Abstract
Preeclampsia (PE) is a pregnancy-related disease with potentially severe consequences with respect to foeto-maternal morbidity and mortality. However, the molecular pathogenesis of PE remains largely unknown. Recent reports have shown that microRNAs (miRNAs or miRs) may play important roles in the development of PE. Analysing the miRNAs in sera from preeclamptic women may improve our understanding of the pathophysiological mechanisms of the disease. The aim of this retrospective study was to identify whether circulating miRNAs were differentially expressed in PE patients compared with controls. Serum samples from 23 women who developed PE were compared with samples from 44 pregnant controls. Seventeen circulating miRNAs previously described in PE were chosen for evaluation of their expression by reverse transcription quantitative polymerase chain reaction (RT-qPCR). In the maternal serum, the miR-210-3p, miR-210-5p, miR-1233-3p, and miR-574-5p levels were found to be significantly higher in the PE patients than in the controls (P<0.05). Using a logistic regression model, we evaluated the discriminant power of those differentially expressed miRNAs, and the combination of miR-210-5p and miR-574-5p yielded an area under the curve of 0.7223 for discriminating PE patients from the controls. In conclusion, the fact that four circulating miRNAs (miR-210-3p, miR-210-5p, miR-1233-3p, and miR-574-5p) were differentially expressed in the sera of women who developed PE compared with controls confirms the possible pathophysiological role of miRNAs in PE.
Keywords: preeclampsia, miRNAs, RT-qPCR, logistic regression model
Introduction
Preeclampsia (PE) is a pregnancy-specific syndrome associated with hypertension and proteinuria or thrombocytopenia, renal insufficiency, impaired liver function, pulmonary oedema, cerebral symptoms, and visual symptoms (1). PE affects 2–5% of pregnancies worldwide (2–4). It can lead to severe maternal and perinatal morbidity, including uterine growth restriction and prematurity. PE is responsible for an elevated perinatal mortality rate and causes 10–15% of maternal deaths (3,5).
The placenta plays a key role in the initiation and progression of the disorders caused by PE (6). As commonly described, PE has a combined genetic, immune, and angiogenic aetiology. The origin of PE seems to be the impaired invasion of maternal spiral arteries by trophoblast cells in the context of local abnormal immune interactions between the fetoplacental unit and mother (7–9). Placental underperfusion induces a chronic hypoxic state (10). Combined with the reoxygenation that occurs, it creates increased local oxidative stress, resulting in apoptosis and necrosis of the placenta. Placental pro-inflammatory and antiangiogenic mediators and apoptotic bodies, released in the maternal circulation, cause maternal systemic endothelial cell dysfunction and systemic inflammation (5,11). Although several hypotheses have been suggested to explain this vascular, multi-systemic pregnancy-related disease, its complete pathogenesis remains poorly understood (12,13), and PE prevention, diagnosis, and treatment remain an important challenge.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs (20–24 nucleotides) that regulate gene expression through post-transcriptional repression or degradation of messenger RNA (14,15). miRNAs are present in animals and plants and have been detected in several human fluids (16). They are critical in many biological processes, including the regulation of cell differentiation, proliferation, apoptosis, angiogenesis, and metabolism (17). Specific plasma-related miRNAs were identified in maternal plasma or serum in normal pregnancies, as well as in pathological pregnancy conditions, such as PE (18,19). Several recent reports described the significant dysregulation of specific miRNA expression in the maternal circulation of PE patients compared with controls (miR-24, miR-26a, miR-29a, miR-103, miR-130b, miR-141, miR-144, miR-152, miR-181a, miR-210, miR-342-3p, miR-520, miR-574-5p, and miR-1233) (20–23).
We hypothesized that the expression levels of specific miRNAs in maternal blood could be dysregulated before the onset of PE and be used to enhance screening, diagnosis, or prognosis. We aimed to analyse, in a case-control study, several miRNAs that were previously described as differentially expressed in PE.
Materials and methods
miRNA selection
After performing a review on PubMed until 26 February, 2015, using the key words, ‘miRNA, pregnancies, preeclampsia, placenta, circulating’, and consulting the miRNA database, 17 miRNAs were selected (Table I).
Table I.
Primers used in quantitative PCR.
