Abstract
Cancer genome sequencing efforts are leading to the identification of genetic mutations in many types of malignancy. However, the majority of these genetic alterations have been considered random passengers that do not directly contribute to tumorigenesis. We have previously conducted a soft agar-based short hairpin RNA (shRNA) screen within colorectal cancer (CRC) candidate driver genes (CAN-genes) using a karyotypically diploid hTERT- and CDK4-immortalized human colonic epithelial cell (HCEC) model and discovered that depletion of 65 of the 151 CAN-genes enhanced anchorage-independent growth in HCECs with ectopic expression of K-RasV12 and/or TP53 knockdown. We now constructed an interaction map of the confirmed CAN-genes with CRC non-CAN-genes and screened for functional tumor suppressors. Remarkably, depletion of 15 out of 25 presumed passenger genes that interact with confirmed CAN-genes (60%) promoted soft agar growth in HCECs with TP53 knockdown compared to only 7 out of 55 (12.5%) of presumed passenger genes that do not interact. We have thus demonstrated a pool of driver mutations among the putative CRC passenger/incidental mutations, establishing the importance of employing biological filters, in addition to bioinformatics, to identify driver mutations.
Keywords: anchorage-independent growth, colon cancer, driver mutations, interaction map
Cancer genome sequencing efforts are leading to the identification of an abundance of mutated genes in many cancer types.1 Although these efforts provide a list and frequency of mutant genes, their functional relevance remains elusive.2 Genes frequently mutated across tumor samples are often considered to be driver mutations whereas those detected at a lower frequency are treated as passenger (or incidental) mutations. Driver mutations are defined as being causally involved in the neoplastic process and are positively selected for during tumorigenesis whereas passenger mutations are thought to provide no selective advantage but are retained by chance during clonal expansion.3 Discriminating between driver and passenger mutations is critical for understanding the signaling pathways and molecular mechanisms underlying cancer initiation, progression, and maintenance. Thus far, the efforts to subdivide driver from passenger mutations have largely, if not entirely, relied upon bioinformatic tools, and have not been subjected to rigorous experimental testing. These frequency-based approaches have raised concerns and debates as to the classification of driver and passenger mutations because of the lack of any experimental evidence.4 Therefore, complementary functional studies are regarded as being critical for validation of driver mutations identified from large-scale genomic screens.5,6
The extreme diversity of mutations present in cancer genomes has contributed to the concept that pathways rather than individual genes govern tumorigenesis. Transcriptomic and proteomic analyses suggest a convergence onto a finite number of pathways that are involved in those mutational and epigenetic events.7 Interactome network analysis has been shown to be a powerful tool in predicting protein functions, delineating pathways involved in human disease, and identifying novel therapeutic targets for drug development.8,9 This approach thereby represents an important step towards a systematic and comprehensive understanding of biological networks.
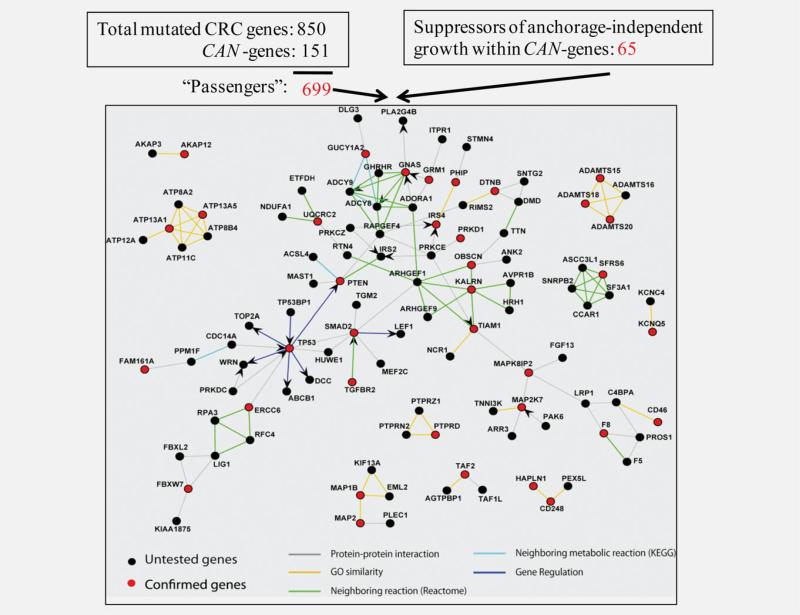
