Abstract
The alarming increase in both the incidence and severity of food allergies has coincided with lifestyle changes in Western societies, such as dietary modifications and increased antibiotic use. These demographic shifts have profoundly altered the co-evolved relationship between host and microbiota, depleting bacterial populations critical for the maintenance of mucosal homeostasis. There is increasing evidence that the dysbiosis associated with sensitization to food fails to stimulate protective tolerogenic pathways, leading to the development of the Type 2 immune responses that characterize allergic disease. Defining the role of beneficial, allergy protective members of the microbiota in the regulation of tolerance to food has exciting potential for new interventions to treat dietary allergies by modulation of the microbiota.
The incidence of allergic diseases has increased dramatically over the last fifty years, particularly in developed countries. The rise in prevalence has occurred in such a short time frame that genetics alone cannot explain it. This same time period has been marked by improvements in sanitation, stark dietary changes, and increased vaccination and antibiotic use in Western societies, all of which have been linked to increasing susceptibility to allergic and autoimmune diseases (1–4). What these lifestyle changes have in common is their ability to alter the populations of commensal microbes (the microbiota) that live in and on our bodies (1). The “hygiene hypothesis” was the first to imply a link between microbes and allergy by suggesting that the lower incidence of allergic diseases in children with older siblings resulted from increased exposure to infectious disease in early childhood (5). This idea was expanded in subsequent epidemiological studies which found that children raised in rural environments had a lower incidence of allergic disease than those in urban settings and had greater environmental exposure to microbial products such as lipopolysaccharide (LPS) (6–10). More recent work has revised the original hygiene hypothesis concept to include increased antibiotic use and vaccination as other lifestyle changes that have reduced childhood infections and altered the microbiota (11). Cesarean birth and formula feeding have also disturbed nature’s co-evolved strategy, altering founder microbial taxa and increasing susceptibility to diseases associated with this “Western” lifestyle (12–14). Consumption of a highly processed modern diet, high in fat, low in fiber, and quite different from that of our ancestors, has had profound consequences for the composition of the intestinal microbiota (15–17). Collectively, these studies suggest that environmental and lifestyle changes have affected the relationship between the commensal microbiota and its human host and contributed to the increasing incidence of allergic disease.
The skin and all mucosal surfaces are populated by a site specific microbiota (18). The microbes present can include bacteria, viruses, bacteriophage, archaea, fungi, parasitic worms and protists (19, 20). Commensal bacteria are the best characterized, particularly in the gastrointestinal tract, where their density increases from an estimated 104 to 108 per milliliter of luminal contents in the small intestine to approximately 1011 organisms per milliliter of luminal content in the colon, the highest bacterial density of any environment analyzed so far on earth (21). In addition to this large community of bacteria, the gastrointestinal tract also contains more immune cells than any other organ. The two are in intimate communication; maintenance of homeostasis between these microbes and the immune system is essential to health. Exciting new research is beginning to identify the mechanisms by which beneficial functions of the microbiota regulate tolerance to dietary antigens (22, 23).
In this review we will discuss the role of the microbiota in maintaining tolerance to food and examine how commensal dysbiosis promotes the development of food allergy. Finally, we will examine the clinical evidence for a role for the microbiota in regulating food allergen sensitization and explore strategies for the development of microbiome-modulating therapeutics to prevent or treat food allergy.