| miRNA | miRBase | Reference Qiagen | Sequence of primer (5′-3′) |
|---|---|---|---|
| hsa-miR-144-3p | MIMAT0000436 | MS00020328 | TACAGTATAGATGATGTACT |
| hsa-miR-29a-3p | MIMAT0000086 | MS00003262 | TAGCACCATCTGAAATCGGTTA |
| hsa-miR-210-3p | MIMAT0000267 | MS00003801 | CTGTGCGTGTGACAGCGGCTGA |
| hsa-miR-210-5p | MIMAT0026475 | MS00045836 | AGCCCCTGCCCACCGCACACTG |
| hsa-miR-1233-3p | MIMAT0005588 | MS00008512 | TGAGCCCTGTCCTCCCGCAG |
| hsa-miR-1233-5p | MIMAT0022943 | MS00045213 | AGTGGGAGGCCAGGGCACGGCA |
| hsa-miR-24-3p | MIMAT0000080 | MS00006552 | TGGCTCAGTTCAGCAGGAACAG |
| hsa-miR-26a-5p | MIMAT0000082 | MS00029239 | TTCAAGTAATCCAGGATAGGCT |
| hsa-miR-130b-3p | MIMAT0000691 | MS00003451 | CAGTGCAATGATGAAAGGGCAT |
| hsa-miR-130b-5p | MIMAT0004680 | MS00008610 | ACTCTTTCCCTGTTGCACTAC |
| hsa-miR-181a-5p | MIMAT0000256 | MS00008827 | AACATTCAACGCTGTCGGTGAGT |
| hsa-miR-181a-3p | MIMAT0000270 | MS00006692 | ACCATCGACCGTTGATTGTACC |
| hsa-miR-342-3p | MIMAT0000753 | MS00004011 | TCTCACACAGAAATCGCACCCGT |
| hsa-miR-574-5p | MIMAT0004795 | MS00043617 | TGAGTGTGTGTGTGTGAGTGTGT |
| hsa-miR-16-2-3p | MIMAT0004518 | MS00008813 | CCAATATTACTGTGCTGCTTTA |
| hsa-miR-124-3p | MIMAT0000422 | MS00006622 | TAAGGCACGCGGTGAATGCC |
| hsa-miR-155-5p | MIMAT0000646 | MS00031486 | TTAATGCTAATCGTGATAGGGGT |
| c-miR-39 | MIMAT0000010 | MS00019789 | TCACCGGGTGTAAATCAGCTTG |
| RNU6-2 | MS00033740 |
RNU6, U6 small nuclear RNA; hsa, Homo sapiens; c, Caenorhabditis elegans.
Study population
In this retrospective study, 67 pregnant women, aged 19–44 years, with a gestational age of 24–36 weeks and 6 days of amenorrhea, were selected among a prospective cohort of pregnant women presenting, at 24 to <37 weeks' gestation, clinical suspicion of, but not manifesting preeclampsia/eclampsia/HELLP syndrome. Suspicion of PE was assessed on the basis of the following criteria: New elevation of blood pressure (<140/90 mmHg), with or without pre-existing essential hypertension; de novo or major proteinuria (<30 mg/dl or <2+ on a urine dipstick); and clinical (epigastric pain, headache, visual disturbances, and excessive weight gain), biological (thrombocytopenia or elevation of transaminases), and ultrasonic (intrauterine growth restriction, elevation of the uterine artery pulsatility index, or uterine artery notch) characteristics favouring PE. Manifestation of PE at selection was an exclusion criterion.
In each patient presenting with the aforementioned signs and symptoms who agreed to participate, blood was collected and further pregnancy follow-up was performed. Retrospectively, the PE and control cohorts were defined based on the diagnosis of PE at delivery.
The present study was approved by the Ethics Committee of the CHR Citadelle Hospital, Liège, Belgium, and informed consent was obtained from all the patients.
Sample collection
Blood samples were immediately centrifuged at 1,000 × g for 15 min, and sediment-free serum samples were collected. The serum aliquots were frozen at −80°C until further analysis and were thawed only once thereafter.
miRNA extraction
Total RNA, including miRNA, was extracted from 200 µl of serum using the miRNeasy Serum/Plasma kit (Qiagen; Hilden, Germany) according to the manufacturer's instructions, and it was then eluted in 30 µl of nuclease-free water. A synthetic spike-in control miRNA (C. elegans miR-39 mimic, Qiagen) was added for subsequent normalisation.
Quantitative polymerase chain reaction (qPCR) for miRNAs
Total miRNA (2 µl) from each sample was reverse-transcribed with the miScript II TR kit (Qiagen) according to the manufacturer's instructions. The miRNAs were subsequently quantified using the miScript SYBR-Green PCR kit (Qiagen) and specific forward primers. The reaction mixture included 2.5 µl of cDNA, 12.5 µl of QuantiTect SYBR-Green PCR Master Mix (Qiagen), 2.5 µl of forward primer, 2.5 µl of miScript Universal Primer (Qiagen), and 5 µl of RNase-free water for a final reaction volume of 25 µl. The thermal cycling consisted of an initial denaturation at 95°C for 15 min, followed by 45 cycles at 95°C for 15 sec, 55°C for 30 sec, and 70°C for 30 sec.
The reactions were run in duplicate. Threshold cycle (Cq) and melting curves were acquired using the quantification and melting curve programs of the LightCycler® 96 Real-Time System (Roche Diagnostics GmbH, Mannheim, Germany). U6 small nuclear RNA (RNU6) and c-miR-39 were used to determine relative miRNA expression with the 2-ΔCq method (24–26). The miRNA-specific primer sequences are listed in Table I.
