Abstract
Soft tissue sarcomas are a heterogeneous group of malignant neoplasms of mesenchymal origin. Partly due to hypoxia, an aggressive and radioresistant phenotype frequently develops, resulting in poorer patient outcome. microRNAs (miRNAs) are tiny, non-coding regulators of gene expression and in situations of cellular stress situations may predict clinical progression and patient outcome. In the present study, hypoxia-associated miR-199a-5p expression in 96 soft tissue sarcoma samples was analysed by reverse transcription-quantitative polymerase chain reaction and associations between miR-199a-5p expression and patient clinicopathological characteristics and survival were measured. Additionally, luciferase reporter assays analyzed the post-transcriptional regulation of hypoxia-associated genes hypoxia-inducible factor 1α (HIF-1α), oxidative stress induced growth inhibitor 2 (OSGIN2) and vascular endothelial growth factor (VEGF) by miR-199a-5p. Survival analyses indicated that low expression of miR-199a-5p was significantly correlated with poorer tumor-specific survival (univariate Cox's-Regression analyses; relative risk=1.92, P=0.029). Furthermore, it was demonstrated that the 3′UTR of HIF-1α and OSGIN2 genes were regulated by miR-199a-5p in-vitro, although the 3′UTR of VEGF was not. To the best of our knowledge, this is the first report demonstrating the regulation of the 3′untranslated region of the OSGIN2 gene by miR-199a-5p and a significant correlation between low miR-199a-5p expression and a poor outcome of patients with soft tissue sarcoma.
Keywords: miR-199a-5p, hypoxia, soft tissue sarcoma, HIF-1α, OSGIN2, survival
Introduction
MicroRNAs (miRNAs) are small (18–25 nt), non-coding RNAs of endogenous origin. To date, >2,500 different mature miRNA species have been detected in humans (1). Following maturation, double-stranded miRNAs are immediately bound to Argonaute proteins and unwound. In the single-stranded conformation, they build up the active component of the RNA-induced silencing complex (RISC) (2,3). miRNAs bind to complementary sequences in the 3′untranslated region (UTR) of their target mRNAs, thus acting as translational repressors. This action is primarily performed by a hexamer sequence at the 5′UTRs called the seed sequence and the subsequent translocation to the P-bodies (processing bodies), which are specialized cellular components for RNA silencing and decay (4,5). Due to the simple target recognition sequence and the vast number of different species of miRNA, it is estimated that ~60% of the human protein-coding genes are regulated post-transcriptionally by miRNAs (6). Due to their high abundance and immense impact on cellular gene expression profiles, miRNAs serve an important role in cellular proliferation and differentiation processes. Furthermore, they contribute to the adaptation to cellular stress, including stress caused by an acidic or hypoxic environment (7,8).
Hypoxia, the state of decreased oxygen saturation in a tissue, is a major problem in the treatment of solid tumors. Due to rapid growth, regions in solid tumors tend to be cut off from oxygen maintenance by blood vessels. The tumor cells must adapt to this new environmental challenge, leading to the clonal selection of successful tumor cells, increased aggressiveness and decreased chemo- and radiosensitivity of the tumor.
There are several studies describing the effect of hypoxia on miRNA expression. Kulshreshthra et al (9,10) have demonstrated that the miRNAs miR-210, miR-23a, miR-24-1, miR-26b and miR-107 are overexpressed in hypoxic conditions, while the miRNAs miR-19a, miR-122a, miR-141, miR-101, miR-186, miR-374, miR-424, miR-422b, miR-320, miR-29b and miR-197 are all downregulated.
The most prominent hypoxia-induced miRNA is the ubiquitously expressed miR-210 (11). In the majority of cell lines studied so far, hypoxia triggers the upregulation of miR-210 (12). Furthermore, high expression of miR-210 is associated with a poorer outcome for patients with mammary carcinoma, head and neck carcinoma, pancreatic adenocarcinoma or soft tissue sarcoma (13–16). Thus, under hypoxic conditions, miR-210 may act as an oncomir, promoting cell survival and adaptation to the changed environment and leading to increased tumor aggressiveness. Hypoxia-inducible miRNAs, including miR-210, may be antagonized by hypoxia-downregulated microRNAs, such as miR-199a. miR-199a is located within a cluster together with miR-214, which is regulated concordantly by diverse transcription factors, including TWIST1 (17). miR-199a-5p regulates hypoxia-inducible factor 1α (HIF-1α) and Sirt mRNA, thus suppressing adaptation to persistent hypoxic conditions (18). Furthermore, expression of the homologous miR-199b is inversely correlated to HIF-1α protein expression in hepatocellular carcinoma (HCC) and its elevated expression predicts an improved outcome for patients with HCC (19). Recently, a self-regulatory network in testicular germ cell tumors was investigated and it was demonstrated that p53 is activated and aids in the positive regulation of miR-199a-2/miR-214 by repressing DNMT1 (20).