Extending the Hygiene Hypothesis to the Microbiota
The pathogenesis of food allergy involves an aberrant type 2 immune response to dietary antigens. The most common allergenic foods are tree nuts, peanuts, milk, eggs, shellfish, fish, wheat and soy, although a great number of other foods can also elicit an allergic response (24). A healthy immune response to food antigens is a state of non-responsiveness, referred to as oral tolerance (25). When oral tolerance is not induced food antigens can instead evoke a response that is characterized by differentiation of naïve T cells into food antigen specific Th2 cells (fTh2), which produce large amounts of IL-4 and IL-13 that drives B cells to produce antigen specific immunoglobulin E (IgE) (26). IgE binds to the surface of mast cells and, upon re-exposure, antigen crosslinking results in mast cell degranulation. The release of mast cell mediators, such as histamine, leads to the symptoms associated with food allergy including anaphylaxis (27). In support of the epidemiological observations attributing a protective role to the microbiota in constraining allergic responses, germ free mice, which are born and raised in a sterile environment, have an exaggerated systemic type 2 immune response characterized by high levels of IgE, and are more susceptible to oral-antigen induced anaphylaxis than mice colonized with a diverse microbiota (28, 29). Early work demonstrated that administration of LPS to germ free mice is sufficient to restore oral tolerance (30). In keeping with this observation mice unable to signal via TLR4, the receptor for LPS, exhibit increased allergen specific IgE and exacerbated anaphylactic symptoms in response to repeated intragastric administration of peanut extract plus cholera toxin when compared to TLR4 sufficient mice (31). TLR4 is one of a group of pattern recognition receptors (PRR) that the immune system uses to detect microbe-derived products, including LPS and DNA. Microbial sensing through TLRs is critical for maintaining intestinal homeostasis and limiting inflammation (32). Oral administration of a broad spectrum antibiotic cocktail evoked food allergen sensitization in TLR4 sufficient mice, suggesting that intestinal bacteria were the source of the TLR4 ligand (31). Recent work by has begun to reveal an even more complex role for LPS in allergic disease (33). Fecal samples were collected during the first three years of life from genetically related, but geographically separated, children at high (Finnish), low (Russian) and transitional (Estonian) risk for the development of autoimmune and allergic disease, including food allergy. The authors found that the low risk Russian children had higher proportions of Bifidobacterium, whereas Finnish and Estonian children had increased abundance of Bacteroides. Surprisingly, metagenomic analysis revealed striking differences in LPS synthesis between the Finnish and Russian cohorts. Russian children had LPS mostly originating from E. coli, whereas the bulk of the LPS in Finnish children originated from Bacteroides. Importantly, the LPS variant produced by Bacteroides was structurally and functionally distinct from E. coli LPS. E. coli LPS is strongly immunostimulatory and chronic exposure results in a refractory state known as endotoxin tolerance (34), which is thought to contribute to the protective effects of the microbiota suggested by the hygiene hypothesis (35). Not only was Bacteroides–derived LPS less immunostimulatory to primary human peripheral blood mononuclear cells (PBMCs) than E. coli LPS, but when PBMCs were treated with the two LPS variants mixed together, the high cytokine production elicited by E. coli LPS was abrogated. These findings raise the possibility that colonization early in life with a low immunostimulatory microbiota can impair aspects of immune education and predispose to inflammatory diseases such as food allergy (35, 36). A better understanding of how various components of the microbiota influence immune system development will inform therapeutic strategies aimed at restoring the benefits conferred by particular microbial communities.
Oral tolerance to food antigens
The gastrointestinal tract is under constant bombardment by microbial and food antigens. The healthy intestinal immune system is therefore, of necessity, geared towards a tolerogenic response characterized by the presence of large numbers of regulatory T cells (Tregs). Originally it was believed that oral tolerance was primarily mediated by the generation of food antigen specific Tregs (37). Antigen encountered in the lamina propria is taken up by a population of CD103+ intestinal dendritic cells (DC) that then migrate to the draining mesenteric lymph nodes (mLN). Within the mLN large amounts of retinoic acid (RA), a vitamin A derivative, and TGF-β produced by both CD103+ DCs and LN stromal cells instruct antigen specific naïve T cells to differentiate into Tregs (38–40). Additionally, RA and TGF-β induce upregulation of the gut homing receptors CCR9 and α4β7 on these newly differentiated Tregs to recruit them back to the intestinal lamina propria (38–40). In the lamina propria these Tregs are expanded by the production of IL-10 from resident CX3CR1 macrophages (41). Some of these newly expanded Tregs may also enter into the circulation to mediate systemic tolerance to orally available antigens (42). In support of this model, in the absence of gut homing, or in animals lacking CX3CR1 macrophages, oral tolerance is abrogated (41). Other work shows that oral antigen exposure induces an allergic phenotype in mice with vitamin A deficiency (43). RA deficient mLN DCs drive naïve T cells towards a pathogenic Th2 phenotype instead of Tregs (43). Collectively, this data supports the concept that food antigen specific Tregs are critical for protection from dietary allergies (41, 42). However, these studies have only shown that antigen specific Tregs, induced by oral administration of model food antigens such as ovalbumin (OVA), induce non-responsiveness to subsequent peripheral immunization (42). As such, it has not been clear if Treg development is a feature common to all food antigens found in a complex diet or whether these food antigen specific Tregs also contribute to intestinal homeostasis. Recent work using germ free mice weaned onto an elemental diet void of antigens has demonstrated that the majority of Tregs in the small intestine are indeed induced in response to food antigens in a complex diet (44). The induction of small intestinal Tregs occurs rapidly following introduction of solid food, and decreases over 4–6 weeks following removal of food antigens (44). Mice weaned onto antigen free diets had a greater proportion of antigen specific T cells differentiating into inflammatory T cells rather than Tregs following oral antigen administration, suggesting that Tregs raised against dietary antigens limit proinflammatory responses to model antigens such as OVA. Food antigen specific Tregs therefore contribute to protection against allergic sensitization to dietary antigens, possibly by reinforcing a tolerogenic environment in the small intestine.