Statistical analysis
Quantitative parameters are presented as the means ± standard errors or medians. The statistical significance of differences in the expression levels of serum miRNAs between PE patients and controls was determined by the non-parametric Mann-Whitney U-Wilcoxon test. To determine the diagnostic accuracy of the miRNA expression levels, we generated receiver operating characteristic (ROC) curves and calculated the area under the ROC curves (AUC) (27). An evaluation of the diagnostic power of multiple combinations of the miRNA serum levels was performed using a logistic regression model. For all the statistical analyses, we used the statistics toolbox of Matlab R2024a software (MathWorks Inc., Natick, MA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics
Table II shows the main features of the patients in each group. The median of gestational ages at inclusion (min-max) of the pregnant women were 32.1 (25.3–36.6) (PE patients) and 31.1 (24.3–36.5) weeks (controls). There were no significant differences between the subjects and controls with respect to gestational age, maternal age, body mass index, smoking status, parity, gestity, ethnicity, personal and family history of PE, previous hypertension, proteinuria, and mean uterine artery pulsatility index. The median of gestational ages (min-max) at delivery were 36.4 (30.0–39.3) (PE patients) and 39.1 (35.2–41.3) weeks (controls) (P<0.0001).
Table II.
Demographic and clinical characteristics of controls and preeclamptic (PE) pregnancies.
| Characteristics | Controls (n=44) | PE (n=23) | P-value |
|---|---|---|---|
| Maternal age (years) | 30 (19–38) | 29 (19–44) | 0.82701 |
| Gestational age at inclusion (weeks) | 31.1 (24.3–36.5) | 32.1 (25.3–36.6) | 0.51313 |
| Gestational age at delivery (weeks) | 39.1 (35.2–41.3) | 36.4 (30.0–39.3) | <0.0001 |
| Body mass index | 27.2 (16.1–39.30) | 27.40 (19–41.5) | 0.85331 |
| Gestity | 2 (1–6) | 2 (1–7) | 0.19814 |
| Parity | 1 (0–5) | 0 (0–3) | 0.13570 |
| Smoking status | |||
| Previous smokers | 6 (13.6%) | 3 (13%) | 0.66976 |
| Smokers | 4 (9%) | 3 (13%) | |
| Previous HTA | 3 (6.8%) | 2 (8.7%) | 0.79403 |
| Family history of PE | 7 (16%) | 6 (26%) | 0.31431 |
| Proteinuria | 36 (81.8%) | 19 (82.6%) | 0.68365 |
| Mean artery pulsatility index | 0.77 (0.36–1.50) | 0.86 (0.37–1.88) | 0.27001 |
| Systolic blood pressure (mmHg) | 129 (105–160) | 134 (120–159) | 0.01763 |
| Diastolic blood pressure (mmHg) | 80 (60–100) | 85 (70–100) | 0.01551 |
Values are presented as median (min-max) or numbers (percentage). Statistical analyses were performed by the Mann-Whitney U-Wilcoxon test.
Differential expression of circulating miRNAs
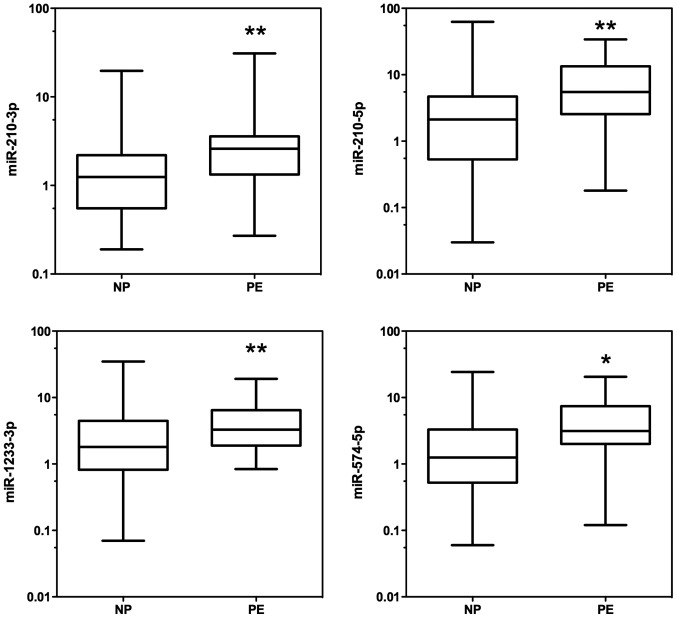
The expression levels of the 17 miRNAs (miR-16-2-3p, miR-24-3p, miR-26a-5p, miR-29a-3p, miR-124-3p, miR-130b-5p miR-130b-3p, miR-144-3p, miR-155-5p, miR-181a-3p, miR-181a-5p, miR-210-3p, miR-210-5p, miR-342-3p, miR-574-5p, miR-1233-3p, and miR-1233-5p) from the serum samples in the two groups (PE patients and controls) were analysed with reverse transcription quantitative polymerase chain reaction (RT-qPCR). According to RT-qPCR analysis, there was a significant upregulation of 4 miRNAs: miR-210-3p, miR-210-5p, miR-1233-3p, and miR-574-5p, in the serum of PE patients compared with controls (P<0.05) (Table III, Fig. 1).
