Abstract
Recent advances in understanding how the mammalian immune system and intestinal microbiota functionally interact have yielded novel insights for human health and disease. Modern technologies to quantitatively measure specific members and functional characteristics of the microbiota in the gastrointestinal tract, along with fundamental and emerging concepts in the field of immunology, have revealed numerous ways in which host-microbiota interactions proceed beneficially, neutrally or detrimentally for mammalian hosts. It is clear that the gut microbiota has a strong influence on the shape and quality of the immune system and correspondingly, the immune system guides the composition and localization of the microbiota. In the following review, we examine the evidence that these interactions encompass homeostasis and inflammation in the intestine and, in certain cases, extraintestinal tissues. Lastly, we discuss translational therapies stemming from research on host-microbiota interactions that could be utilized for the treatment of chronic inflammatory diseases.
1. Introduction
The human body hosts a remarkable variety (1) and quantity (2) of microorganisms collectively referred to as the microbiota. The microbiota encompasses archaea, bacteria, eukarya and viruses, which form a complex ecosystem thought to have co-evolved with mammalian hosts over time. Commensal bacteria are the most well-defined member of the microbiota and amongst the various body surfaces where commensal bacteria reside, the gastrointestinal (GI) tract contains the highest densities, which are estimated to range between 1011 to 1014 cells per gram of luminal content (3). This enormous cellular and genetic component of the human body is now well recognized to provide indispensible functions in digestion, nutrition status and protection against invasive pathogens (4).
The mammalian immune system is also significantly enriched in the GI tract and engages in a complex dialogue with the microbiota in order to maintain a state of homeostasis that is mutually beneficial. For example, the requirement for microbiota in the proper development of the immune system was first demonstrated in animals reared in microorganism-free environments, known as germ-free. Germ-free animals display a variety of intestinal immune defects including impaired development of gut-associated lymphoid tissues, lower amounts of secreted immunoglobulin, and also reduced intraepithelial CD8+ T cells (5). Additionally, evidence has supported the notion of the gut microbiota as having a strong influence over the development of the immune system outside of the intestine (6). In germ-free mice, splenic CD4+ T helper (Th) cells are skewed towards the Th2 cell subset, and promote enhanced allergic responses and type 2 immunity (6). Germ-free mice also have decreased total numbers of peripheral CD4+ T cells, including both Th17 cells (7) and regulatory T cell (Treg) compartments (8, 9). Conversely, the intestinal immune system also actively shapes the composition and compartmentalization of the microbiota through various mechanisms (10-13). Overall, these observations demonstrate that the colonizing microbiota and host immune system have a complex, dynamic, and reciprocal dialogue.
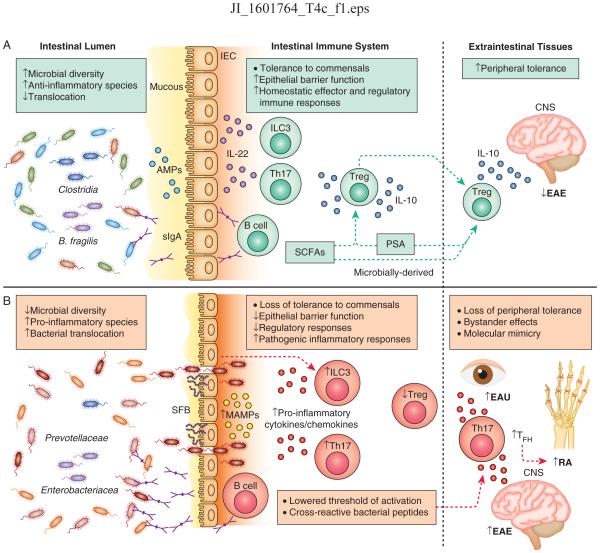
Members of the microbiota are recognized by the innate immune system through their conserved pathogen-associated molecular patterns, referred to herein as microbe-associated molecular patterns (MAMPs) (14) to encompass such ligands in normally non-pathogenic organisms of the microbiota. MAMPs are recognized by germline-encoded pattern recognition receptors (PRRs) distributed spatiotemporally across various cell types and tissues. Despite this ability to directly respond to microbiota-derived signals, several features of the immune system act in cooperation with the intestinal barrier to protect the body from opportunistic pathogens and to limit the immune system from over-reacting to beneficial microbiota in the gut (Fig. 1A). Such features include the following: a thick mucus lining the lumen of the gut epithelial cells which physically excludes most microorganisms (15), secreted IgA which recognizes and binds microbe-specific epitopes and facilitates their removal (16), and secreted anti-microbial peptides (AMPs) that directly neutralize micro-organisms (17, 18). In addition to their pathogen-protective effects, these features help to maintain sequestration of the microbiota, thus reducing the likelihood of the mammalian immune system mounting an over-reactive response to commensal bacteria. However, when the epithelial barrier is compromised due to chemical, pathogenic or inflammatory insults, the immune system must also deal with the resulting influx of commensal and opportunistic microorganisms. In most contexts, the immune system responds appropriately to protect the host from invasive microbes while afterwards maintaining long-term tolerance to the largely beneficial members of the microbiota. Not surprisingly, sustained breakdown of the intestinal barrier is linked to several chronic inflammatory diseases, although the mechanisms are still being determined (19, 20) (Fig. 1B).
Figure 1. Host-microbiota interactions underlie homeostasis and inflammation in the intestine and extraintestinal tissues.
