and Proportion Estimation for Bseq-SC
To estimate the proportions of alpha, beta, gamma, delta, acinar and duct cells we formulated a ‘basis’ matrix composed of the characteristic expression profiles for each of the estimated cell types. The matrix was generated based upon the single-cell data using only healthy donors (Table S1). Different cell types have different transcriptomic activity, the heterogeneity of which is also present in bulk tissue gene expression data, yet is hidden due to the count summarization across all cells. Information on cell-type transcriptomic expression variability is captured by single-cell measurements as shown by the variability of the average number of counts (prior to transcripts per million (TPM) normalization) in cells across cell types (Figure S9A). To reflect this heterogeneity in the basis matrix and avoid estimation biases, we multiplied the TPM profiles of the cells within a cell type by their mean total counts. These re-scaled cell expression profiles were then averaged first within each islet and then across islets. This resulted in a matrix of genes × cell types. The deconvolution basis matrix was defined as the sub-matrix formed by a set of cell type-specific marker genes (Figure S9B). Genes were selected based on three conditions: 1) high expression (fraction of total TPM above 0.5), 2) high variation (Fano factor above mean-dependent threshold), and 3) cell-type restricted (p < 10−5, defined by a Kolmogorov-Smirnov test). This resulting basis matrix was then applied for estimating cell proportion of each sample using CIBERSORT (Newman et al., 2015). Differences in cell type proportions between groups was assessed using Wilcoxon test applied to the arcsine-transformed estimated proportion matrix (p value cutoff 0.05).
Bseq-SC Cell Type-Specific Differential Expression and Statistical Adjustment of Cell Type Proportions
We integrated bulk gene expression data with the estimated proportion using csSAM (Shen-Orr et al., 2010), which we applied to compare islets from normoglycemic patients (HBA1C < 6) to hyperglycemic patients (sub-groups 6 ≤ HBA1C < 6.5 and HBA1C ≥ 6.5). We performed two separate cell-type dependent gene expression analyses. First, we repeated the original bulk data analysis (Fadista et al., 2014) using edgeR (Robinson et al., 2010) which includes gender and age as unique covariates, and tested differences in either hyperglycemic sub-group (F-test). This was performed on all genes, after removal of pseudogenes, mitochondrial and ribosomal genes. Then we expanded this model and statistically adjusted the bulk expression profile of each sample for variation in cell-type proportions, so as to allow for differential expression analysis of bulk samples independent of the biases introduced by variation in between sample cell-type proportions. To limit the number of covariates, we only included the estimated proportions of alpha, beta, acinar and ductal cells, which were the three cell-types whose proportion distribution we observed to be most variable between normoglycemic and hyperglycemic subjects. Second, we used the csSAM methodology to estimate the average cell-type expression profile of each sample and perform differential cell-type specific expression profiling between groups (hyperglycemic versus normoglycemic). In order to increase statistical power, we applied these this analysis on a set of 366 genes defined based on the first analysis, as those whose p value after proportion adjustment was ≤ 0.01 and lower than before adjustment. The six ER-stress genes were added to this list. Markers used to estimate cell type proportions were excluded and we used an FDR cutoff of 0.05 and 0.1 for alpha and beta cells respectively, selecting genes that were exclusively identified in either cell type.
Quantification and Statistical Analysis
Statistical parameters including the exact value of R2 and p values are reported in the Figures and the Figure Legends. For the de-convolution analysis, genes were judged to be statistically significant when p < 0.01 and cell-type specific differentially expressed having FDR < 0.05.
Data and Software Availability
Software
The Bseq-SC software is available as an R package at http://github.com/shenorrlab/bseq-sc.
Data Resources
The accession number for the data reported in this paper is NCBI GEO: GSE84133.
Supplementary Material
Figure S1. A transcriptomic map of the human and mouse pancreas, Related to figure 1
(A) Same as Figure 1C. tSNE plots for the dataset derived from each of the four donors and two sacrificed mice.