Table III.
Circulating miRNA expression levels in controls and preeclamptic pregnancies.
| miRNA | Controls (n=44) | PE (n=23) | P-value (Wilcoxon) |
|---|---|---|---|
| miR-210-3p | 2.25±0.51 | 4.10±1.30 | 0.005 |
| miR-210-5p | 5.46±1.78 | 9.18±2.00 | 0.006 |
| miR-1233-3p | 3.73±0.96 | 4.97±0.98 | 0.021 |
| miR-574-5p | 2.47±0.59 | 5.84±1.30 | 0.005 |
| miR-144-3p | 2.47±0.49 | 3.56±1.05 | 0.187 |
| miR-29a-3p | 2.84±0.46 | 2.85±0.30 | 0.129 |
| miR-1233-5p | 3.23±0.86 | 2.91±0.42 | 0.129 |
| miR-24-1-3p | 2.99±0.46 | 3.15±0.35 | 0.088 |
| miR-26a-5p | 2.63±0.36 | 2.41±0.33 | 0.736 |
| miR-130b-3p | 3.55±0.69 | 3.22±0.40 | 0.352 |
| miR-130b-5p1 | 4.39±0.87 | 4.92±0.92 | 0.113 |
| miR-181a-5p | 2.04±0.27 | 2.27±0.32 | 0.222 |
| miR-181a-3p | 2.63±1.17 | 2.28±0.61 | 0.113 |
| miR-342-3p | 3.06±0.39 | 3.18±0.34 | 0.282 |
| miR-16-2-3p | 2.14±0.36 | 3.27±0.90 | 0.124 |
| miR-124-3p | 3.51±0.72 | 2.97±0.35 | 0.362 |
| miR-155-5p | 3.58±0.62 | 3.76±0.65 | 0.402 |
Figure 1.
miRNAs differentially expressed in the sera of control pregnant women (NP) and preeclamptic women (PE). The expression of levels of miR-210-3p, miR-210-5p, miR-1233-3p and miR-574-5p were higher in preeclamptic women. Data are presented by box-and-whisker plots. Boxes indicate the 25th and 75th percentiles. The solid line within the box indicates the median value. *P<0.05 and **P<0.001.
Assessment of the screening value of circulating miRNAs in PE
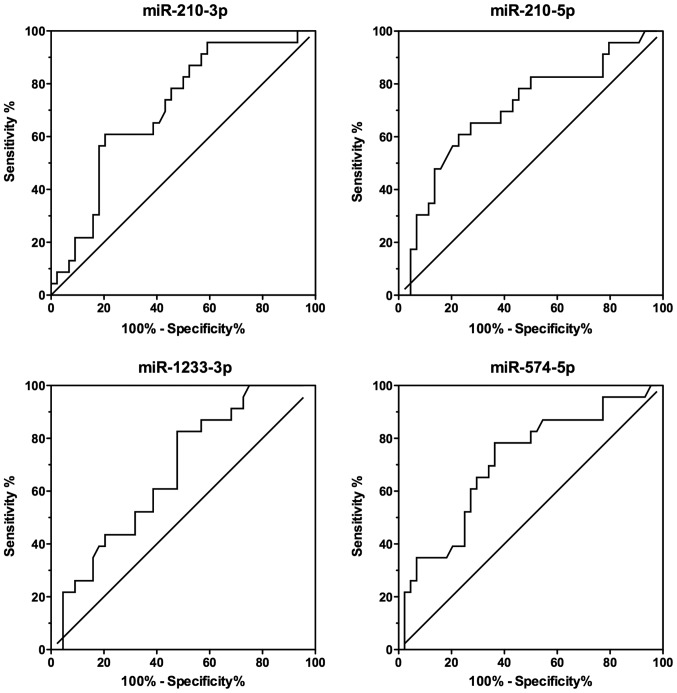
We constructed and examined the ROC curves of the 4 miRNAs that were upregulated in PE serum to evaluate their potential utility as biomarkers (Fig. 2). The AUCs of miR-210-3p, miR-210-5p, miR-1233-3p, and miR-574-5p were 0.7090 [95% confidence interval (CI), 0.582–0.836], 0.7040 (95% CI, 0.568–0.840), 0.673 (95% CI, 0.543–0.803), and 0.710 (95% CI, 0.578–0.842), respectively (Table IV). To improve the diagnostic power of the miRNA expression levels, we applied a logistic regression model to the data from PE patients and controls. Different combinations were added to the model, and a maximum AUC of 0.7223 was found for miR-210-5p and miR-574-5p (Fig. 3). The fitted logistic regression model is as follows:
Figure 2.