Initial efforts to sequence the colorectal cancer (CRC) genome detected 151 highly mutated candidate genes (CAN-genes) and ~700 mutations occurring at a lower frequency (passenger mutations).10 CAN-genes were determined primarily based on the total number of mutations per nucleotide sequenced.10 With few exceptions, almost all of these mutations occur at very low frequencies. Anchorage-independent growth is considered one of the hallmarks of cancer progression and one of the most reliable markers for in vitro transformation as well as in vivo human tumorigenic and metastasis potential.11 We previously functionally interrogated CRC mutations by conducting a soft agar-based short hairpin RNA (shRNA) screen within the cohort of CAN-genes. Knockdown of 65 of the 151 CAN-genes enhanced anchorage-independent growth in hTERT and CDK4 immortalized karyotypically diploid human colonic epithelial cells (HCECs) either expressing a shRNA targeting TP53 and/or oncogenic K-RasV12.12 We have now probed the extent of proclaimed passenger mutations and delineated the important pathways involved by constructing an interaction map of all 65 confirmed driver mutations with the additional less frequently mutated genes (Fig. 1). We then screened for novel tumor suppressors of anchorage-independent growth within the cohort of passenger mutations. We found that knockdown of those passenger mutated genes that interacted with the confirmed CAN-genes enhanced anchorage-independent growth at a significantly higher frequency as compared with those that did not interact. Additionally, there were more tumor suppressors than predicted by current statistical models even among the noninteracting low frequency mutated genes compared with random genes not involved in CRC. This study has identified a pool of novel driver mutations causally involved in anchorage-independent growth among the putative passenger mutations, demonstrating the need for biological functional assays to distinguish driver from passenger mutations. These studies may lead to better predictive models and identification of novel therapeutic targets.
Figure 1.
Discovering novel tumor suppressors from passenger mutations by interaction mapping. Data on the interaction of confirmed CAN-genes (red nodes) and less frequently mutated genes (black nodes) are from Wood et al. Interactions are colored according to type of interactions shown in the color key. (Adapted from Fig. 4 in Ref. 12.)
Material and Methods
Cells
HCEC growth media and tissue culture conditions are described elsewhere.13 HCECs isolated from normal colonic biopsies were immortalized by successive infections of CDK4 and hTERT followed by selection with respective antibiotics—G418 (250 μg/ml) and blastocidin (2.5 μg/ml). shRNAs against p53 were introduced with retroviruses and p53 knockdown efficiency was verified as described elsewhere.12 Human colon cancer cell lines (HCT116, DLD-1, and RKO) and virus-producing cell lines (293FT, Phoenix A) were cultured in basal medium supplemented with 10% serum. The identity of all cell lines was verified by DNA fingerprinting.
Viral transduction
One microgram of shRNA together with 1 μg of helper plasmids (0.4 μg pMD2G and 0.6 μg psPAX2) were transfected into 293FT cells with Polyjet reagent (SignaGen). Viral supernatants were collected 48 hr after transfection and cleared through 0.45-μm filter. Cells were infected with viral supernatants containing 4 μg/ml polybrene (Sigma) at multiplicity of infection (MOI) of ~1. Successfully infected cells were selected with 1 μg/ml puromycin for 3 days. Clone IDs for each shRNA used in this study are listed in Supporting Information Table S1.
Quantitative reverse transcription PCR
Total RNA was isolated from cells using RNeasyMinikit (Qiagen, Chatsworth, CA) according to the manufacturer's protocol. Then 1 μg RNA was converted to cDNA using a First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN). Real-time quantitative PCR reactions were set up in triplicate with Ssofast Master Mix (Biorad) and run on a LightCycler® 480 (Roche). All the primers (Sigma) used in this study are listed in Supporting Information Table S2.