(B) Histogram of number of transcripts detected per cell.
(C) Histogram of number of genes detected per cell.
(D) Fraction cell type per sample for human (left) and mouse (right). For each donor the bar stack indicates the fraction cells annotated to each of the cell types.
Figure S2. INS/ins1 defines the beta cells cluster in mouse and human, Related to figure 1. tSNE scatter plots of donor 1 (left) and mouse 1 (right) colored by the expression of INS and ins1, respectively.
Figure S3. Beta cells expression are highly correlated between mice, Related to figure 2. Plot comparing the average expression (log10, tpm) of beta cells of mouse 1 and 2.
Figure S4. A transcriptomic markers of Schwann cells in the human pancreas, Related to figure 3. Violin plots for expression of genes enriched in Schwann cells relative to their expression in other cell types. Gene specific to this population are associated with a neural progenitor role.
Figure S5. Testing for structured variation in cell population using PCA, Related to figure 5.
(A) PCA for each cell type.
(B) PCA for alpha cells with total transcript count indicated by color, and correlation betweenPC2 and total transcript count.
(C) PCA for ductal cells with total transcript count indicated by color.
(D) Histogram of the PCA loadings of the ductal cells with the 85 genes indicated.
Figure S6. Subpopulation of the ductal cells in the human pancreas, Related to figure 5. Heatmaps showing genes that are differentially expressed across PC1 for each of three donors including all the genes names.
Figure S7. Subpopulation of the ductal cells in the mouse pancreas, Related to figure 5. Heatmaps showing genes that are differentially expressed across PC1.
Figure S8. Heterogeneity of beta cells reveals unfolded protein response, Related to figure 6. Heatmaps showing genes that are differentially expressed across PC1 including all the genes names.
Figure S9. Properties of BSeq-SC deconvolution, Related to figure 7.
(A) Pancreatic cell types are not equal with respect to their transcriptomic activity. The average number of transcript in a given cell (y-axis) varies significantly depending across cell types (x-axis).
(B) Deconvolution basis matrix composed of the expression profiles of marker genes. The heatmap shows the deconvolution matrix used to estimate the proportions of alpha, beta, gamma, delta, acinar and ductal cells in bulk samples. The rows were z-score-transformed to highlight the cell type-specificity of each marker.
(C) FDR plot from BSeq-SC cell type-specific analysis, revealing up-regulation in alpha cells and down-regulation beta-cells of hyperglycemic patients. The x-axis shows the number of genes called up-regulated (left) and down-regulated (right) at a given FDR cutoff (y-axis).
(D) Complete list of genes found to be exclusively dys regulated in either alpha (blue) or beta cells (purple). Barplot shows estimated cell type-specific effect size (x-axis) for each gene (y-axis).
Table S1. Ages, BMI, and sex of human donors, Related to figure 1
Table S2. Marker genes used in analysis are indicated for each cell type, Related to figure 1
Table S3. Transcription factor described in Figure 3C whose expression profiles are described cited in the literature, Related to figure 3.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| C-peptide antibody GN-ID4 | University of Iowa, Department of Biology | Cat#AB_2255626; RRID: AB_2255626 |
| FFAR4/GPR120 antibody | Novus Biologicals | Cat#NBP1-00858; RRID: AB_1503311 |
| AlexaFluor488 goat anti-rat | Life Technologies | Cat#ab150113; RRID: AB_2576208 |
| AlexaFluor594 goat anti-rabbit | Life Technologies | Cat#ab150080; |
| Conjugated MUC1 antibody | Life Technologies | Cat#ab196189 |
| CFTR anti mouse primary | Life Technologies | Cat#ab2784; RRID: AB_303297 |
| Alexa Fluor 568 Anti-mouse | Life Technologies | Cat#ab175701 |
| Critical Commercial Assays | ||
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat#RS-122-2101 |
| Deposited Data | ||
| Raw data files for RNA sequencing | NCBI Gene Expression | GSE84133 |
| Experimental Models: Organisms/Strains | ||
| Human samples | Prodo/NDRI | N/A |
| Mouse C57BL/6 abd ICR | Melton Lab | N/A |
| Software and Algorithms | ||
| Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Bowtie | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/index.shtml |
| inDrop pipeline | Klein et al., 2015 | N/A |
| Kallisto | Hensman et al., 2015 | https://pachterlab.github.io/kallisto/ |
| Bseq-SC | This study | http://github.com/shenorrlab/bseq-sc |
| CIBERSORT | Newman et al., 2015 | https://cibersort.stanford.edu/ |
| csSAM | Shen-Orr et al., 2010 | https://cran.r-project.org/web/packages/csSAM/index.html |
| edgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
Highlights.