Receiver operating characteristic (ROC) curves of different microRNAs.
Table IV.
ROC results for several miRs.
| miRs | Area under the ROC curve (SE) | 95% CI | P-value |
|---|---|---|---|
| miR-210-3p | 0.7090 (0.06496) | 0.5816–0.8363 | 0.005 |
| miR-210-5p | 0.7041 (0.06918) | 0.5684–0.8397 | 0.006 |
| miR-1233-3p | 0.6729 (0.06632) | 0.5429–0.8029 | 0.021 |
| miR-574-5p | 0.7100 (0.06717) | 0.5783–0.8417 | 0.005 |
CI, confidence interval; SE, standard error; miRs, microRNAs.
Figure 3.
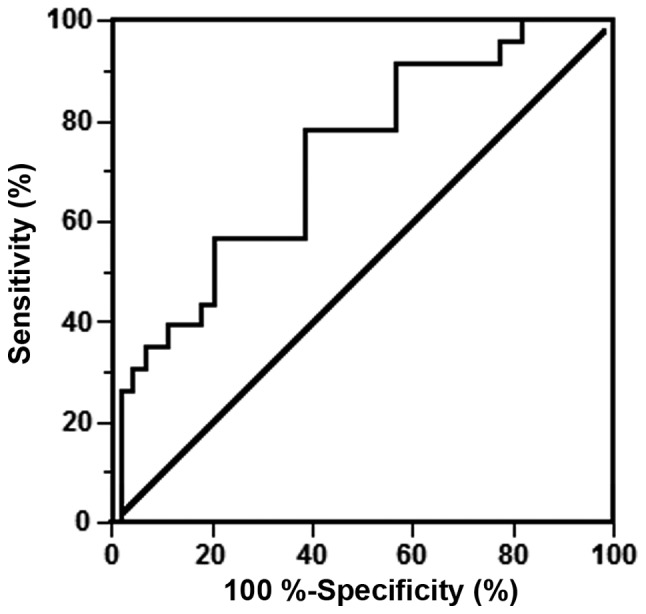
Receiver operating characteristic (ROC) curves of combined microRNAs. Logistic regression model.
where P(Y=1) is the probability that the sample is a preeclamptic sample.
Discussion
Recently, the aberrant expression of miRNAs has been documented in different pregnancy-related conditions, such as PE, ectopic pregnancy, gestational diabetes mellitus, small for gestational age, recurrent pregnancy loss, and preterm delivery (28). These findings may be useful to improve our understanding of the underlying pathophysiology of these conditions.
In the present study, we confirmed the differential expression of several miRNAs that were identified in previous studies (20–23). Of the 17 miRNAs selected, miR-210-3p, miR-210-5p, miR-1233-3p, and miR-574-5p were upregulated in the sera of women who later developed PE compared with the controls who did not develop PE according to RT-qPCR. By performing logistic regression analysis of the differentially expressed miRNAs, we found that the combination of miR-210-5p and miR-574-5p had the best diagnostic power with an AUC of 0.7223.
In our selection of miRNAs based on the data published in previous studies, we found very little overlap between the reported studies. This may be due to differences in the sample collection (sera versus plasma) or methods used for the analyses (RT-qPCR versus microarrays). Even results obtained from the same platform using products from different companies may be responsible for this low correlation of results (29,30). RT-qPCR is often considered as the ‘gold standard’ for the detection and quantification of gene expression. As a result, we chose to validate previously identified miRNA results using this procedure. However, one important step of RT-qPCR is normalisation of the miRNA expression level. Using improper reference genes for normalisation can result in evaluation bias during data analysis. To the best of our knowledge, there has been no consensus regarding a reliable housekeeping miRNA for analysing circulating miRNAs. Therefore, it becomes difficult to directly compare the results from different studies. In the present study, we used a double normalisation procedure, including the use of the synthetic c-miR-39 and RNU6B. The same amount of c-miR-39 was added to each sample before the extraction procedure, and the c-miR-39 expression level reflected differences at the same time in the extraction, reverse transcription, and PCR amplification procedures (31). However, the use of RNU6B complements technical bias with biological bias. Although RNU6B does not necessarily represent the ‘best’ reference, it has been previously used to analyse miRNAs in the sera of preeclamptic women (23,32).
The selection of the patients included in different studies may also introduce some bias. To identify new biomarkers for PE, a common strategy is to analyse a few samples or a pool of samples when performing miRNA profiling. This step is further followed by a validation procedure, frequently RT-qPCR, with additional samples from PE cases and controls. Usually, the second step does not completely validate the profiling step (18,20,21). Thus, in order to evaluate previously differentially reported miRNAs in PE, we performed a case-control study with 23 PE patients and 44 controls. The two groups were found to be comparable with respect to their maternal age, gestational age at inclusion, gestity, and parity. However, the retrospective nature of the investigation was one limitation of the study. A second limitation is that we were unable to separately analyse early- and late-onset PE due to the small sample size.