In the current study, the impact of miR-199a-5p expression on the clinical parameters and outcome of patients with soft tissue sarcoma was evaluated. Several studies have highlighted the association of miR-199a-5p downregulation with the cellular hypoxia response (18,21–23). Furthermore, hypoxia is an important factor in sarcomagenesis and the increasing aggressiveness of sarcoma; therefore, the present study evaluated the impact of miR-199a-5p on hypoxia-related genes by retesting the regulation of HIF-1α and VEGF through miR-199a-5p and additionally proposing oxidative stress induced growth inhibitor 2 (OSGIN2) as a novel miR-199a-5p target gene.
Materials and methods
Patients
The cohort consisted of 96 patients with soft tissue sarcoma who underwent surgical tumor resection between March 1998 and April 2001 in the Department of Surgery, University of Leipzig (Leipzig, Germany). None of the patients had received adjuvant treatment prior to resection. Patients and tissue samples have been described in previous studies (24,25). Tissues were snap-frozen in liquid nitrogen immediately following resection and stored at −80°C. The present study was approved by the local ethics committee of the Martin Luther University Halle-Wittenberg. All patients gave written informed consent.
RNA isolation
RNA was isolated using TriZol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA USA) according to the manufacturer's protocol. Briefly, the tissue was cut in a cryotome and 20–30 slices (30-µm thick) were lysed in TriZol reagent. The tissue protein and DNA were precipitated by centrifugation. DNAse I (Qiagen GmbH, Hilden, Germany) digestion was performed on the flow-through for 30 min to eliminate remaining traces of DNA. RNA was precipitated by isopropanol (Sigma-Aldrich, St. Louis, Mo, USA) overnight at −20°C and washed twice with ice-cold ethanol prior to elution in RNAse-free water.
miRNA synthesis and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
A total of 10 ng RNA was used for cDNA synthesis of miR-199a-5p and U18 small nucleolar (sno) RNA (reference gene) using stem-loop primers of the TaqMan® microRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). cDNA synthesis was performed by MuLV Reverse Transcriptase kit (Fermentas; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. qPCR was performed with the HotStartTaq DNA Polymerase kit (Qiagen GmbH) on a Rotorgene Cycler (LTF Labortechnik, Wasserburg, Germany). U18 snoRNA values were uniform and served as an internal reference. Cq values were obtained and transformed via the ΔΔCq method (26) and the lowest Cq value served as a calibrator (27).
Luciferase reporter assay
3′UTR regions of HIF-1α, OSGIN2 and VEGF were amplified using the following primers: HIF-1α, forward, 5′-AAACTCGAGTGGCATGTAGACTGCTGGGGCAA-3′ and reverse, 5′-AAACTCGAGTGGCTACCACGTACTGCTGGCAA-3′; OSGIN2: forward, 5′-AAACTCGAGTGGGGTTTTGCAGTGTACTGGCT-3′ and reverse, 5′-AAACTCGATGGACCCCACCCCCAGTTATACA-3′; VEGF forward, 5′-AAACTCGAGTGGACCACACCATCACCATCGAC-3′ and reverse, 5′-AAACTCGAGTGGCGTCTGACCTGGGGTAGAGA-3′. The 3′UTR sequences with the putative miRNA binding sites were cloned into a psiCheck™-2 vector carrying a renilla luciferase and a constitutively expressed firefly luciferase gene (Promega Corporation, Madison, WI, USA) by applying restriction digestion with XhoI (Fermentas; Thermo Fisher Scientific, Inc.) and ligation with Ligase I (Fermentas; Thermo Fisher Scientific, Inc.). Cells from the human osteosarcoma SAOS-2 cell line were cultivated for 24 h in Dulbecco's modified Eagle's medium (DMEM) in 5% CO2, transfected with psiCheck2-3′UTR-constructs and mimics for miR-199a-5p (Ambion; Thermo Fisher Scientific, Inc.). miR-199a-5p mimics were transfected as double-stranded RNA according to the manufacturer's protocol, with the complementary miR-199a-3p strand inactivated by appropriate chemical modifications. Luciferase activity measurements with the Dual-Glo® Luciferase assay (Promega Corporation) were performed after 24 h in a Tecan X50 reader (Tecan Austria GmbH, Grödig, Austria), to determine relative chemiluminescence. Firefly luciferase activity served as an internal reference. Relative values were standardized on the luminescence values of the non-mimic transfected control.