We report over 12,000 individual pancreatic cell transcriptomes in human and mouse
We detect novel expression of TFs, signaling receptors, and medically relevant genes
We identify subpopulations and heterogeneity within pancreatic cell types
We deconvolve bulk gene expression samples using the single-cell data
Acknowledgments
M.B. was supported by an EMBO short-term fellowship (ASTF 474-2015). We would like to thank Yaron Fuchs's lab at the Technion for assistance with the immunohistochemistry experiments and Virginia Savova for preparing a distributable version of the sequence analysis pipeline. A.M.K. is a founder of OneCell Bio.
Footnotes
Supplemental Information: Supplemental Information includes nine figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2016.08.011.
Author Contributions: M.B. and I.Y. conceived the study. A. Veres prepared the human samples. J.H.R. prepared the mouse samples. M.B, A. Veres, and S.L.W. performed the inDrop method. M.B., A. Veres, S.L.W., A.F., and I.Y. analyzed the data. M.B., A. Vetere, and B.W. performed the immunohistochemistry experiments. R.G., M.B., and S.S.S.-O. developed the Bseq-SC method. M.B., A. Veres, A.F., R.G., S.S.S.-O., and I.Y. drafted the manuscript, on which all authors commented. A.M.K., D.A.M., and I.Y. coordinated the project.
References
- Benner C, van der Meulen T, Cacéres E, Tigyi K, Donaldson CJ, Huising MO. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014;15:620. doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett DM, Nowosielska A, Afik S, Pechhold S, Cura AJ, Kennedy NJ, Kim S, Kucukural A, Davis RJ, Kent SC, et al. Novel Observations From Next-Generation RNA Sequencing of Highly Purified Human Adult and Fetal Islet Cell Subsets. Diabetes. 2015;64:3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Hrvatin SSŠ, Schuetz C, Bonal C, Rezania A, Melton DA. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol. 2012;30:261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonal C, Herrera PL. Genes controlling pancreas ontogeny. Int J Dev Biol. 2008;52:823–835. doi: 10.1387/ijdb.072444cb. [DOI] [PubMed] [Google Scholar]
- Brissova M, Fowler MJ, Nicholson WE, Chu A, Hirshberg B, Harlan DM, Powers AC. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J Histochem Cytochem. 2005;53:1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- Buchholz M, Kestler HA, Holzmann K, Ellenrieder V, Schneiderhan W, Siech M, Adler G, Bachem MG, Gress TM. Transcriptome analysis of human hepatic and pancreatic stellate cells: organ-specific variations of a common transcriptional phenotype. J Mol Med. 2005;83:795–805. doi: 10.1007/s00109-005-0680-2. [DOI] [PubMed] [Google Scholar]
- Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren PO, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci USA. 2006;103:2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA maging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C, Schug J, Lin CF, Canaday PS, Fox AJ, Smirnova O, Bonnah R, Streeter PR, Stoeckert CJ, Jr, Kaestner KH, Grompe M. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011;54:2832–2844. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drel VR, Mashtalir N, Ilnytska O, Shin J, Li F, Lyzogubov VV, Obrosova IG. The leptin-deficient (ob/ob) mouse: a new animal model of peripheral neuropathy of type 2 diabetes and obesity. Diabetes. 2006;55:3335–3343. doi: 10.2337/db06-0885. [DOI] [PubMed] [Google Scholar]
- Drucker DJ. The role of gut hormones in glucose homeostasis. J Clin Invest. 2007;117:24–32. doi: 10.1172/JCI30076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, Storm P, Osmark P, Ladenvall C, Prasad RB, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube D, Bohn R. The microanatomy of human islets of Langerhans, with special reference to somatostatin (D-) cells. Arch Histol Jpn. 1983;46:327–353. doi: 10.1679/aohc.46.327. [DOI] [PubMed] [Google Scholar]