A) At homeostasis, gut bacteria are compartmentalized within the lumen through exclusion by the mucous, neutralization by anti-microbial peptides (AMPs) produced by intestinal epithelial cells (IECs), and release of secretory IgA (sIgA) from intestinal resident B cells. In response to various cues, ILC3s and Th17 cells in the intestine produce IL-22 which acts on IECs to promote compartmentalization of the microbiota. Regulatory T cells (Treg) produce IL-10 and are induced by microbially-derived short chain fatty acids (SCFAs) and polysaccaride A (PSA) or by the bacterial species B. fragilis and Clostridia. Intestinal activation of Tregs can protect against neuroinflammation in the central nervous system (CNS) during experimental autoimmune encephalitis (EAE). B) During chronic intestinal inflammation, loss of intestinal barrier function results in bacterial translocation across the epithelium, release of commensally derived microbe-associated molecular patterns (MAMPs), pro-inflammatory cytokine and chemokine activation, and Th17 and B cell responses. Specific bacteria exacerbate intestinal inflammation including Prevotellaceae, Enterobacteriacea and the Th17-inducing segmented filamentous bacteria (SFB). Loss of tolerance to self antigens can occur due to lowered thresholds for auto-activation (‘bystander effect’) that can mediate autoimmunity in extraintestinal tissues. Bystander effects initiate and exacerbate Th17-mediated inflammation in mouse models of EAE and rheumatoid arthritis (RA). In RA, Th17 and TFH responses aid in auto-antibody production in secondary lymph nodes. Licensing of cross-reactive T cell responses that recognize microbially-derived peptides and react to self-peptides can also initiate autoimmunity in extraintestinal tissues, exemplified in a mouse model of experimental autoimmune uveitis (EAU).
In this review, we assess how functional interactions between the mammalian immune system and the microbiota in the gut can drive inflammatory diseases both locally and systemically. Comparatively, we also examine settings in which host-microbiota interactions can prevent or constrain autoimmune disease. Lastly, we briefly discuss the evidence advocating for the therapeutic modulation of host immune factors as well as the manipulation of microbiota or microbiota-derived biomolecules for treating chronic inflammatory disease.
2: Aberrant host-microbiota interactions underlie intestinal inflammation
Inflammatory bowel disease (IBD) comprises a family of chronic inflammatory disorders of the GI tract. In the clinic, IBD is frequently diagnosed as either Crohn’s disease (CD), affecting any part of the GI tract, or ulcerative colitis (UC) in which pathology is restricted primarily to the colon (20). As with most complex diseases, IBD is thought to occur from a combination of genetic (21, 22), environmental, and life-style associated risk factors (23) that culminate in dysregulated host innate and adaptive immune responses to the intestinal microbiota. Despite the complexity of IBD’s etiology, the host-microbiota interactions that drive disease pathogenesis are becoming better understood through studies in human IBD patients and animal models of intestinal damage and inflammation.
Genetic analyses have identified loss-of-function mutations and polymorphisms in key immune tolerance-related genes and immune response elements that can lead to early-onset IBD or increase disease-susceptibility in adulthood (21, 22). Many primary immunodeficiencies first manifest in the GI tract (24). For example, individuals with loss-of-function mutations in IL-10/IL-10R signaling present with very early-onset IBD due to their incapacity to regulate inflammatory immune responses to commensal bacteria in the GI tract (25-27). Indeed, several other primary immune-deficiencies, such as combined T and B-cell deficiencies, are linked to early-onset gastrointestinal disorders (24), and many more are continually being identified through whole exome sequencing (28). Genome-wide association studies (GWAS) have also identified a number of polymorphisms that are associated with an increased susceptibility to developing IBD in early-life or adulthood (21, 22). The susceptibility loci include genes and gene-pathways involved in intestinal barrier function, innate immune recognition, adaptive immunity, and cellular homeostasis (21). For example, genetic alterations in NOD2, an intracellular PRR for bacterial peptidoglycans, confer increased susceptibility to developing CD in adulthood (29, 30). To extend the genetic evidence, many of the identified genetic alterations found in IBD patients can phenocopy aspects of human disease when experimentally induced in animal models (31). This supports the concept that diverse impairments of hematopoietic and non-hematopoietic cell signaling pathways underlie abnormal host-microbiota interactions.
Environmental and lifestyle risk factors also play a role in disease development (23) as evidenced by modest disease concordance between monozygotic twins who develop UC and CD in adulthood (32). For example, diet is implicated to play causative and preventative roles in IBD through various mechanisms (33). Of note, short- and long-term dietary patterns can modify the composition of the gut microbiota (34-36). Diet’s link to IBD, among other environmental risk factors that alter the gut microbiota such as antibiotic use, has motivated investigation of associations between microbial ‘dysbiosis’ and intestinal inflammation. Dysbiosis is defined as a microbial imbalance resulting in a shift (i.e. loss or outgrowth of a species) and overall reduction of microbial diversity. Using 16S sequencing of gut fecal content, it is observed that IBD patients have reduced colonic microbial diversity and a detectable shift in bacterial enterotypes as compared to healthy individuals (37). In a cohort of CD patients, for example, Frank et al. detected a relative decrease of Firmicutes and Bacteroides in the intestinal microbiota as well as a relative increase in the pro-inflammatory bacteria Enterobacteriacea relative to controls (38) (Fig. 1B). Additionally, fecal metabolite analysis has revealed a decrease in butyrate producing bacterium in CD patients (39, 40). However, so far, no single bacterial strain or combinations of strains have been shown to directly cause or prevent IBD in humans.