Western blot analyses
For the analysis of miR-199a-5p-mediated translation inhibition of the selected target genes, two sarcoma cell lines were chosen as representative sarcoma systems. Rhabdomyosarcoma (RD) and SAOS-2 cells were transfected with miR-199a-5p mimics or scrambled RNA (Ambion; Thermo Fisher Scientific, Inc.), respectively and cultivated for 24 h under hypoxic or normoxic conditions in DMEM in 5% CO2. Cells were harvested on ice immediately following reoxygenation using a cell scraper and protein was isolated according to the RIPA method. For western blot analyses, 20 ng of whole protein was separated on a sodium dodecyl sulphate-polyacrylamide gel electrophoresis gel (4–12%; Thermo Fisher Scientific, Inc.), blotted on a polyvinylidene fluoride membrane and blocked with 5% dry milk/phosphate-buffered saline (Carl Roth GmbH & Co. KG, Karlsruhe, Germany). Membranes were incubated with murine anti-human OSGIN2 (dilution 1:500; #ab88829; Abcam, Cambridge, UK), murine anti-human HIF-1α (dilution 1:1,000; #610958; BD Transduction Laboratories™; BD Biosciences) or murine anti-β-actin antibody (dilution 1:2,000; #A2228; Sigma-Aldrich) and tagged with a secondary horseradish peroxidase-coupled antibody (dilution 1:10,000; #P0260; Dako, Glostrup, Denmark). Detection was performed with enhanced chemiluminescent Detection substrate (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 (IBM SPSS, Armonk, NY, USA). In detail, bivariate correlations (Spearman-Rho), Kaplan-Meier analyses, uni- and multivariate Cox's regression analyses (the latter was adjusted to tumor entity, tumor localisation, resection type and tumor stage) were performed to evaluate possible correlations between miR-199a-5p and clinical parameters and outcome. To evaluate the luciferase reporter assay, unpaired Student's t-tests were performed. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-199a-5p is associated with clinical and molecular factors in soft tissue sarcoma
miR-199a-5p expression was quantified in 96 patients with soft tissue sarcoma: 37 of these patients experienced ≥1 relapses, 29 experienced metastasis and 50 died of the tumor (Table I). Bivariate correlation analyses according to Spearman-Rho were performed to detect significant associations between miR-199a-5p expression and relevant clinical or molecular factors (Table II). Intriguingly, it was demonstrated that miR-199a-5p expression is associated with the number and interval of relapses (rs=0.39, P=0.00009 and rs=0.52, P=3.6×10−8, respectively) as well as the tumor-specific survival time (rs=0.36, P=0.00024). There was an association between miR-199a-5p expression and OSGIN2 mRNA expression, although this was not statistically significant (rs=−0.23; P=0.09). Furthermore, there was a correlatiojn between miR-199a-5p and HIF-1α mRNA expression (r =0.27, P=0.018). These findings may suggest the existence of a feedback-loop, induced by increased HIF-1α levels, leading to a downregulation through miR-199a.
Table I.
Clinical and histopathological parameters and miR-199a-5p expression level.
| Patient characteristic | No. cases | miR-199a-5p ≤2.24 | miR-199a-5p >2.24 | χ2 test (P-value) |
|---|---|---|---|---|
| Total | 96 | |||
| Gender | ||||
| Male | 47 | 12 | 35 | |
| Female | 49 | 21 | 28 | n.s. |
| Histological subtype | ||||
| LS | 18 | 7 | 11 | |
| FS | 7 | 1 | 6 | |
| RMS | 7 | 3 | 4 | |
| LMS | 17 | 9 | 8 | |
| NS | 11 | 4 | 7 | |
| Syn | 7 | 0 | 7 | |
| NOS | 22 | 7 | 15 | |
| Other | 7 | 1 | 6 | n.s. |
| Tumor size | ||||
| T1 | 21 | 3 | 18 | |
| T2 | 75 | 29 | 46 | n.s. |
| Tumor gradea | ||||
| I | 13 | 6 | 7 | |
| II | 41 | 9 | 32 | |
| III | 42 | 17 | 25 | n.s. |
| Tumor stageb | ||||
| I | 10 | 4 | 6 | |
| II | 42 | 10 | 32 | |
| III | 34 | 12 | 22 | |
| IV | 10 | 6 | 4 | n.s. |
| Complete resection | ||||
| Radical (R0) | 62 | 20 | 42 | |
| Not radical (R1) | 34 | 12 | 22 | n.s. |
| Location | ||||
| Extremities | 61 | 18 | 43 | |
| Trunk wall | l1 | 3 | 8 | |
| Head/neck | 3 | 1 | 2 | |
| Abdomen/retro-peritoneum | 19 | 9 | 10 | |
| Multiple locations | 2 | 1 | 1 | n.s. |
| Number of relapses | ||||
| 0 | 59 | 22 | 37 | |
| 1 | 17 | 5 | 12 | |
| >2 | 20 | 5 | 15 | n.s. |
| Metastasis | ||||
| Yes | 29 | 10 | 19 | |
| No | 67 | 22 | 45 | n.s. |
| Patient status | ||||
| Alive | 46 | 11 | 35 | |
| Succumbed | 50 | 21 | 29 | n.s. (P=0.06) |
According to van Unnik et al, 1993 (50).
According to the Union for International Cancer Control guidelines (51). LS, liposarcoma; FS, fibrosarcoma; RMS, rhabdomyosarcoma; LMS, leiomyosarcoma; NS, neuronal sarcoma; Syn, synovial sarcoma; NOS, not other specified; tumor size, T1 ≤5 cm in diameter, T2 >5 cm in diameter; n.s., not significant.
Table II.