- Haber PS, Keogh GW, Apte MV, Moran CS, Stewart NL, Crawford DH, Pirola RC, McCaughan GW, Ramm GA, Wilson JS. Activation of pancreatic stellate cells in human and experimental pancreatic fibrosis. Am J Pathol. 1999;155:1087–1095. doi: 10.1016/S0002-9440(10)65211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensman J, Papastamoulis P, Glaus P, Honkela A, Rattray M. Fast and accurate approximate inference of transcript expression from RNA-seq data. Bioinformatics. 2015;31:3881–3889. doi: 10.1093/bioinformatics/btv483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann W, Hauser F. The P-domain or trefoil motif: a role in renewal and pathology of mucous epithelia? Trends Biochem Sci. 1993;18:239–243. doi: 10.1016/0968-0004(93)90170-r. [DOI] [PubMed] [Google Scholar]
- Holliday ND, Watson SJ, Brown AJH. Drug discovery opportunities and challenges at g protein coupled receptors for long chain free Fatty acids. Front Endocrinol (Lausanne) 2012;2:112. doi: 10.3389/fendo.2011.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro H, Steward MC, Naruse S, Ko SBH, Goto H, Case RM, Kondo T, Yamamoto A. CFTR functions as a bicarbonate channel in pancreatic duct cells. J Gen Physiol. 2009;133:315–326. doi: 10.1085/jgp.200810122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia. 2008;56:1552–1565. doi: 10.1002/glia.20761. [DOI] [PubMed] [Google Scholar]
- Johnson KH, O'Brien TD, Hayden DW, Jordan K, Ghobrial HK, Mahoney WC, Westermark P. Immunolocalization of islet amyloid polypeptide (IAPP) in pancreatic beta cells by means of peroxidase-antiperoxidase (PAP) and protein A-gold techniques. Am J Pathol. 1988;130:1–8. [PMC free article] [PubMed] [Google Scholar]
- Kimmel RA, Meyer D. Molecular regulation of pancreas development in zebrafish. Methods Cell Biol. 2010;100:261–280. doi: 10.1016/B978-0-12-384892-5.00010-4. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang H, Xing Y, Brown LN, Samuvel DJ, Panganiban CH, Havens LT, Balasubramanian S, Wegner M, Krug EL, Barth JL. Neural stem/progenitor cell properties of glial cells in the adult mouse auditory nerve. Sci Rep. 2015;5:13383. doi: 10.1038/srep13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SSF, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levetan CS, Pierce SM. Distinctions between the islets of mice and men: implications for new therapies for type 1 and 2 diabetes. Endocr Pract. 2013;19:301–312. doi: 10.4158/EP12138.RA. [DOI] [PubMed] [Google Scholar]
- Li W, Cavelti-Weder C, Zhang Y, Clement K, Donovan S, Gonzalez G, Zhu J, Stemann M, Xu K, Hashimoto T, et al. Long-term persistence and development of induced pancreatic beta cells generated by lineage conversion of acinar cells. Nat Biotechnol. 2014;32:1223–1230. doi: 10.1038/nbt.3082. [DOI] [PubMed] [Google Scholar]
- Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, Berishvili E, Bock C, Kubicek S. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti P, Bugliani M, Lupi R, Marselli L, Masini M, Boggi U, Filipponi F, Weir GC, Eizirik DL, Cnop M. The endoplasmic reticulum in pancreatic beta cells of type 2 diabetes patients. Diabetologia. 2007;50:2486–2494. doi: 10.1007/s00125-007-0816-8. [DOI] [PubMed] [Google Scholar]
- Mastracci TL, Sussel L. The endocrine pancreas: insights into development, differentiation, and diabetes. Wiley Interdiscip Rev Dev Biol. 2012;1:609–628. doi: 10.1002/wdev.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J, Rodríguez-Seguí S, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka T, Asilmaz E, Hu J, Dishinger JF, Kurpad AJ, Elias CF, Li H, Elmquist JK, Kennedy RT, Kulkarni RN. Disruption of leptin receptor expression in the pancreas directly affects beta cell growth and function in mice. J Clin Invest. 2007;117:2860–2868. doi: 10.1172/JCI30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC. Pancreas and beta-cell development: from the actual to the possible. Development. 