Several animal studies have demonstrated that dysbiosis can drive inflammatory pathogenesis in the intestine. These studies involve the demonstration that a pro-inflammatory consortium of microbiota, generally resulting from immune impairment, can transfer disease-phenotype to healthy wild-type (WT) recipient animals. For example, one of the first animal studies to implicate an IBD-causative consortium of bacteria was performed through the horizontal transfer of microbiota from a spontaneous model of colitis (mice deficient in both T-bet and Rag2, referred to as TRUC) into a healthy recipient mouse resulting in transfer of colitis (41). Garrett et al. later identified more specifically that K. pneumonia and P. mirabilis, which grow out in TRUC mice, act in conjunction with the presence of normal gut flora to drive the colitogenic effect upon transfer (42). In another model, NLRP6 inflammasome-deficient mice displayed spontaneous colitis that was transferrable to WT neonates or adults via cross-fostering or cohousing (43). Prevotellaceae was implicated as the primary driver of the inflammatory effect in this study (43). In another study, Chamaillard et al. found that mice with a deficiency in NOD2 had a dysbiotic consortium of microbiota that could transfer colitis to healthy recipient mice (44). These studies underscore that genetic disruption of immune-pathways in the intestine is sufficient to initiate colitogenic-inducing microbial dysbiosis. These microbial consortiums can then transfer disease even in the context of a functional immune system.
Specific microbial species have been characterized by their capacity to provoke inflammatory responses in the intestine. For example, segmented filamentous bacteria (SFB) have been found to preferentially induce the differentiation of pro-inflammatory CD4+ Th17 cells in the lamina propria of the ileum through the TLR5 innate pathway, serum ameloid A (SAA) and direct epithelial cell adhesion (45, 46) (Fig. 1B). Recently, it was found that a number of bacterial species exhibiting epithelial cell adhesion (such as C. rodentium, EHEC, and C. albicans) could similarly induce Th17 cells in the intestine (47). Epithelial adhesion was an indispensible growth-characteristic for this effect as adhesion-defective bacterial mutants failed to induce Th17 cell responses (47). Finally, Helicobacter hepaticus has also been demonstrated to induce pro-inflammatory innate lymphoid cell (ILC) and Th17 cell responses through the induction of cytokines IL-1β and IL-23 in the colon (48). Thus, the induction of pro-inflammatory immune cells and their subsequent effector functions underlie some of the pathogenic effects of these bacterial members (Fig. 1B). It is important to note that ILCs and Th17 cells also mediate protection from pathogens and proper containment of commensal bacteria, thus promoting homeostasis in the gut (49), which will be examined in section four of this review. Furthermore, SFB has been well characterized to significantly induce Th17 cell responses, but it has also been demonstrated to promote the development of intestinal Tregs (8). Thus, a comprehensive analysis of microbiota-induced responses should carefully be considered, and single species cannot always be defined solely as pro- or anti-inflammatory. Moreover, dysbiotic blooms of bacterial species in the gut, such as Enterobacteriaceae, can be secondary to intestinal inflammation from various insults (50) which may owe to the unique ability of these bacteria to feed off the by-products of host inflammation (51). Such observations obscure cause-and-effect relationships between microbial dysbiosis and inflammation.
In summary, IBD arises from a framework of genetic predispositions, environmental risk factors, and dysbiotic microbiota that underpin its chronic nature. Continued interrogation of the complexity of host-microbiota interactions that promote or constrain IBD in the various contexts of human disease should yield more rationally informed preventative or therapeutic approaches.
3: Involvement of the gut microbiota in initiating or exacerbating extraintestinal inflammatory diseases
In parallel with research on how host-microbiota interactions drive inflammatory diseases in the intestine, mounting evidence suggests that these interactions also impact systemic inflammatory disease (52-55) (Fig. 1B). Indeed, IBD patients frequently have extraintestinal disease manifestations involving the joints, skin and eyes (56). Multi-disease cohort GWAS studies have revealed significant overlap in genetic susceptibility loci for IBD with a variety of inflammatory diseases involving extraintestinal tissues (21, 57). Additionally, similar to UC, some autoimmune diseases, such as rheumatoid arthritis (RA), show less disease concordance between monozygotic twins than in other autoimmune diseases (58). This suggests a strong role for environmental factors in disease development. Alterations in the gut microbiota are also associated with several chronic inflammatory diseases outside of the intestine (5). While causal relationships have not yet been demonstrated, these observations provoke the theory that disrupted host-microbiota interactions in the gut, arising from genetic or environmental perturbations, may underlie or impact the course of systemic autoimmune diseases.
Several research efforts in animal models have illuminated how the gut microbiota can have a causative or protective role in autoimmune disease outside of the intestine. In the following sections we categorize such evidence into two, non-mutually exclusive groupings: bystander effects (antigen-nonspecific) and molecular mimicry (antigen-specific). While these have already been proposed and demonstrated as ways in which infectious pathogens, such as viruses, may lead to chronic autoimmune disease in humans (59) less research has elucidated mechanisms in which resident or transient microbiota can have similar roles in systemic inflammatory diseases.