Soft agar assay
Stably knockdown cells were suspended in 0.375% Noble agar (Difco, Detroit, MI) in supplemented basal medium at two densities (1,000 and 2,000) and overlaid on 0.75% Noble agar in 24-well plates. Each density was seeded in triplicate and each assay was performed at least twice using cells from different cell suspensions at different times. Colony formation efficiency was calculated by average number of colonies counted per well divided by number of seeded cells. Nonsilencing shRNA expressing cells were seeded with each assay at the same density to be used as normalization control to correct for plate-to-plate variations. Colonies larger than 0.1 mm were measured and counted after 3 weeks growth and the average of those counts was used. Data was plotted as fold change compared to nonsilencing shRNA expressing cells. GraphPad Prism 5 (GraphPad Software) was used to plot data and perform two tailed Student's t-tests.
Network construction
Network construction was done in the NetWalker suite (Komurov et al., submitted) (http://netwalkersuite.org), using the comprehensive network of biomolecular relationships of protein–protein, gene regulation, neighboring, and metabolic reactions described earlier.14
Results
A large number of putative passenger mutated genes enhanced anchorage-independent growth upon depletion
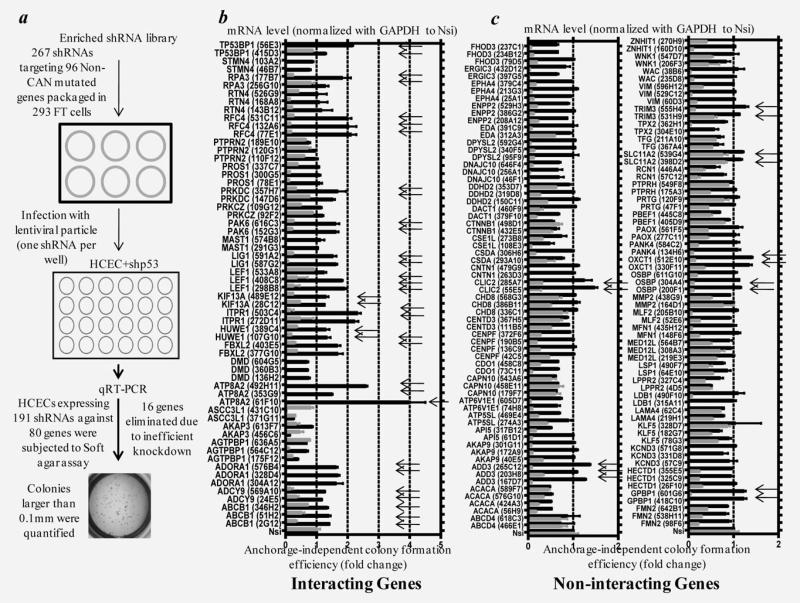
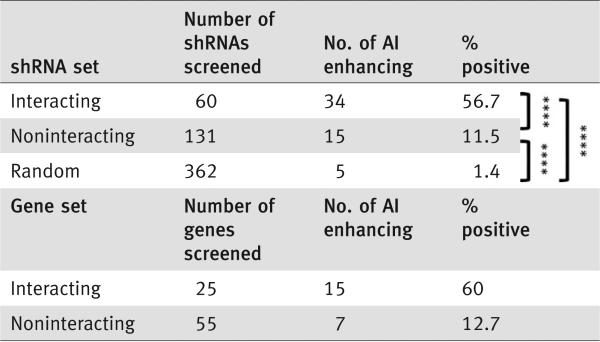
To discover novel tumor suppressors within the putative passenger genes, we first mapped the confirmed CAN-genes (from our previous screen that enhanced anchorage-independent growth upon depletion) onto the human protein interaction network. We then identified which of ~700 passenger CRC mutations had direct interactions with the confirmed CAN-genes (Fig. 1). The soft agar-based shRNA screen was performed in a one-shRNA-one-well format using hTERT and CDK4 immortalized diploid HCECs with TP53 knockdown. TP53 is one of the most frequent mutations found in CRC. This premalignant HCEC derivative was not tumorigenic,12 thus providing an ideal sensitized background that might allow for discovery of novel tumor suppressors. In the present studies, we tested 25 less frequently mutated genes that interacted with at least one confirmed CAN-gene within the interaction map and 55 putative passenger genes that did not. Knockdown efficiency for all the tested genes was confirmed using quantitative real-time PCR except for those with nondetectable mRNA levels. Genes with at least two shRNAs that reduced mRNA levels were subjected to the soft agar assay and colony formation efficiency. Results were quantified, after normalizing to nonsilencing shRNA controls (Fig. 2a). We discovered that knockdown of 15 out of 25 less frequently mutated genes that interacted with confirmed CAN-genes (60%) promoted soft agar growth (Fig. 2b) whereas knockdown of 7 out of 55 less frequently mutated genes that did not interact with any of the confirmed CAN-genes enhanced soft agar growth (12.7%) (Fig. 2c). In our previous screen, we tested 362 random shRNAs and only 5 of them enhanced soft agar growth, thus giving a nonspecific background rate of 1.4%.12 Statistical analyses using Fisher's exact test revealed that the percentage of shRNAs scoring positive within either interacting (56.7%) or noninteracting (11.5%) category was higher than that of the random shRNAs, and a higher fraction of the interacting passenger mutated genes were causally involved in anchorage-independent growth (Table 1). Almost all of the scored hits exhibited a good correlation between knockdown levels and soft-agar growth enhancement, suggesting a dosage effect of these tumor suppressors. This supports the recently proposed continuum model of tumor suppression.15 In addition, the stable knockdown HCECs expressing shRNAs against the most potent hits also exhibited enhanced invasion through Matrigel® (Supporting Information Fig. S1a). We examined the relevance of these observations to authentic cancer cell