Bivariate correlations (Spearman's Rho test) between miR-199a-5p expression in tumor tissues and different clinical and molecular factors of soft tissue sarcoma patients.
| Parameter | rs | P-value | n |
|---|---|---|---|
| Age | −0.21 | 0.04a | 98 |
| Tumor-specific survival time | 0.36 | 0.00024a | 98 |
| Number of relapses | 0.39 | 0.00009a | 98 |
| Time interval of relapse | 0.52 | (3.6×10−8)a | 98 |
| HIF-1α mRNA | 0.266 | 0.018a | 85 |
P<0.05, indicating a statistically significant value. HIF-1α, hypoxia-inducible factor 1α.
Survival analyses
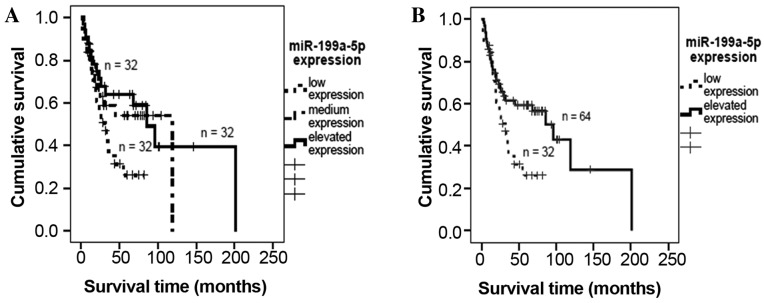
miR-199a-5p expression was transformed according to a tercentile distribution (low expression, <2.24; medium expression, 2.24–10.3; elevated expression, >10.4). A Kaplan-Meier analysis indicated that low miR-199a-5p expression was associated with decreased patient survival time, although this association was not significant (P=0.07, mean survival times: Low expression, 38.3 months; medium expression, 72.2 months; elevated expression, 105.9 months; Fig. 1A). To investigate the effect of lower levels of miR-199a-5p expression in detail, the groups of patients exhibiting medium and elevated expression were combined. It was demonstrated that lower miR-199a-5p expression was associated with a significant decrease in survival time (P=0.026; median survival time for low expression, 38.3 months; for elevated expression, 95.6 months; Fig. 1B).
Figure 1.
Survival analyses for miR-199a-5p expression in patients with soft tissue sarcoma. Kaplan-Meier analysis demonstrated decreased survival rates of patients with low miR-199a-5p expression compared to patients with elevated miR-199a-5p expression. Univariate Cox's regression analyses confirmed the negative effect of low miR-199a-5p expression on disease-specific survival (RR=1.9; P=0.029). miR-199a-5p, microRNA-199a-5p; RR, relative risk.
Furthermore, a univariate Cox's regression analysis demonstrated that patients with low miR-199a expression exhibited significantly poorer outcomes and were 1.92 times more likely to experience tumor-associated mortality [P=0.029; 95% confidence interval (CI), 1.07–3.44]. However, in a multivariate Cox's regression analysis (adjusted to resection type, tumor stage, entity and localisation), no correlation was detected between miR-199a-5p expression and tumor-specific survival (P=0.66; relative risk=1.16; 95% CI, 0.6–2.22).
OSGIN2 and HIF-1α are target-genes of miR-199a-5p in vitro
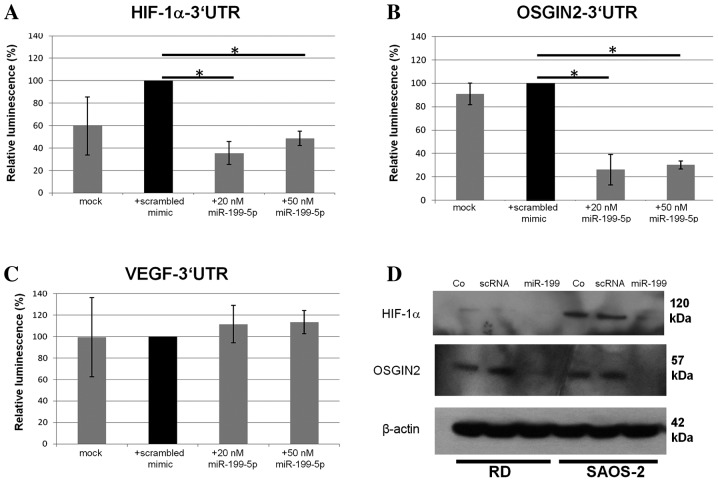
To analyse possible linkages between miR-199a expression and hypoxia-related target genes, an in silico database search for putative 3′-UTR mRNA interactions of miR-199a was performed. Beside others, TargetScan (www.targetscan.org) suggested HIF-1α, VEGF and OSGIN2 mRNA as putative target genes of miR-199a-5p (Fig. 2). The 3′UTR sequences of these genes, including the predicted miR-199a-5p target binding site, were cloned into psiCheck2 vectors and luciferase reporter assays were performed to validate regulation by miR-199a-5p. Co-transfection of SAOS-2 cells with 20 or 50 nM miR-199a-5p mimics induced a significant downregulation of HIF-1α and OSGIN2 3′UTR coupled luciferase expression (Fig. 3A and B; 35.4 and 26.2% respectively, compared to controls treated with scrambled mimics; P=0.008 and P=0.01, respectively; Student's t-test). However, no significant change in VEGF-3′UTR coupled luciferase expression (105.6% compared to control, Fig. 3C) was detected.