2007;134:427–438. doi: 10.1242/dev.02770. [DOI] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Ongen H, Irminger JC, Bosco D, Berney T, Antonarakis SE, Halban PA, Dermitzakis ET. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro MC, Sarangi F, Yang C, Holland A, Elefanty AG, Stanley EG, Greiner DL, Keller G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Reports. 2015;4:591–604. doi: 10.1016/j.stemcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Unger RH. Functional subdivision of islets of Langerhans and possible role of D cells. Lancet. 1975;2:1243–1244. doi: 10.1016/s0140-6736(75)92078-4. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JA, Lebrun-Julien F, Suter U. Molecular mechanisms regulating myelination in the peripheral nervous system. Trends Neurosci. 2012;35:123–134. doi: 10.1016/j.tins.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O'Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, et al. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RB, O'Donnell AC, Stamateris RE, Ha B, McCloskey KM, Reynolds PR, Arvan P, Alonso LC. Insulin demand regulates β cell number via the unfolded protein response. J Clin Invest. 2015;125:3831–3846. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol. 2013;25:571–578. doi: 10.1016/j.coi.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T, Sarwal MM, Davis MM, Butte AJ. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward MC, Ishiguro H, Case RM. Mechanisms of bicarbonate secretion in the pancreatic duct. Annu Rev Physiol. 2005;67:377–409. doi: 10.1146/annurev.physiol.67.031103.153247. [DOI] [PubMed] [Google Scholar]
- Stone VM, Dhayal S, Brocklehurst KJ, Lenaghan C, örhede Winzell M, Hammar M, Xu X, Smith DM, Morgan NG. GPR120 (FFAR4) is preferentially expressed in pancreatic delta cells and regulates somatostatin secretion from murine islets of Langerhans. Diabetologia. 2014;57:1182–1191. doi: 10.1007/s00125-014-3213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten LJP, Hinton JE. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- van der Meulen T, Xie R, Kelly OG, Vale WW, Sander M, Huising MO. Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PLoS ONE. 2012;7:e52181. doi: 10.1371/journal.pone.0052181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen T, Donaldson CJ, Cáceres E, Hunter AE, Cowing-Zitron C, Pound LD, Adams MW, Zembrzycki A, Grove KL, Huising MO. Urocortin3 mediates somatostatin-dependent negative feedback control of insulin secretion. Nat Med. 2015;21:769–776. doi: 10.1038/nm.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walpita D, Hasaka T, Spoonamore J, Vetere A, Takane KK, Fomina-Yadlin D, Fiaschi-Taesch N, Shamji A, Clemons PA, Stewart AF, et al. A human islet cell culture system for high-throughput screening. J Biomol Screen. 2012;17:509–518. doi: 10.1177/1087057111430253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb DC, Lowe ME. Human pancreatic digestive enzymes. Dig Dis Sci. 2007;52:1–17. doi: 10.1007/s10620-006-9589-z. [DOI] [PubMed] [Google Scholar]
- Xie R, Everett LJ, Lim HW, Patel NA, Schug J, Kroon E, Kelly OG, Wang A, D'Amour KA, Robins AJ, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Kim J, Ni M, Wei Y, Okamoto H, Lee J, Adler C, Cavino K, Murphy AJ, Yancopoulos GD, et al. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc Natl Acad Sci USA. 2016;113:3293–3298. doi: 10.1073/pnas.1602306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KH, Ko SH, Cho JH, Lee JM, Ahn YB, Song KH, Yoo SJ, Kang MI, Cha BY, Lee KW, et al. Selective beta-cell loss and alpha-cell expansion in patients with type 2 diabetes mellitus in Korea. J Clin Endocrinol Metab. 2003;88:2300–2308. doi: 10.1210/jc.2002-020735. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Manchado ABM, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A transcriptomic map of the human and mouse pancreas, Related to figure 1
(A) Same as Figure 1C. tSNE plots for the dataset derived from each of the four donors and two sacrificed mice.