Microbiota-mediated tolerance to food antigens
In addition to dietary antigens the intestinal immune system must also maintain a tolerogenic response to the microbiota resident within the gut lumen. Components of the microbiota strongly induce colonic Tregs, as demonstrated by a deficit in Tregs in the colonic lamina propria of germ free mice which increase in frequency following colonization (45, 46). Colonic Treg induction has been attributed to the Clostridia, a class of mucosa-associated Firmicutes, (45, 47). Spore-forming Clostridia isolated from both mouse and human feces strongly induce colonic Tregs, (45, 47). More recently, however, it has been suggested that colonic Tregs can be induced by other members of the microbiota as well (48). It is unclear whether there is a common mode of action of Treg induction between these diverse bacterial groups, or whether they stimulate specific TLRs. It is also not known whether all (or most) Tregs induced by commensal bacteria bear bacteria specific T-cell receptors (TCRs) (49). Moreover, there is evidence that bacteria-induced Tregs also contribute to tolerance towards other antigens, including those from food. Indeed, Clostridia-induced Treg expansion was associated with protection from food allergen sensitization (22, 45, 47). As there is heterogeneity within the intestinal Treg population it is possible that particular bacteria may induce specific populations of Tregs that have different functions for maintaining homeostasis (50). Kim et al observed that antibiotic treatment markedly reduces RORγt+ Tregs in the colonic lamina propria, suggesting that these Tregs are bacteria dependent, whereas mice fed an antigen-free diet had a selective reduction of RORγt- Tregs in the small intestine indicating these are food antigen dependent (44). This finding supports previous literature that commensal bacteria induce a population of RORγt+ Tregs in the colon (48, 51). DCs from the small intestine and colon migrate to anatomically different mLNs and induce immunologically distinct T cell responses (52, 53). Colonic and small intestinal DCs differ both phenotypically and functionally, which may reflect the different antigenic burdens encountered by DCs at these physiologically distinct sites along the GI tract (52, 53). This finding may help to explain why bacteria and food antigens induce distinct populations of Tregs that differ in both location and phenotype. Evidence that bacteria-mediated Treg expansion can protect from food allergen sensitization (22, 45, 47) suggests that food and bacteria derived Tregs work cooperatively to mediate oral tolerance and protect from food allergen sensitization (Figure 1). It is possible that bacteria specific Tregs migrate to the small intestinal lamina propria (perhaps through recirculation via the mLN) and secrete IL-10 to reinforce the tolerogenic environment (54). Indeed, a population of RORγt+ Tregs is present within the small intestine, albeit at a lower frequency than in the colon, and is reduced following antibiotic treatment, supporting the idea that microbe-driven Tregs can also localize at this site (44). In light of this, the tolerogenic environment maintained by bacteria-derived Tregs likely contributes to the production of food antigen specific Tregs rather than pro-allergic Th2 cells upon subsequent exposure to food antigens. In addition, RORγt+ Tregs are found in small numbers in other body sites, including the lung, spleen and skin (55), suggesting that these cells disseminate throughout the body. This may help to explain the role of the intestinal microbiota in protection from other allergic diseases including asthma.
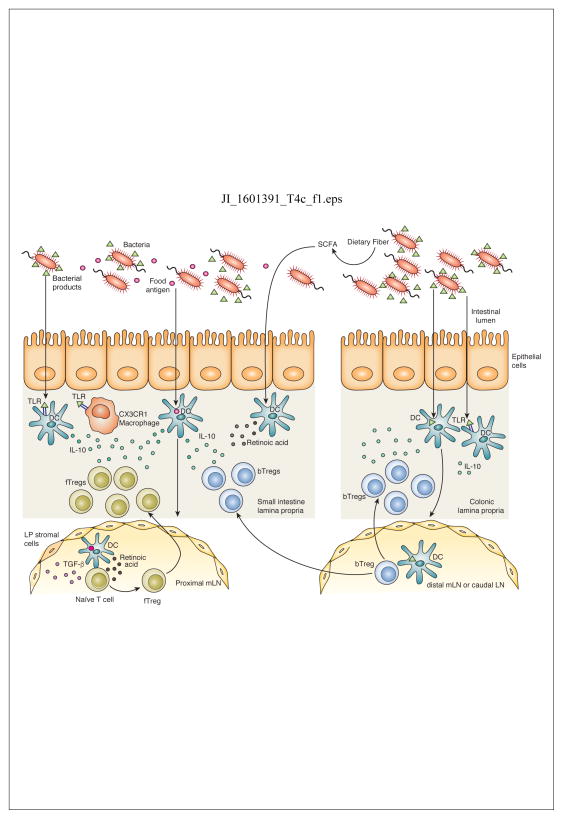
Figure 1. Induction of tolerance to food and bacterial antigens in the intestine.