Figure 2.
In silico analyses of miR-199a-5p target sequence binding. The overlapping sequences of mi-R-199a-5p and (A) HIF-1α, (B) OSGIN2 and (C) VEGF are presented according to TargetScan annotation (www.targetscan.org). The complimentary seed sequences are indicated by boxes. HIF-1α, hypoxia-inducible factor 1α OSGIN2, oxidative stress induced growth inhibitor 2; VEGF, vascular endothelial growth factor.
Figure 3.
Luciferase reporter assays with reporter constructs containing (A) HIF-1α (B) OSGIN2 and (C) VEGF 3′UTRs. Relative luminescence coefficients were standardized on a scrambled mimic transfected control. Co-transfection with 20 nM miR-199a-5p yielded a significant downregulation of the luciferase-coupled HIF-1α-3′UTR construct (35.4%, P=0.041, Student's t-test) and the OSGIN2-3′UTR construct (26.2%; P=0.029, Student's t-test, error bars defining standard derivation) compared with controls (n=3; *P<0.05). No such downregulation was observed in VEGF 3′UTRs. (D) Western blot analyses presented a downregulation of HIF-1α and OSGIN2 expression following miR-199a-5p (20 nM) overexpression. HIF-1α, hypoxia-inducible factor 1α OSGIN2, oxidative stress induced growth inhibitor 2; VEGF, vascular endothelial growth factor; UTR, untranslated region; scRNA, scrambled RNA; RD, Rhabdomyosarcoma cells.
Furthermore, western blot analyses of HIF-1α and OSGIN2 protein expression following miR-199a-5p transfection were performed in RD and SAOS-2 cells under hypoxic conditions. HIF-1α and OSGIN2 proteins were expressed in hypoxic conditions, whereas no visible bands were detectable under normoxic conditions (data not shown). In hypoxic conditions, transfection of miR-199a-5p mimics resulted in the downregulation of HIF-1α and OSGIN2 protein in comparison to untreated control cells or cells treated with negative mimics only (Fig. 3D). Taken together, these results suggest that miR-199a-5p downregulation contributes to the adaptation of cells to hypoxia.
Discussion
The present study evaluated the impact of miR-199a-5p on the survival of patients with soft tissue sarcoma and validated HIF-1α and OSGIN2 as miR-199a-5p target genes. Interestingly, OSGIN2 was linked for the first time to miR-199a-5p regulation and may be an interesting target for analyses of sarcomagenesis and progression, tumor hypoxia and patient outcome.
The effect of the expression of several well-known cancer genes on the mRNA and protein level is well known, however, the data about the impact of most types of miRNA on the outcome of patients with soft tissue sarcoma remains scarce. To the best of our knowledge, the present study is first to report a 1.92-fold increased risk of tumor-associated mortality for soft tissue sarcoma patients (33% percentile) with low miR-199a-5p expression (univariate Cox's regression analysis, P=0.029; 95% CI: 1.07–3.44). Following multivariate Cox's Regression analysis, no effect of miR-199a-5p expression on patients' survival was detected, however, non-parametric correlation analyses demonstrated a correlation with tumor entity (P=0.014, Kruskal-Wallis test). This is concordant with the results of a previous study by Guled et al (28), describing a diagnostic role of miR-199-5p for the identification of undifferentiated pleomorphic sarcoma compared to leiomyosarcoma. It has also been demonstrated that low miR-199a-5p expression is associated with a poorer outcome in serous ovarian carcinoma (29), non-small cell lung carcinoma measured in patients' sera (30) and renal cell carcinoma (31). Concordantly, decreased expression of the complementary miRNA sequence miR-199a-3p is associated with a shorter time to recurrence in hepatocellular carcinoma (32). Furthermore, multivariate Cox's regression analyses demonstrated that low miR-199a-3p expression was significantly associated with poorer overall survival in patients with osteosarcoma (33). However, previous studies have linked the elevated expression of miR-199a-3p with poorer overall patient survival in colorectal cancer (34) and esophageal adenocarcinoma (35), pointing towards a differential expression signature and possibly altered regulation mechanisms of miR-199a processing and RISC incorporation in tumor cells.