lines by overexpressing the gene (ITPR1) whose knockdown had produced the greatest effect on invasion through extracellular matrices in our test cell lines. Ectopically expressed wild type ITPR1 cDNA reduced invasion efficiency in all three colon cancer cell lines tested (Supporting Information Fig. S1b). Sequencing analysis of these three cancer cell lines revealed that there are no nonsynonymous mutations within the coding sequence fragments where the mutations in those CRC patients have been identified. One possibility is that other related protein components within this pathway are mutated, rendering these cell lines sensitive to the restoration of ITPR1 function. Future experiments will further investigate the tumor-suppressive function of this gene in CRC.
Figure 2.
Identification of novel driver mutations among putative passenger mutations. (a) Schematic representation of the overall screening strategy. HCECs expressing shRNA against p53 were infected with lentiviral shRNA constructs in a “one-shRNA-one-well” format, knockdown efficiency were measured by quantitative RT-PCR and colonies were quantified after 3 weeks. (b) Quantitative validation of shRNA knockdown efficiency (gray bars) and soft agar growth enhancement (black bars) for passenger mutated genes that interact with confirmed CAN-genes in shTP53 expressing HCECs. (c) Same analysis as in b for passenger mutated genes that did not interact with confirmed CAN-genes. “One-fold” represents no change compared to nonsilencing shRNAs for both mRNA levels and anchorage independent growth. Each black bar represents nine data points (triplicates from three separate experiments). Arrows denote shRNAs that enhance anchorage-independent growth in a statistically significant manner (two tailed Student's t-test, as compared to nonsilencing, mean ± s.e.m., p < 0.05). Nsi, nonsilencing.
Table 1.
Summary of low frequency mutated genes scored positive in the screen
Expression of shRNAs against “passenger” mutated genes that interact with confirmed CAN-genes enhanced anchorage-independent growth at higher fraction than those that do not interact in HCECs with TP53 knockdown. Genes scored with at least two different shRNAs are considered as positive hits. Data for random shRNAs were from our previous screen.12 Higher fraction of interacting passenger mutated genes scored positive than noninteracting ones. The percentage of positive hits in both interacting and noninteracting categories is higher than that of the background rate. Fisher's exact test, ****p < 0.0001. AI: anchorage-independent growth.
Newly identified driver mutations are highly represented across CRC tumor samples
A closer analysis of the genomic-wide sequenced CRC tumor samples from Wood et al.10 revealed that on average, at least 9.4% of all mutated genes in any individual colon cancer tumor are involved in anchorage independent growth (Supporting Information Table S3) and this percentage could be underestimated considering that our approach is not at all exhaustive. Furthermore, analysis of the most recently available Cancer Genome Atlas (TCGA) CRC dataset (http://www.cbioportal.org/public-portal/) showed that the 22 newly identified suppressor genes are altered in 29% of all 193 cases (Supporting Information Table S4). Of note, ITPR1, one of the most potent hits scored in our screen, is altered in 5.7% of all CRC cases, higher than almost all the other cancer types, supporting its important role in CRC tumorigenesis (Supporting Information Table S5). Most of the mutations identified in CRC patients occur within the six functional domains in the coding sequence and are predicted to have medium or high functional impact (http://www.cbioportal.org/public-portal/). These analyses suggest that a large fraction of mutations present within a tumor are causally involved in tumorigenesis rather than incidental events and each of these mutations might provide an additive fitness for precancerous cells to progress into a more malignant state.