In certain contexts, gut microbiota can exert an ‘adjuvant’ effect in the priming of autoreactive adaptive immune responses. Animal models support the concept that microbiota provide a necessary bystander role in initiating autoimmune diseases in extraintestinal sites (Fig. 1B). For example, several groups have demonstrated that mice treated with either antibiotics (60) or reared in germ-free conditions (61) show reduced induction of experimental autoimmune encephalitis (EAE). Recolonization of germ-free mice with SFB alone induces Th17 cells in the gut, and enhance neurodegeneration in the central nervous system (CNS) upon active EAE induction (61). SFB can have a similar impact, although through a different mechanism, to induce autoimmunity in a mouse model of spontaneous autoimmune arthritis (K/BxN) (62). Germ-free K/BxN mice are resistant to the development of arthritis, mainly due to reduced systemic germinal center formation and subsequent loss of autoantibody production. Recolonization with SFB restores autoimmune arthritis through the activation of Th17 cells in the intestine which then traffic to the spleen to aid in germinal center formation and the production of autoantibodies that mediate disease (62). More recently, SFB colonization in K/BxN mice was found to induce the activation of follicular helper T cells (Tfh) in the PPs, which subsequently egress to the spleen and aid in the production of auto-antibodies (63). A unique variation of the bystander model has also been demonstrated by Campisi et al., in which Citrobacter rodentium infection causes self-antigen release from apoptotic host cells, which are then processed and presented by APCs alongside bacterial peptides and results in the licensing of auto-reactive Th17 cells (64). This reinforces the concept that severe inflammation in the intestine, arising from infection in this case, can provide the initiating conditions for the development of local and systemic autoimmunity.
Molecular mimicry, or an adaptive response that recognizes and responds to both foreign-derived non-self and self-antigens, has also been proposed as a general mechanism for autoimmunity (65, 66). Understandably, cross-reactivity of adaptive responses has direct benefit to the host when it results in broader protection to phylogenically related pathogens, but in contrast, can adversely result in an inappropriate response to self-antigens. Using mouse models, several groups have provided evidence that microbiota-derived antigens may provide the ‘antigenic basis’ for the initiation of systemic autoimmune disease (55). One seminal observation that molecular mimicry to common microbial peptides can induce autoimmunity was performed in a series of experiments demonstrating that structurally related microbial peptides could active MBP-specific T cells (in the Ob TCR-DR2b mouse model), which then induce neurodegeneration in mice (67). More recently, Horai et al. has shown that gut microbiota can provide the antigenic material for cross-reactivity to a self-antigen (68). Using a spontaneous model of autoimmune uveitis (TCR-Tg for the retinal protein IRBP), the authors found that IRBP-specific CD4+ T cells are first activated in the gut, migrate to the eye, and drive pathogenic autoimmune uveitis (68). Interestingly, Kadowaki et al. demonstrated that myelin protein (MOG)-specific CD4+ intraepithelial lymphocytes (IELs) can be activated and proliferate in response to gut antigens. In this context, CD4+ T cells differentiate into a regulatory Th17 cell phenotype that expresses CTLA4 and TGFBR1 (69). Upon transfer into WT mice, MOG-specific CD4+ IELs infiltrate the CNS and upregulate LAG3 expression where they reduce neuro-inflammation (69). These experiments demonstrate that molecular mimicry can activate both pro-inflammatory and immunoregulatory pathways that influence autoimmune disease in extraintestinal sites.
Despite the correlative evidence that commensal bacteria can directly initiate autoimmunity in extraintestinal tissues, no single bacteria or consortia of bacteria have been specifically identified in humans yet. Additional research and experimental tools are warranted to clarify the mechanisms of bystander effects and molecular mimicry in extraintestinal autoimmune disease settings. Recently published works have demonstrated systematic approaches to identify commensal bacteria that incite colitis (70) or diet-dependent enteropathy (71). These approaches consist of sequencing IgA-targeted bacterial taxa (IgA-Seq) from the fecal microbiota then validating the bacterium’s immunological impact in gnotobiotic mice, which may prove useful for the identification of specific intestinal commensal bacteria involved in autoimmune diseases outside of the gut. Furthermore, stratification of IgA-Seq into T-cell independent and T-cell dependent IgA production mechanisms (72) may help to delineate bacterial members that have the strongest capacity to invoke auto-reactive T-cell responses.
4: Protective host-microbiota interactions can prevent or diminish local and systemic inflammatory responses
In addition to the detrimental outcomes of host-microbiota interactions, attention has also focused on interactions that lead to beneficial physiological states associated with mammalian health. Recently identified mechanisms in which microbiota-derived metabolites, regulatory immune cell-types, systemic immunoglobulin and intrinsic immune functions coordinate to enforce peripheral tolerance have been elucidated from experimental models of inflammatory diseases.
The presence of microbiota in the gut of conventionalized mice was shown to have an essential role in generating the CD4+ Foxp3+ Treg compartment (73), a key cellular mediator of regulatory immune responses. Several research groups have identified specific species of commensal bacteria, or consortia of commensal bacteria, that regulate the number, quality and TCR repertoire of intestinal Tregs. As reviewed recently (74), it is clear that many members of the commensal microbiota have a Treg inducing capacity, but some of the well-documented examples include Bacteroides fragilis (75) and Clostridia (9) (Fig. 1A). Recently, Faith et al. devised a systematic approach to elucidate combinations of human-associated microbial species that can promote intestinal Treg responses in gnotobiotic mice (76). This method may permit the identification of new immuno-regulatory phenotypes that are associated with particular members of the human microbiota.