Several studies have described miR-199a as a type of miRNA associated with hypoxia-adaptation (36–39). It was demonstrated that miR-199a-5p downregulation is AKT-dependent (39,40) and may be antagonized by the β-adrenergic receptor (40). In the current study, in order to facilitate luciferase reporter assays, the interactions between miR-199a-5p and the 3′UTRs of three putative hypoxia-related target genes, namely HIF-1α, OSGIN2 and VEGF, were investigated. These target genes were chosen following a literature search demonstrating the potential link between miR-199a-5p expression and hypoxia. The results of the present study demonstrated that miR-199a-5p post-transcriptionally regulated HIF-1α and OSGIN2, but not VEGF. Downregulation of miR-199a-5p in cardiac myocytes mimics hypoxia preconditioning by lowering the miR-199a-5p-mediated suppression of HIF-1α mRNA translation (18). Furthermore, ethanol downregulates miR-199a expression in liver-sinusoidal endothelial liver cells independently of hypoxic stress, and this downregulation coincides with an increase in HIF-1α and Endothelin-1 mRNA (41). By contrast, overexpression of miR-199a-5p and miR-199a-3p has been detected in advanced liver fibrosis in samples from mouse and human tissue (42). On a molecular level miR-199a-5p targets the prominent oncogenes ERBB2 and ERBB3 directly in ovarian and lung carcinoma cell lines (43). Additionally, it was observed that miR-199a-5p overexpression in ovarian carcinoma cells significantly decreased their ability to induce angiogenesis, an effect mediated via ERBB2 and ERBB3 signalling (21). Moreover, overexpression of miR-199a-5p in multiple myeloma cells significantly impaired migration by downregulating the expression of adhesion molecules, including vascular cell adhesion molecule-1 (VCAM-1) and intracellular adhesion molecule 1 (ICAM-1) (39). By contrast, the complementary miR-199a-3p transcript regulates the tumor-associated mammalian target of rapamycin (mTOR) and c-Met in hepatocellular carcinoma cells and its overexpression restores hypoxia and doxorubicin sensitivity (32). Recently, Kinose et al (44) demonstrated that miR-199a-3p serves as tumor suppressor gene in ovarian cancer by directly repressing its target gene c-Met and subsequently inhibiting proliferation, adhesion and invasiveness (44). The present study validated miR-199a-5p as regulator of HIF-1α, but not VEGF. These results are concordant with those from previous studies demonstrating that VEGF is an indirect target gene of miR-199a, which is regulated by HIF-1α and ERBB2/3 downregulation rather than by direct miRNA-medicated VEGF translation inhibition and mRNA destabilization (21).
Furthermore, the results of the present study suggested that OSGIN2 is a putative target of miR-199a-5p and suggested a possible functional link between a hypoxic tumor environment and the miRNA-induced derepression of stress genes protecting tumor as well as normal cells from stress-induced cell death. OSGIN2 is localized on chromosome 8q21.3 in the neighbourhood of Nijmegen breakage syndrome gene 1 (45). The transcript is translated into a protein 505 amino acids long and there is an additional shorter transcript variant translated to a protein with a distinct N-terminus (45). OSGIN2 is a still poorly characterized homolog of OSGIN1, with 49% identity and 62% similarity (46). A previous study demonstrated that OSGIN2 was upregulated in the liver biopsies of transplants exhibiting initial poor graft function (47). Furthermore, in a high-throughput assessment for known and novel breast cancer candidate genes, OSGIN2 was mapped to a chromosomal region demonstrating a significant amplification gain in breast cancer cell lines and tumor tissue (48). Additionally, it has been identified that OSGIN2 is overexpressed following activation of PGC-1 related coactivator, a protein maintaining mitochondrial homeostasis and linking mitochondrial status to cell cycle (49). However, the physiological function of OSGIN2, and its potential association with cancer genesis and progression remain elusive and further studies are necessary to clarify the impact of this gene on tumor hypoxia.
In conclusion, the present study identified a correlation between low miR-199a-5p expression and a poorer outcome of patients with soft tissue sarcoma. Low miR-199a-5p expression increased the risk of tumor-associated mortality by 1.92-fold. Furthermore, HIF-1α and OSGIN2 may be target genes of miR-199a-5p. However, the precise role of miR-199a-5p in cancer remains ambiguous, due to its diverging functions in different cell types and stress situations. Thus, further studies, particularly on the regulation and impact of this hypoxia-associated miRNA are required.
Acknowledgements
The authors wish to thank Ms. Gabriele Thomas and Ms. Katrin Theile for their excellent technical assistance. Thomas Greither's work was supported by a junior research group grant of Wilhelm-Roux program of the Medical faculty of Martin Luther University Halle-Wittenberg (grant no. FKZ 25/43).