Subnetworks involving DNA repair pathways and cyclic nucleotide metabolic process are enriched within the network map
Considering that PTEN, TP53, and FBXW7 are among the most frequent mutations in colon cancer, we focused on the interacting passenger mutations that link these genes. Surprisingly, we found that all the interacting less frequently mutated genes for which adequate shRNAs were available within this small subnetwork scored positive in our screen (Supporting Information Fig. S2). Four of them (RPA3, RFC, LIG1, and HUWE1), together with TP53 and ERCC6 have established roles in DNA repair. We next placed all the positive hits from both of our screens as the central nodes and explored additional interactors among those less frequently mutated CRC genes by extending the interaction map using the same network analysis (Supporting Information Fig. S3). Enrichment analysis for all the genes within this extended map using Molecular Signature Database showed that 7 out of all the enriched pathways are related to this process, such as double-strand break repair, homologs recombination repair, and nucleotide excision repair (Supporting Information Table S6). Although it is well established that deficiency in these pathways contribute to tumorigenesis,16–18 it remains largely controversial whether these proteins function as tumor suppressors.19–21 Thus, our study provides the first direct evidence for the suppressive role of these proteins in tumorigenesis.
Functional enrichment analysis for all the genes within this extended interaction map revealed the enhancement for various aspects of biological processes among which cyclic nucleotide metabolic process is the most highly enriched (Supporting Information Table S7). All the tested genes involved in this process (GNAS, ADCY9, ADORA1, and GUCY1A2) scored positive in our screen, supporting the important role of cyclic nucleotide processes in colon tumor development. These two sub-networks (TP53 and cyclic nucleotides) are representative of the central components of the regulatory network underlying CRC tumorigenesis (Supporting Information Fig. S4). We then performed mutual exclusivity/co-occurrence analysis based on TCGA CRC dataset using odds ratio and Fisher's exact tests. We found that there was a strong tendency towards co-occurrence for most pairs of the genes present in DNA repair subnetwork (p < 0.05), suggesting that these genes harbor additive effects in tumor development and disruption of various aspects of DNA repair processes might contribute to tumorigenesis, or they may play a role in other pathways beyond DNA repair. In contrast, no such strong tendency was observed among the genes implicated in cyclic nucleotide metabolic process. Notably, GNAS and GHRHR tended to be mutually exclusive with the other five genes within this subnetwork (Supporting Information Tables 8A and B). Similar analysis performed for another small transcriptional control central hub consisting of TP53, TGFBR2, and SMAD2, which were among the most frequent mutations present in CRC and HUWE1, a newly identified driver mutation from our screen showed that TGFBR2 and TP53 tended to be mutually exclusive whereas TGFBR2 and HUWE1 tended to co-occur in CRC tumor samples (Supporting Information Table 8C). Taken together, these observations indicate the important roles of DNA repair pathways, cyclic nucleotide metabolism and many other biological processes in the regulation of colon tumor development and revealed molecular relationships among the protein components within the representative central subnetworks. These results provide a platform for future system-level analyses as well as detailed mechanistic investigations.
Discussion
This study functionally interrogated the putative passenger mutated genes in the CRC genome by using a relevant biological transformation assay in combination with network analysis. We identified a number of novel driver tumor suppressors among the genes that are otherwise considered passengers. We discovered that driver mutations are enriched in the putative passenger mutations interacting with confirmed CAN-genes, and there are far more “drivers” even among the noninteracting “passengers.” Additionally, we demonstrated that a subset of newly identified candidate tumor suppressors are also involved in invasion through the basement membrane and thus may play important roles in multiple stages of CRC tumorigenesis. These multifunctional low frequency mutated genes may represent a pool of novel potential drug targets that were previously unknown or misclassified as passenger mutations. For example, ADORA1, a newly identified tumor suppressor from the present study, has recently been shown to suppress CW2 human colonic cancer growth by inducing apoptosis.22 Thus ADORA1 might be a novel drug target considering the availability of highly selective and potent agonists against this adenosine receptor.23 In conclusion, the observation that a large fraction of the low frequently mutated genes have been misclassified as passengers reveals that frequency-based biostatistic models performed poorly for parsing driver mutations and demonstrate the need for implementing biologically relevant functional filters to distinguish between driver and passenger mutations instead of solely depending on bioinformatics.