There is currently no consensus regarding a set of differentially expressed miRNAs in PE that may be used as biomarkers. Our approach was to collect several potentially interesting miRNAs that were previously described in the literature and test them in our set of case-control samples.
Notably, miR-210, which is one of the more commonly recurring miRNAs overexpressed in PE (22,32–36), was also elevated in our PE cohort. This particular miR-210 has been identified as a hypoxia-induced miRNA in many cell types and tissues and is consistently upregulated by hypoxia in both physiological and malignant conditions (37,38). Several target genes with roles in mitochondrial metabolism, angiogenesis, and DNA repair have been identified, and most of them may be involved in the pathogenesis of PE. However, it remains unclear whether the differential expression of miR-210 is the consequence or origin of PE even though this particular miRNA has been described as a serum biomarker for PE (32).
There was also a higher level of circulating miR-1233 in the group of women who developed PE compared with the group of women with a normal pregnancy. Our results confirmed those of Ura et al (23), who were the first researchers to identify a potential role for miR-1233, which has already been described in renal carcinoma, in predicting PE. It is important to note that both miR-1233 and miR-210, which are upregulated in PE, are also upregulated in renal cell carcinoma. Of note, in patients with renal cell carcinoma, only miR-1233 was found to be increased (39). Information on the function of miR-1233, located on chromosome 15q14, is incomplete. Several potential target genes have been predicted using TargetScan 6.0 (40), but they have not been validated. The expression level of miR-574-5p was also elevated in the blood of women with PE (21). However, the expression of miR-574-5p has been poorly investigated.
Although there are many studies on PE screening, a biologically predictive and prognostic test with high accuracy for routine clinical use has yet to be identified (41–43). The most promising strategies are multiparametric approaches that include a combination of maternal factors, biophysical tests (mean blood pressure, uterine artery Doppler), and biochemical markers (44–50). Sequential multiparametric testing is superior to screening in the first trimester alone (51). Nevertheless, a panel of tests is necessary to effectively identify women early in pregnancy who are at the highest risk for developing PE.
Although the prevalence of PE is relatively low and miRNA analysis is technically challenging, further studies with larger cohorts are mandatory. Discriminating between the different phenotypes of PE is also important because they may involve different sets of miRNA expression.
Furthermore, patients were sampled before the onset of PE and delivery (mean:one month before). Indeed, cases were enrolled when there was suspicion of PE, but manifest PE was an exclusion criterion. This protocol may partially explain the reason for only 4 of the 17 selected miRNAs being significantly upregulated.
The fact that four circulating miRNAs (miR-210-3p, miR-210-5p, miR-1233-3p, and miR-574-5p) were differentially expressed in the sera of women who later developed PE compared with women who did not develop PE, confirms the possible pathophysiological role of miRNAs in PE, as previously suggested (19,52,53). Particularly, exploring the role of miR-1233 and miR-574 in PE could enrich our understanding of this disease.
Future research on the biological pathway of circulating miRNAs may enhance our understanding of the pathogenesis of PE and aid in the development of new biomarkers for clinical application. Using these miRNAs in a multiparametric test may also provide a new clinical strategy for identifying women who are at risk for developing PE.
Acknowledgements
The authors acknowledge Nathalie Lefin and Sophie Kessler for their excellent technical assistance. C.M. is a Research Associate from the Fonds de la Recherche Scientifique-FNRS (F.R.S.-FNRS, Belgium). This study was supported by grants from the Fonds de la Recherche Scientifique-FNRS (F.R.S.-FNRS, Belgium), the Fonds spéciaux de la Recherche (University of Liège), the Fonds Léon Fredericq (University of Liège), the Direction Générale Opérationnelle de l'Economie, de l'Emploi et de la Recherche from the S.P.W. (Région Wallonne, Belgium).