In parallel with the identification of groups or specific members of the microbiota, more reductionist approaches have identified common microbiota-derived molecules or metabolites that stimulate Treg responses. To date, the best-characterized molecules by which the microbiota promote Treg differentiation are the bacterial-derived polysaccharide A (PSA) and short chain fatty acids (SCFAs) (Fig. 1A). PSA was the first documented microbiota-derived molecule that directs the development of a ‘balanced’ T-cell compartment in mice (6). Later, it was found that colonization of mice with PSA-sufficient strains of the commensal Bacteroides fragilis or purified PSA protect against experimentally-induced colitis and that the protective effect required the presence of a functional IL-10 producing CD4+ T cell compartment (6). Furthermore, prophylactic or therapeutic administration of PSA could protect mice from the induction of EAE, which was dependent on an IL-10 producing Treg population (77). This suggests that the effects of PSA-mediated Treg induction can have a systemic influence over peripheral tolerance in tissues outside of the intestine. SCFAs are another group of immunoregulatory molecules that are primarily derived from the microbiota-mediated digestion of dietary fiber and also promote the differentiation of peripheral Tregs (78-80). Interestingly, SCFAs such as acetate can be detected in the blood circulation (81), suggesting that microbiota derived SCFAs can have far-ranging effects outside of the intestine.
Along with the well-documented role of Tregs, other recently identified cell types and cytokine-cytokine receptor pathways that connect hematopoietic and non-hematopoietic cells also mediate tolerance at the intestinal barrier. For example, ILCs maintain dialogue with intestinal epithelial cells (IECs), intestinal dendritic cells, and other cell types to coordinate protective and regulatory immune responses (82, 83). Of note, group 3 ILCs (ILC3s), can modify the composition and anatomical localization of the microbiota. ILC3s respond to a variety of inflammatory cytokines (i.e. IL-1β, IL-23, IL-6) and microbiota-derived metabolites (i.e. aryl hydrocarbon receptor (AhR) ligands) (83). Following activation, ILC3s produce multiple effector molecules including the cytokine IL-22 which acts directly on IECs to produce AMPs, increase mucous production from goblet cells and increase fucosylation of the mucous (84-86). During steady state, the physiological outcome of this response is to maintain proper localization and composition of commensal bacteria (87, 88). In response to breakdown of the intestinal barrier from various insults, such as infection with C. rodentium, this pathway reinforces compartmentalization of the pathogen to prevent its systemic dissemination from the intestine (89, 90).
Recently, it was demonstrated that indole derivatives of microbiota-derived tryptophan in the diet can target AhR in Th17 cells and ILC3s to promote production of IL-22 (91). Mice deficient for the adaptor protein CARD9 develop spontaneous colitis and display a loss of bacterial species able to convert tryptophan into ligands for AhR. Supplementation of three Lactobacillus strains capable of metabolizing tryptophan into AhR ligands protected CARD9−/− mice against inflammation in the colon (91). These results further the evidence that particular metabolites in the gut microenvironment are key to the proper regulation of gut homeostasis. Notably, ILC3s can also directly limit microbiota-specific T cell responses to maintain intestinal homeostasis through antigen presentation on MHCII (92, 93). Future investigation of ILC3s and other regulatory pathways that influence host-microbiota interactions in the gastrointestinal tract could provoke the development of novel treatment options for inflammatory disease.
Systemic immunoglobulin responses to commensal microbiota have also been found to be essential for the maintenance of beneficial host-microbiota interactions. For example, maternally acquired IgA and IgG in neonatal mice leads to dampened T-dependent immune responses against commensal bacteria (94). Additionally, systemic IgG responses to gram-negative bacterial commensals, acquired early and over the course of life, were shown to provide cross-protection to gram-negative pathogens such as E. coli and Salmonella in mice (95). These observations reinforce the concept that microbial composition and the timing of host-commensal interactions provide the foundation for balanced immunity in the intestine. Lastly, the microbiota has also been observed to promote its own compartmentalization within the intestinal lumen as well as provide protection to the host from pathogens (96, 97). Recent identification of the ‘gut-vascular barrier’ system in the small intestine (98), which restricts dissemination of gut bacteria, should provide a new therapeutic framework for constraining disease manifestations due to bacterial translocation across the intestinal epithelium.
5: Manipulation of intestinal microbiota and host-microbiota interactions to treat inflammatory diseases.
Established immunosuppressive medicines, such as glucocorticoids, still represent effective front-line therapies to treat inflammatory disease (99) but have clear disadvantages as a long-term treatment option. Not surprisingly, the medical and biotechnology sectors have taken an interest in translating knowledge of host-microbiota interactions into better standards of care and medicines to treat autoimmune diseases. For example, therapies aiming to adoptively transfer Tregs or promote their in vivo induction in patients are being explored as therapeutic approaches for IBD (100). Additionally, monoclonal antibodies have been developed to block cytokine pathways implicated in chronic inflammation. For instance, blockade of the Th1 and Th17 cell pathways by targeting the shared anti-p40 subunit of IL-12/IL-23 with the monoclonal antibody Ustekinumab is rapidly advancing through clinical trials as a novel treatment for moderate-to-severe CD (101, 102). ILCs have also received increasing attention as novel targets for the treatment of inflammatory diseases given some of their analogous signaling pathways with T cells (103). Clinical investigation on how specific subsets of ILCs respond to approved monoclonal antibody therapies that target cytokine/cytokine receptor pathways, or novel small molecules targeting transcriptional regulators, will extend and refine the paradigm of ILC involvement in provoking and resolving inflammatory disease in the intestine and in extraintestinal tissues (104-106).