Supplementary Material
What's new?
Although the field of cancer genomics has produced long lists of mutated genes from many types of cancer, researchers have assumed that most of these are “passenger” mutations that aren't directly involved in tumorigenesis. However, by using RNA interference to inactivate particular genes and mapping their interactions with known “driver” mutations (i.e. those that lead to malignant growth), the authors were able to uncover a number of potential tumor-suppressors among genes that had previously been dismissed. Combining functional assays with biological filters may lead to better predictive models and more potential therapeutic targets than bioinformatics alone.
Acknowledgements
We thank Michael A. White and Ugur Eskiocak for valuable discussions.
Grant sponsor: CPRIT Training; Grant number: RP101496; Grant sponsor: NASA; Grant numbers: NNX09AU95G, NNX11AC15G, NNX11AC54G
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 2.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parmigiani G, Boca S, Lin J, et al. Design and analysis issues in genome-wide somatic mutation studies of cancer. Genomics. 2009;93:17–21. doi: 10.1016/j.ygeno.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frohling S, Scholl C, Levine RL, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–13. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–63. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu ET. Functional genomics of cancer. Curr Opin Genet Dev. 2008;18:251–6. doi: 10.1016/j.gde.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Chautard E, Thierry-Mieg N, Ricard-Blum S. Interaction networks: from protein functions to drug discovery. A review. Pathol Biol (Paris) 2009;57:324–33. doi: 10.1016/j.patbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Tewari M, Hu PJ, Ahn JS, et al. Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-beta signaling network. Mol Cell. 2004;13:469–82. doi: 10.1016/s1097-2765(04)00033-4. [DOI] [PubMed] [Google Scholar]
- 10.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 11.Mori S, Chang JT, Andrechek ER, et al. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene. 2009;28:2796–805. doi: 10.1038/onc.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskiocak U, Kim SB, Ly P, et al. Functional parsing of driver mutations in the colorectal cancer genome reveals numerous suppressors of anchorage-independent growth. Cancer Res. 2011;71:4359–65. doi: 10.1158/0008-5472.CAN-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roig AI, Eskiocak U, Hight SK, et al. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138:1012–21. e1–5. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 14.Komurov K. Modeling community-wide molecular networks of multicellular systems. Bioinformatics. 2012;28:694–700. doi: 10.1093/bioinformatics/btr718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–9. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–54. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 17.Hung RJ, Hall J, Brennan P, et al. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–42. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 18.Jiricny J, Marra G. DNA repair defects in colon cancer. Curr Opin Genet Dev. 2003;13:61–9. doi: 10.1016/s0959-437x(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 19.Ljuslinder I, Golovleva I, Palmqvist R, et al. LRIG1 expression in colorectal cancer. Acta Oncol. 2007;46:1118–22. doi: 10.1080/02841860701426823. [DOI] [PubMed] [Google Scholar]
- 20.Givalos N, Gakiopoulou H, Skliri M, et al. Replication protein A is an independent prognostic indicator with potential therapeutic implications in colon cancer. Mod Pathol. 2007;20:159–66. doi: 10.1038/modpathol.3800719. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Seth AK, Aplin AE. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol Cancer Res. 2006;4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 22.Saito M, Yaguchi T, Yasuda Y, et al. Adenosine suppresses CW2 human colonic cancer growth by inducing apoptosis via A(1) adenosine receptors. Cancer Lett. 2010;290:211–5. doi: 10.1016/j.canlet.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Lohse MJ, Klotz KN, Schwabe U, et al. 2-Chloro-N6-cyclopentyladenosine: a highly selective agonist at A1 adenosine receptors. Naunyn Schmiedebergs Arch Pharmacol. 1988;337:687–9. doi: 10.1007/BF00175797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.