Glossary
Abbreviation
- PE
preeclampsia
References
- 1.American College6 of Obstetricians and Gynecologists, corp-author. Task Force on Hypertension in Pregnancy: Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists' Task Force on Hypertension in Pregnancy. Obstet Gynecol. 2013;122:1122–1131. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- 2.Redman CW, Sargent IL. Latest advances in understanding preeclampsia. Science. 2005;308:1592–1594. doi: 10.1126/science.1111726. [DOI] [PubMed] [Google Scholar]
- 3.Duley L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol. 2009;33:130–137. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Report of the National High Blood Pressure Education Program Working Group on High Blood Pressure in Pregnancy. Am J Obstet Gynecol. 2000;183:S1–S22. doi: 10.1067/mob.2000.107928. [DOI] [PubMed] [Google Scholar]
- 5.Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet. 2005;365:785–799. doi: 10.1016/S0140-6736(05)71003-5. [DOI] [PubMed] [Google Scholar]
- 6.Huppertz B. Placental origins of preeclampsia: Challenging the current hypothesis. Hypertension. 2008;51:970–975. doi: 10.1161/HYPERTENSIONAHA.107.107607. [DOI] [PubMed] [Google Scholar]
- 7.Foidart JM, Noël A, Chantraine F, Lorquet S, Petit P, Munaut C, Berndt S, Pequeux C, Schaaps JP. Defective placental implantation and its effects on maternal endothelial function. Bull Acad Natl Med. 2009;193:1059–1064. discussion 1064–1066, 1067–1068. (In French) [PubMed] [Google Scholar]
- 8.Foidart JM, Schaaps JP, Chantraine F, Munaut C, Lorquet S. Dysregulation of anti-angiogenic agents (sFlt-1, PLGF, and sEndoglin) in preeclampsia-a step forward but not the definitive answer. J Reprod Immunol. 2009;82:106–111. doi: 10.1016/j.jri.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Redman CW, Sargent IL, Staff AC. IFPA Senior Award Lecture: Making sense of pre-eclampsia - two placental causes of preeclampsia? Placenta. 2014;35:S20–S25. doi: 10.1016/j.placenta.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Munaut C, Lorquet S, Pequeux C, Blacher S, Berndt S, Frankenne F, Foidart JM. Hypoxia is responsible for soluble vascular endothelial growth factor receptor-1 (VEGFR-1) but not for soluble endoglin induction in villous trophoblast. Hum Reprod. 2008;23:1407–1415. doi: 10.1093/humrep/den114. [DOI] [PubMed] [Google Scholar]
- 11.Roberts JM, Hubel CA. The two stage model of preeclampsia: Variations on the theme. Placenta. 2009;30:S32–S37. doi: 10.1016/j.placenta.2008.11.009. (Suppl A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redman CW, Sargent IL. Immunology of pre-eclampsia. Am J Reprod Immunol. 2010;63:534–543. doi: 10.1111/j.1600-0897.2010.00831.x. [DOI] [PubMed] [Google Scholar]
- 13.Dechend R, Staff AC. Placenta messages to the mother: Not just debris. Hypertension. 2012;59:191–193. doi: 10.1161/HYPERTENSIONAHA.111.184861. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai EC. Micro RNAs are complementary to 3 UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 16.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong SG, Lee WH, Kodo K, Wu JC. MicroRNA-mediated regulation of differentiation and trans-differentiation in stem cells. Adv Drug Deliv Rev. 2015;88:3–15. doi: 10.1016/j.addr.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura K, Miura S, Yamasaki K, Higashijima A, Kinoshita A, Yoshiura K, Masuzaki H. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem. 2010;56:1767–1771. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 19.Tsochandaridis M, Nasca L, Toga C, Levy-Mozziconacci A. Circulating microRNAs as clinical biomarkers in the predictions of pregnancy complications. BioMed Res Int. 2015;2015:294954. doi: 10.1155/2015/294954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ge Q, Guo L, Lu Z. Maternal plasma miRNAs expression in preeclamptic pregnancies. Biomed Res Int. 2013;2013:970265. doi: 10.1155/2013/970265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu L, Zhou H, Lin H, Qi J, Zhu C, Gao Z, Wang H. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction. 2012;143:389–397. doi: 10.1530/REP-11-0304. [DOI] [PubMed] [Google Scholar]
- 22.Gunel T, Zeybek YG, Akçakaya P, Kalelioğlu I, Benian A, Ermis H, Aydınlı K. Serum microRNA expression in pregnancies with preeclampsia. Genet Mol Res. 2011;10:4034–4040. doi: 10.4238/2011.November.8.5. [DOI] [PubMed] [Google Scholar]
- 23.Ura B, Feriotto G, Monasta L, Bilel S, Zweyer M, Celeghini C. Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan J Obstet Gynecol. 2014;53:232–234. doi: 10.1016/j.tjog.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Kok MG, Halliani A, Moerland PD, Meijers JC, Creemers EE, Pinto-Sietsma SJ. Normalization panels for the reliable quantification of circulating microRNAs by RT-qPCR. FASEB J. 2015;29:3853–3862. doi: 10.1096/fj.15-271312. [DOI] [PubMed] [Google Scholar]