In parallel to therapeutic strategies of directly modulating host immune-factors, recent approaches have utilized mechanistic understanding of intestinal microbiota, or microbiota-derived products, for the development of novel treatments (107-109). Among others, proposed and emerging therapies include modification of diet (81, 110), more targeted antibiotics to preserve microbiota integrity (111), supplementation of immunoregulatory metabolites, administration of live biotherapeutic products (112), and fecal microbiota transplant therapy (FMT) (113-116). Although FMT studies have reported promising results in pre-clinical and clinical-settings for infectious diseases such as C. difficile, preliminary clinical trials of FMT for IBD have revealed limited efficacy (114, 117). To date, no microbiota-based or microbiota-derived medical products have been approved by governmental regulatory agencies for the prevention or treatment of inflammatory disease.
Among other challenges faced (107), a prerequisite for translational development includes distinguishing the temporal and kinetic influences of the microbiota on the host immune system (118). Animal studies emphasize that the functions of several immune cell types arise during a critical ‘window of opportunity’ in infancy (118). For example, a subset of RORγt+ Tregs arise in the colon early after birth and prior to weaning (119-121). Additionally, germ free mice display increased frequencies of invariant natural killer T (iNKT) cells in the colon, which predisposes the mice to environmentally triggered colitis (122). Hyper-responsive iNKT responses in GF mice are reversible through administration of a normal microbiota, or B. fagilis-derived antigens, but only during the first two weeks of life (122). Nevertheless, many beneficial influences of the microbiota and microbiota-derived biomolecules on immune cell subsets are age-independent (118) and conceivably represent the most appropriate treatment options for reversing immune defects present in adulthood.
Conclusions
Interrogation of host-microbiota interactions in the intestine has revealed unexpected and novel insights for human health and disease. Human genetic, epidemiologic, and microbial analysis, paired with complementary animal disease models, support the concept that disrupted host-microbiota interactions with the immune system underlie the chronic nature of many inflammatory diseases. Furthermore, rationally designed therapies that modulate or reestablish beneficial interactions are a promising approach for the treatment of intestinal and extraintestinal inflammatory diseases. Emergent technologies aim to focus research efforts on identifying the scope and relevance of these interactions more systematically and unambiguously.
Acknowledgements
We thank members of the Sonnenberg laboratory for discussions and critical reading of the manuscript.
Funding and financial support
Research in the Sonnenberg laboratory is supported by the National Institutes of Health (DP5OD012116, R01AI123368, R21DK110262 and U01AI095608), the NIAID Mucosal Immunology Studies Team (MIST), the Crohn’s and Colitis Foundation of America, the Searle Scholars Program, and the American Asthma Foundation Scholar Award.
References
- 1.Human Microbiome Project, C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14:660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 11.Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol Rev. 2014;260:35–49. doi: 10.1111/imr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM. Host adaptive immunity alters gut microbiota. ISME J. 2015;9:770–781. doi: 10.1038/ismej.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolle L, Tran HQ, Etienne-Mesmin L, Chassaing B. Policing of gut microbiota by the adaptive immune system. BMC Med. 2016;14:27. doi: 10.1186/s12916-016-0573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 15.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mantis NJ, Rol N, Corthesy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 18.Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity. 2015;42:28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 19.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 21.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu TC, Stappenbeck TS. Genetics and Pathogenesis of Inflammatory Bowel Disease. Annu Rev Pathol. 2016;11:127–148. doi: 10.1146/annurev-pathol-012615-044152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal S, Mayer L. Diagnosis and treatment of gastrointestinal disorders in patients with primary immunodeficiency. Clin Gastroenterol Hepatol. 2013;11:1050–1063. doi: 10.1016/j.cgh.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hatscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah N, Kammermeier J, Elawad M, Glocker EO. Interleukin-10 and interleukin-10-receptor defects in inflammatory bowel disease. Curr Allergy Asthma Rep. 2012;12:373–379. doi: 10.1007/s11882-012-0286-z. [DOI] [PubMed] [Google Scholar]
- 27.Engelhardt KR, Grimbacher B. IL-10 in humans: lessons from the gut, IL-10/IL-10 receptor deficiencies, and IL-10 polymorphisms. Curr Top Microbiol Immunol. 2014;380:1–18. doi: 10.1007/978-3-662-43492-5_1. [DOI] [PubMed] [Google Scholar]
- 28.Kelsen JR, Baldassano RN, Artis D, Sonnenberg GF. Maintaining intestinal health: the genetics and immunology of very early onset inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2015;1:462–476. doi: 10.1016/j.jcmgh.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 30.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 31.Mizoguchi A, Takeuchi T, Himuro H, Okada T, Mizoguchi E. Genetically engineered mouse models for studying inflammatory bowel disease. J Pathol. 2016;238:205–219. doi: 10.1002/path.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spehlmann ME, Begun AZ, Burghardt J, Lepage P, Raedler A, Schreiber S. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14:968–976. doi: 10.1002/ibd.20380. [DOI] [PubMed] [Google Scholar]