Glossary
Abbreviations
- HIF-1α
hypoxia-inducible factor 1α
- OSGIN2
oxidative stress induced growth inhibitor 2
- VEGF
vascular endothelial growth factor
- RR
relative risk of tumor-related death
- p
probability
- CI
95% confidence interval
- rS
Spearman's rank correlation coefficient
- h
hour
- UTR
untranslated region
References
- 1.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y, Kim VN. MicroRNA factory: RISC assembly from precursor microRNAs. Mol Cell. 2012;46:384–386. doi: 10.1016/j.molcel.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emde A, Hornstein E. miRNAs at the interface of cellular stress and disease. EMBO J. 2014;33:1428–1437. doi: 10.15252/embj.201488142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15:667–671. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 10.Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA expression: The hypoxic component. Cell Cycle. 2007;6:1426–1431. doi: 10.4161/cc.6.12.4410. [DOI] [PubMed] [Google Scholar]
- 11.Huang X, Zuo J. Emerging roles of miR-210 and other non-coding RNAs in the hypoxic response. Acta Biochim Biophys Sin (Shanghai) 2014;46:220–232. doi: 10.1093/abbs/gmt141. [DOI] [PubMed] [Google Scholar]
- 12.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life. 2011;63:94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. Hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 14.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, Homer J, Corbridge R, Cox G, West CM, et al. Hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 15.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 16.Greither T, Würl P, Grochola L, Bond G, Bache M, Kappler M, Lautenschläger C, Holzhausen HJ, Wach S, Eckert AW, Taubert H. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int J Cancer. 2012;130:1230–1235. doi: 10.1002/ijc.26109. [DOI] [PubMed] [Google Scholar]
- 17.Baumgarten A, Bang C, Tschirner A, Engelmann A, Adams V, von Haehling S, Doehner W, Pregla R, Anker MS, Blecharz K, et al. TWIST1 regulates the activity of ubiquitin proteasome system via the miR-199/214 cluster in human end-stage dilated cardiomyopathy. Int J Cardiol. 2013;168:1447–1452. doi: 10.1016/j.ijcard.2012.12.094. [DOI] [PubMed] [Google Scholar]
- 18.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Song B, Song W, Liu J, Sun A, Wu D, Yu H, Lian J, Chen L, Han J. Underexpressed microRNA-199b-5p targets hypoxia-inducible factor-1α in hepatocellular carcinoma and predicts prognosis of hepatocellular carcinoma patients. J Gastroenterol Hepatol. 2011;26:1630–1637. doi: 10.1111/j.1440-1746.2011.06758.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen BF, Suen YK, Gu S, Li L, Chan WY. A miR-199a/miR-214 self-regulatory network via PSMD10, TP53 and DNMT1 in testicular germ cell tumor. Sci Rep. 2014;4:6413. doi: 10.1038/srep06413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Jing Y, Li W, Qian X, Xu Q, Li FS, Liu LZ, Jiang BH, Jiang Y. Roles and mechanism of miR-199a and miR-125b in tumor angiogenesis. PLoS One. 2013;8:e56647. doi: 10.1371/journal.pone.0056647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding G, Huang G, Liu HD, Liang HX, Ni YF, Ding ZH, Ni GY, Hua HW. MiR-199a suppresses the hypoxia-induced proliferation of non-small cell lung cancer cells through targeting HIF1α. Mol Cell Biochem. 2013;384:173–180. doi: 10.1007/s11010-013-1795-3. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Lei S, Long J, Liu X, Wu Q. MicroRNA-199a-5p inhibits tumor proliferation in melanoma by mediating HIF-1α. Mol Med Rep. 2016;13:5241–5247. doi: 10.3892/mmr.2016.5202. [DOI] [PubMed] [Google Scholar]
- 24.Kappler M, Köhler T, Kampf C, Diestelkötter P, Würl P, Schmitz M, Bartel F, Lautenschläger C, Rieber EP, Schmidt H, et al. Increased survivin transcript levels: An independent negative predictor of survival in soft tissue sarcoma patients. Int J Cancer. 2001;95:360–363. doi: 10.1002/1097-0215(20011120)95:6<360::AID-IJC1063>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 25.Würl P, Kappler M, Meye A, Bartel F, Köhler T, Lautenschläger C, Bache M, Schmidt H, Taubert H. Co-expression of survivin and TERT and risk of tumour-related death in patients with soft-tissue sarcoma. Lancet. 2002;359:943–945. doi: 10.1016/S0140-6736(02)07990-4. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 28.Guled M, Pazzaglia L, Borze I, Mosakhani N, Novello C, Benassi MS, Knuutila S. Differentiating soft tissue leiomyosarcoma and undifferentiated pleomorphic sarcoma: A miRNA analysis. Genes Chromosomes Cancer. 2014;53:693–702. doi: 10.1002/gcc.22179. [DOI] [PubMed] [Google Scholar]
- 29.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 30.Sanfiorenzo C, Ilie MI, Belaid A, Barlési F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Si T, Liu C, Xu K, Gui Y. Association of miR-199a expression with clinicopathologic characteristics and prognosis of renal cell carcinoma. Nan Fang Yi Ke Da Xue Xue Bao. 2012;32:1568–1571. (In Chinese) [PubMed] [Google Scholar]