- 27.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem. 2013;46:953–960. doi: 10.1016/j.clinbiochem.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Gelfond JA, McManus LM, Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 2009;10:407. doi: 10.1186/1471-2164-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato F, Tsuchiya S, Terasawa K, Tsujimoto G. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS One. 2009;4:e5540. doi: 10.1371/journal.pone.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts TC, Coenen-Stass AML, Wood MJA. Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One. 2014;9:e89237. doi: 10.1371/journal.pone.0089237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anton L, Olarerin-George AO, Schwartz N, Srinivas S, Bastek J, Hogenesch JB, Elovitz MA. miR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. Am J Pathol. 2013;183:1437–1445. doi: 10.1016/j.ajpath.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261.e1–261.e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204:178.e12–178.e21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muralimanoharan S, Maloyan A, Mele J, Guo C, Myatt LG, Myatt L. miR-210 modulates mitochondrial respiration in placenta with preeclampsia. Placenta. 2012;33:816–823. doi: 10.1016/j.placenta.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Fei M, Xue G, Zhou Q, Jia Y, Li L, Xin H, Sun S. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: New insights into molecular mechanisms for the disease. J Cell Mol Med. 2012;16:249–259. doi: 10.1111/j.1582-4934.2011.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 38.Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel G, von Ruecker A, Müller SC, et al. MicroRNAs in renal cell carcinoma: Diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6:e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 41.Grill S, Rusterholz C, Zanetti-Dällenbach R, Tercanli S, Holzgreve W, Hahn S, Lapaire O. Potential markers of preeclampsia-a review. Reprod Biol Endocrinol. 2009;7:70. doi: 10.1186/1477-7827-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meads CA, Cnossen JS, Meher S, Juarez-Garcia A, ter Riet G, Duley L, Roberts TE, Mol BW, van der Post JA, Leeflang MM, et al. Methods of prediction and prevention of pre-eclampsia: systematic reviews of accuracy and effectiveness literature with economic modelling. Health Technol Assess. 2008;12:1–270. doi: 10.3310/hta12060. [DOI] [PubMed] [Google Scholar]
- 43.Zakiyah N, Postma MJ, Baker PN, van Asselt AD. IMPROvED Consortium: Pre-eclampsia diagnosis and treatment options: A review of published economic assessments. Pharmacoeconomics. 2015;33:1069–1082. doi: 10.1007/s40273-015-0291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon LC, Stratieva V, Piras S, Piri S, Nicolaides KH. Hypertensive disorders in pregnancy: Combined screening by uterine artery Doppler, blood pressure and serum PAPP-A at 11–13 weeks. Prenat Diagn. 2010;30:216–223. doi: 10.1002/pd.2440. [DOI] [PubMed] [Google Scholar]
- 45.Poon LC, Maiz N, Valencia C, Plasencia W, Nicolaides KH. First-trimester maternal serum pregnancy-associated plasma protein-A and pre-eclampsia. Ultrasound Obstet Gynecol. 2009;33:23–33. doi: 10.1002/uog.6280. [DOI] [PubMed] [Google Scholar]
- 46.Scazzocchio E, Figueras F, Crispi F, Meler E, Masoller N, Mula R, Gratacos E. Performance of a first-trimester screening of preeclampsia in a routine care low-risk setting. Am J Obstet Gynecol. 2013;208:203.e1–203.e10. doi: 10.1016/j.ajog.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 47.Park FJ, Leung CH, Poon LC, Williams PF, Rothwell SJ, Hyett JA. Clinical evaluation of a first trimester algorithm predicting the risk of hypertensive disease of pregnancy. Aust N Z J Obstet Gynaecol. 2013;53:532–539. doi: 10.1111/ajo.12126. [DOI] [PubMed] [Google Scholar]
- 48.Akolekar R, Syngelaki A, Poon L, Wright D, Nicolaides KH. Competing risks model in early screening for preeclampsia by biophysical and biochemical markers. Fetal Diagn Ther. 2013;33:8–15. doi: 10.1159/000341264. [DOI] [PubMed] [Google Scholar]
- 49.Akolekar R, Syngelaki A, Sarquis R, Zvanca M, Nicolaides KH. Prediction of early, intermediate and late pre-eclampsia from maternal factors, biophysical and biochemical markers at 11–13 weeks. Prenat Diagn. 2011;31:66–74. doi: 10.1002/pd.2660. [DOI] [PubMed] [Google Scholar]
- 50.Foidart JM, Munaut C, Chantraine F, Akolekar R, Nicolaides KH. Maternal plasma soluble endoglin at 11–13 weeks' gestation in pre-eclampsia. Ultrasound Obstet Gynecol. 2010;35:680–687. doi: 10.1002/uog.7621. [DOI] [PubMed] [Google Scholar]
- 51.Kusanovic JP, Romero R, Chaiworapongsa T, Erez O, Mittal P, Vaisbuch E, Mazaki-Tovi S, Gotsch F, Edwin SS, Gomez R, et al. A prospective cohort study of the value of maternal plasma concentrations of angiogenic and anti-angiogenic factors in early pregnancy and midtrimester in the identification of patients destined to develop preeclampsia. J Matern Fetal Neonatal Med. 2009;22:1021–1038. doi: 10.3109/14767050902994754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lycoudi A, Mavreli D, Mavrou A, Papantoniou N, Kolialexi A. miRNAs in pregnancy-related complications. Expert Rev Mol Diagn. 2015;15:999–1010. doi: 10.1586/14737159.2015.1053468. [DOI] [PubMed] [Google Scholar]
- 53.Morales Prieto DM, Markert UR. MicroRNAs in pregnancy. J Reprod Immunol. 2011;88:106–111. doi: 10.1016/j.jri.2011.01.004. [DOI] [PubMed] [Google Scholar]