- 33.Lee D, Albenberg L, Compher C, Baldassano R, Piccoli D, Lewis JD, Wu GD. Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology. 2015;148:1087–1106. doi: 10.1053/j.gastro.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank DN, Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimoto T, Imaeda H, Takahashi K, Kasumi E, Bamba S, Fujiyama Y, Andoh A. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. J Gastroenterol Hepatol. 2013;28:613–619. doi: 10.1111/jgh.12073. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, Wang G, Xia B. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014;52:398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, Gaillot O, Schreiber S, Lemoine Y, Ryffel B, Hot D, Nunez G, Chen G, Rosenstiel P, Chamaillard M. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, Bonneau R, Littman DR. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nunez G, Camp JG, Hattori M, Umesaki Y, Honda K. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, Maloy KJ. IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. J Exp Med. 2012;209:1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Baumler AJ. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferreira CM, Vieira AT, Vinolo MA, Oliveira FA, Curi R, Martins Fdos S. The central role of the gut microbiota in chronic inflammatory diseases. J Immunol Res. 2014;2014:689492. doi: 10.1155/2014/689492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho JT, Chan GC, Li JC. Systemic effects of gut microbiota and its relationship with disease and modulation. BMC Immunol. 2015;16:21. doi: 10.1186/s12865-015-0083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zarate-Blades CR, Horai R, Caspi RR. Regulation of Autoimmunity by the Microbiome. DNA Cell Biol. 2016;35:455–458. doi: 10.1089/dna.2016.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vavricka SR, Schoepfer A, Scharl M, Lakatos PL, Navarini A, Rogler G. Extraintestinal Manifestations of Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21:1982–1992. doi: 10.1097/MIB.0000000000000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richard-Miceli C, Criswell LA. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med. 2012;4:6. doi: 10.1186/gm305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bogdanos DP, Smyk DS, Rigopoulou EI, Mytilinaiou MG, Heneghan MA, Selmi C, Gershwin ME. Twin studies in autoimmune disease: genetics, gender and environment. J Autoimmun. 2012;38:J156–169. doi: 10.1016/j.jaut.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Getts DR, Chastain EM, Terry RL, Miller SD. Virus infection, antiviral immunity, and autoimmunity. Immunol Rev. 2013;255:197–209. doi: 10.1111/imr.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 61.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;1(108 Suppl):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, Umesaki Y, Wu HJ. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer's Patch T Follicular Helper Cells. Immunity. 2016;44:875–888. doi: 10.1016/j.immuni.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campisi L, Barbet G, Ding Y, Esplugues E, Flavell RA, Blander JM. Apoptosis in response to microbial infection induces autoreactive TH17 cells. Nat Immunol. 2016;17:1084–1092. doi: 10.1038/ni.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oldstone MB. Molecular mimicry and autoimmune disease. Cell. 1987;50:819–820. doi: 10.1016/0092-8674(87)90507-1. [DOI] [PubMed] [Google Scholar]
- 66.Yurkovetskiy LA, Pickard JM, Chervonsky AV. Microbiota and autoimmunity: exploring new avenues. Cell Host Microbe. 2015;17:548–552. doi: 10.1016/j.chom.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harkiolaki M, Holmes SL, Svendsen P, Gregersen JW, Jensen LT, McMahon R, Friese MA, van Boxel G, Etzensperger R, Tzartos JS, Kranc K, Sainsbury S, Harlos K, Mellins ED, Palace J, Esiri MM, van der Merwe PA, Jones EY, Fugger L. T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity. 2009;30:348–357. doi: 10.1016/j.immuni.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 68.Horai R, Zarate-Blades CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, Jittayasothorn Y, Chan CC, Yamane H, Honda K, Caspi RR. Microbiota-Dependent Activation of an Autoreactive T Cell Receptor Provokes Autoimmunity in an Immunologically Privileged Site. Immunity. 2015;43:343–353. doi: 10.1016/j.immuni.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kadowaki A, Miyake S, Saga R, Chiba A, Mochizuki H, Yamamura T. Gut environment-induced intraepithelial autoreactive CD4(+) T cells suppress central nervous system autoimmunity via LAG-3. Nat Commun. 2016;7:11639. doi: 10.1038/ncomms11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, Ashorn P, Dewey KG, Houpt ER, Hsieh CS, Gordon JI. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, Antonopoulos DA, Bendelac A. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strauch UG, Obermeier F, Grunwald N, Gurster S, Dunger N, Schultz M, Griese DP, Mahler M, Scholmerich J, Rath HC. Influence of intestinal bacteria on induction of regulatory T cells: lessons from a transfer model of colitis. Gut. 2005;54:1546–1552. doi: 10.1136/gut.2004.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanoue T, Atarashi K, Honda K. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol. 2016;16:295–309. doi: 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- 75.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–495. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- 78.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 81.Richards JL, Yap YA, McLeod KH, Mackay CR, Marino E. Dietary metabolites and the gut microbiota: an alternative approach to control inflammatory and autoimmune diseases. Clin Transl Immunology. 2016;5:e82. doi: 10.1038/cti.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. 2016;17:765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- 84.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pham TA, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP, Keane JA, Page AJ, Kumasaka N, Kane L, Mottram L, Harcourt K, Hale C, Arends MJ, Gaffney DJ, P. Sanger Mouse Genetics. Dougan G, Lawley TD. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, Setoyama H, Imaoka A, Uematsu S, Akira S, Domino SE, Kulig P, Becher B, Renauld JC, Sasakawa C, Umesaki Y, Benno Y, Kiyono H. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, Tardif MR, Sathaliyawala T, Kubota M, Farber DL, Collman RG, Shaked A, Fouser LA, Weiner DB, Tessier PA, Friedman JR, Kiyono H, Bushman FD, Chang KM, Artis D. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, Wang K, Ziegler CG, Goc J, Shima T, Umesaki Y, Sartor RB, Sullivan KV, Lawley TD, Kunisawa J, Kiyono H, Sonnenberg GF. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity. 2016;44:634–646. doi: 10.1016/j.immuni.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 90.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, Sokol H. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, Wherry EJ, Koni PA, Bushman FD, Elson CO, Eberl G, Artis D, Sonnenberg GF. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, Eberl G, Baldassano RN, Laufer TM, Elson CO, Sonnenberg GF. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, Barton GM. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Nunez G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knoop KA, McDonald KG, Kulkarni DH, Newberry RD. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016;65:1100–1109. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spadoni I, Zagato E, Bertocchi A, Paolinelli R, Hot E, Di Sabatino A, Caprioli F, Bottiglieri L, Oldani A, Viale G, Penna G, Dejana E, Rescigno M. A gut-vascular barrier controls the systemic dissemination of bacteria. Science. 2015;350:830–834. doi: 10.1126/science.aad0135. [DOI] [PubMed] [Google Scholar]
- 99.Coutinho AE, Chapman KE. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol Cell Endocrinol. 2011;335:2–13. doi: 10.1016/j.mce.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Geem D, Harusato A, Flannigan K, Denning TL. Harnessing regulatory T cells for the treatment of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1409–1418. doi: 10.1097/MIB.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O'Toole A, Moss AC. Optimizing Biologic Agents in Ulcerative Colitis and Crohn's Disease. Curr Gastroenterol Rep. 2015;17:32. doi: 10.1007/s11894-015-0453-1. [DOI] [PubMed] [Google Scholar]
- 102.Simon EG, Ghosh S, Iacucci M, Moran GW. Ustekinumab for the treatment of Crohn's disease: can it find its niche? Therap Adv Gastroenterol. 2016;9:26–36. doi: 10.1177/1756283X15618130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goldberg R, Prescott N, Lord GM, MacDonald TT, Powell N. The unusual suspects--innate lymphoid cells as novel therapeutic targets in IBD. Nat Rev Gastroenterol Hepatol. 2015;12:271–283. doi: 10.1038/nrgastro.2015.52. [DOI] [PubMed] [Google Scholar]
- 104.Perry JS, Han S, Xu Q, Herman ML, Kennedy LB, Csako G, Bielekova B. Inhibition of LTi cell development by CD25 blockade is associated with decreased intrathecal inflammation in multiple sclerosis. Sci Transl Med. 2012;4:145ra106. doi: 10.1126/scitranslmed.3004140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, Chapman A, Smith CH, Di Meglio P, Nestle FO. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol. 2014;134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Withers DR, Hepworth MR, Wang X, Mackley EC, Halford EE, Dutton EE, Marriott CL, Brucklacher-Waldert V, Veldhoen M, Kelsen J, Baldassano RN, Sonnenberg GF. Transient inhibition of ROR-gammat therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med. 2016;22:319–323. doi: 10.1038/nm.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Olle B. Medicines from microbiota. Nat Biotechnol. 2013;31:309–315. doi: 10.1038/nbt.2548. [DOI] [PubMed] [Google Scholar]
- 108.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scott KP, Antoine JM, Midtvedt T, van Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis. 2015;26:25877. doi: 10.3402/mehd.v26.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Olendzki BC, Silverstein TD, Persuitte GM, Ma Y, Baldwin KR, Cave D. An anti-inflammatory diet as treatment for inflammatory bowel disease: a case series report. Nutr J. 2014;13:5. doi: 10.1186/1475-2891-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yao J, Carter RA, Vuagniaux G, Barbier M, Rosch JW, Rock CO. A Pathogen-Selective Antibiotic Minimizes Disturbance to the Microbiome. Antimicrob Agents Chemother. 2016;60:4264–4273. doi: 10.1128/AAC.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ross JJ, Boucher PE, Bhattacharyya SP, Kopecko DJ, Sutkowski EM, Rohan PJ, Chandler DK, Vaillancourt J. Considerations in the development of live biotherapeutic products for clinical use. Curr Issues Mol Biol. 2008;10:13–16. [PubMed] [Google Scholar]
- 113.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9:88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 114.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, Lee CH. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology. 2015;149:102–109. doi: 10.1053/j.gastro.2015.04.001. e106. [DOI] [PubMed] [Google Scholar]
- 115.Anderson JL, Edney RJ, Whelan K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment Pharmacol Ther. 2012;36:503–516. doi: 10.1111/j.1365-2036.2012.05220.x. [DOI] [PubMed] [Google Scholar]
- 116.Suskind DL, Brittnacher MJ, Wahbeh G, Shaffer ML, Hayden HS, Qin X, Singh N, Damman CJ, Hager KR, Nielson H, Miller SI. Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn's disease. Inflamm Bowel Dis. 2015;21:556–563. doi: 10.1097/MIB.0000000000000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rossen NG, Fuentes S, van der Spek MJ, Tijssen JG, Hartman JH, Duflou A, Lowenberg M, van den Brink GR, Mathus-Vliegen EM, de Vos WM, Zoetendal EG, D'Haens GR, Ponsioen CY. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology. 2015;149:110–118. doi: 10.1053/j.gastro.2015.03.045. e114. [DOI] [PubMed] [Google Scholar]
- 118.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, Lee JY, Lee M, Surh CD. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016;351:858–863. doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- 120.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, Busslinger M, Cerf-Bensussan N, Boneca IG, Voehringer D, Hase K, Honda K, Sakaguchi S, Eberl G. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 121.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]