- 32.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 33.Tian R, Xie X, Han J, Luo C, Yong B, Peng H, Shen J, Peng T. miR-199a-3p negatively regulates the progression of osteosarcoma through targeting AXL. Am J Cancer Res. 2014;4:738–750. [PMC free article] [PubMed] [Google Scholar]
- 34.Wan D, He S, Xie B, Xu G, Gu W, Shen C, Hu Y, Wang X, Zhi Q, Wang L. Aberrant expression of miR-199a-3p and its clinical significance in colorectal cancers. Med Oncol. 2013;30:378. doi: 10.1007/s12032-012-0378-6. [DOI] [PubMed] [Google Scholar]
- 35.Feber A, Xi L, Pennathur A, Gooding WE, Bandla S, Wu M, Luketich JD, Godfrey TE, Litle VR. MicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. Ann Thorac Surg. 2011;91:1523–1530. doi: 10.1016/j.athoracsur.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y, Zhu Y, Wang X, Gong J, Hu C, Guo B, Zhu B, Li Y. Temporal regulation of HIF-1 and NF-κB in hypoxic hepatocarcinoma cells. Oncotarget. 2015;6:9409–9419. doi: 10.18632/oncotarget.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi HP, Subramanian IV, Schnettler EK, Ghosh G, Rupaimoole R, Evans C, Saluja M, Jing Y, Cristina I, Roy S, et al. Dynamin 2 along with microRNA-199a reciprocally regulate hypoxia-inducible factors and ovarian cancer metastasis. Proc Natl Acad Sci USA. 2014;111:5331–5336. doi: 10.1073/pnas.1317242111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuno S, Bogaard HJ, Gomez-Arroyo J, Alhussaini A, Kraskauskas D, Cool CD, Voelkel NF. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1α expression in lungs from patients with COPD. Chest. 2012;142:663–672. doi: 10.1378/chest.11-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raimondi L, Amodio N, Di Martino MT, Altomare E, Leotta M, Caracciolo D, Gullà A, Neri A, Taverna S, D'Aquila P, et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: In vitro and in vivo anti-tumor activity. Oncotarget. 2014;5:3039–3054. doi: 10.18632/oncotarget.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rane S, He M, Sayed D, Yan L, Vatner D, Abdellatif M. An antagonism between the AKT and beta-adrenergic signaling pathways mediated through their reciprocal effects on miR-199a-5p. Cell Signal. 2010;22:1054–1062. doi: 10.1016/j.cellsig.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microrNA-199. J Immunol. 2009;183:5232–5243. doi: 10.4049/jimmunol.0901564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami Y, Toyoda H, Tanaka M, Kuroda M, Harada Y, Matsuda F, Tajima A, Kosaka N, Ochiya T, Shimotohno K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One. 2011;6:e16081. doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He J, Xu Q, Jing Y, Agani F, Qian X, Carpenter R, Li Q, Wang XR, Peiper SS, Lu Z, et al. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO Rep. 2012;13:1116–1122. doi: 10.1038/embor.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinose Y, Sawada K, Nakamura K, Sawada I, Toda A, Nakatsuka E, Hashimoto K, Mabuchi S, Takahashi K, Kurachi H, et al. The hypoxia-related microRNA miR-199a-3p displays tumor suppressor functions in ovarian carcinoma. Oncotarget. 2015;6:11342–11356. doi: 10.18632/oncotarget.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tauchi H, Matsuura S, Isomura M, Kinjo T, Nakamura A, Sakamoto S, Kondo N, Endo S, Komatsu K, Nakamura Y. Sequence analysis of an 800-kb genomic DNA region on chromosome 8q21 that contains the Nijmegen breakage syndrome gene, NBS1. Genomics. 1999;55:242–247. doi: 10.1006/geno.1998.5657. [DOI] [PubMed] [Google Scholar]
- 46.Ong CK, Ng CY, Leong C, Ng CP, Foo KT, Tan PH, Huynh H. Genomic structure of human OKL38 gene and its differential expression in kidney carcinogenesis. J Biol Chem. 2004;279:743–754. doi: 10.1074/jbc.M308668200. [DOI] [PubMed] [Google Scholar]
- 47.Defamie V, Cursio R, Le Brigand K, Moreilhon C, Saint-Paul MC, Laurens M, Crenesse D, Cardinaud B, Auberger P, Gugenheim J, et al. Gene expression profiling of human liver transplants identifies an early transcriptional signature associated with initial poor graft function. Am J Transplant. 2008;8:1221–1236. doi: 10.1111/j.1600-6143.2008.02249.x. [DOI] [PubMed] [Google Scholar]
- 48.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar AF, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raharijaona M, Le Pennec S, Poirier J, Mirebeau-Prunier D, Rouxel C, Jacques C, Fontaine JF, Malthiery Y, Houlgatte R, Savagner F. PGC-1-related coactivator modulates mitochondrial-nuclear crosstalk through endogenous nitric oxide in a cellular model of oncocytic thyroid tumours. PLoS One. 2009;4:e7964. doi: 10.1371/journal.pone.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Unnik JA, Coindre JM, Contesso C, Lutter CE Albus, Schiodt T, Sylvester R, Thomas D, Bramwell V, Mouridsen HT. Grading of soft tissue sarcomas: Experience of the EORTC soft tissue and bone sarcoma group. Eur J Cancer 29A. 1993:2089–2093. doi: 10.1016/0959-8049(93)90039-I. [DOI] [PubMed] [Google Scholar]
- 51.Weiss SW, Goldblum JR, editors. Enzinger and Weiss's Soft Tissue Tumors. 5th. Mosby Elsevier; Philadelphia, PA: 2008. Malignant soft tissue tumors of uncertain type; pp. 3–8. [Google Scholar]