(B) Histogram of number of transcripts detected per cell.
(C) Histogram of number of genes detected per cell.
(D) Fraction cell type per sample for human (left) and mouse (right). For each donor the bar stack indicates the fraction cells annotated to each of the cell types.
Figure S2. INS/ins1 defines the beta cells cluster in mouse and human, Related to figure 1. tSNE scatter plots of donor 1 (left) and mouse 1 (right) colored by the expression of INS and ins1, respectively.
Figure S3. Beta cells expression are highly correlated between mice, Related to figure 2. Plot comparing the average expression (log10, tpm) of beta cells of mouse 1 and 2.
Figure S4. A transcriptomic markers of Schwann cells in the human pancreas, Related to figure 3. Violin plots for expression of genes enriched in Schwann cells relative to their expression in other cell types. Gene specific to this population are associated with a neural progenitor role.
Figure S5. Testing for structured variation in cell population using PCA, Related to figure 5.
(A) PCA for each cell type.
(B) PCA for alpha cells with total transcript count indicated by color, and correlation betweenPC2 and total transcript count.
(C) PCA for ductal cells with total transcript count indicated by color.
(D) Histogram of the PCA loadings of the ductal cells with the 85 genes indicated.
Figure S6. Subpopulation of the ductal cells in the human pancreas, Related to figure 5. Heatmaps showing genes that are differentially expressed across PC1 for each of three donors including all the genes names.
Figure S7. Subpopulation of the ductal cells in the mouse pancreas, Related to figure 5. Heatmaps showing genes that are differentially expressed across PC1.
Figure S8. Heterogeneity of beta cells reveals unfolded protein response, Related to figure 6. Heatmaps showing genes that are differentially expressed across PC1 including all the genes names.
Figure S9. Properties of BSeq-SC deconvolution, Related to figure 7.
(A) Pancreatic cell types are not equal with respect to their transcriptomic activity. The average number of transcript in a given cell (y-axis) varies significantly depending across cell types (x-axis).
(B) Deconvolution basis matrix composed of the expression profiles of marker genes. The heatmap shows the deconvolution matrix used to estimate the proportions of alpha, beta, gamma, delta, acinar and ductal cells in bulk samples. The rows were z-score-transformed to highlight the cell type-specificity of each marker.
(C) FDR plot from BSeq-SC cell type-specific analysis, revealing up-regulation in alpha cells and down-regulation beta-cells of hyperglycemic patients. The x-axis shows the number of genes called up-regulated (left) and down-regulated (right) at a given FDR cutoff (y-axis).
(D) Complete list of genes found to be exclusively dys regulated in either alpha (blue) or beta cells (purple). Barplot shows estimated cell type-specific effect size (x-axis) for each gene (y-axis).
Table S1. Ages, BMI, and sex of human donors, Related to figure 1
Table S2. Marker genes used in analysis are indicated for each cell type, Related to figure 1
Table S3. Transcription factor described in Figure 3C whose expression profiles are described cited in the literature, Related to figure 3.