Food antigen is taken up by dendritic cells (DCs) in the small intestine which migrate to the proximal mesenteric lymph node (mLN). TGF-β and retinoic acid produced by LN stromal cells and DCs induce differentiation of food antigen specific Tregs (fTregs) and upregulation of gut homing molecules. fTregs migrate back to the small intestine lamina propria where TLR signaling by bacterial products induces production of IL-10 by resident CX3CR1 macrophages and DCs that supports Treg expansion and IL-10 production. Bacterial products are also taken up by colonic DCs that migrate to the distal mLN and caudal LN to induce differentiation of bacterial specific Tregs (bTreg). Although predominant in the colon, bTregs also migrate to the small intestine where they release IL-10 to maintain the tolerogenic immune environment. Fermentation of dietary fiber to short chain fatty acids (SCFAs) may enhance retinoic acid production by DCs and promote Treg differentiation. TLR signaling by bacterial products such as LPS induces a tolerogenic phenotype in colonic and small intestinal DCs that promotes differentiation of Tregs.
Other studies are beginning to identify the signaling pathways for bacteria mediated Treg induction important for preventing allergic sensitization to food. Several reports have implicated microbial sensing by host TLRs in the induction of colonic Tregs that maintain tolerogenic responses (46, 56). Mice lacking MyD88, an adaptor molecule for downstream TLR signaling, develop more severe intestinal inflammation in response to epithelial damage induced by dextran sodium sulfate, suggesting an important role for microbial sensing in limiting inflammatory responses (32). TRAF6 is another important adaptor molecule for TLR signaling that acts downstream of MyD88 to activate transcription factors such as NFκB to induce cytokine production (57). Mice with a specific deletion of TRAF6 in CD11c+ antigen presenting cells (Traf6ΔCD11c) have reduced numbers of Tregs in their small intestine and present with a spontaneous type 2 inflammation at this site (58). Exacerbation of the inflammatory phenotype in germ free Traf6ΔCD11c mice further supports a role for microbe induced Tregs in preventing pathogenic type 2 intestinal inflammation (58) (59). Interestingly, this phenotype was specific for the small intestine as no overt inflammation was observed in the colon. This may be due to the different physiological functions of the small and large intestine since the large intestine functions primarily to reabsorb water while the small intestine encounters the bulk of the food antigens. As such, the tissue selective inflammation observed in this model may indicate that the inflammatory response occurs as a result of a breakdown in tolerance to food antigens.
In addition to microbial ligands that can be sensed via PRRs, commensal bacteria release metabolites such as short chain fatty acids (SCFAs) upon fermentation of insoluble dietary fibers (4). SCFAs, including acetate, propionate and butyrate have been demonstrated to have immunogenic activity both locally, within the intestine, and systemically (23, 60–62). SCFAs are utilized by colonocytes as an energy source (63, 64), but also have immunomodulatory properties by signaling through G-protein coupled receptors (GPCRs) (65–67) and act to inhibit histone deactelylases (HDACs) (68). One possible consequence of dysbiosis for the development of allergic responses may result from reduced levels of SCFAs (69). There is a body of literature supporting the idea that low levels of SCFAs are associated with an allergic phenotype and that increasing SCFA levels can ameliorate disease (61, 63, 70). One mechanism by which SCFAs protect against allergic disease is through the induction of colonic Tregs (60, 62, 68). The addition of SCFAs to the drinking water of germ free mice resulted in an increased abundance of colonic Tregs and protected against colonic inflammation (62). Recent work showed that mice fed a high fiber diet had increased SCFA levels and were protected against food allergy, in part through enhanced induction of Tregs in the mLN (23). DCs isolated from the MLN of the high fiber diet fed mice had increased retinal dehydrogenase (RALDH) activity when compared to controls, suggesting a link between the protective effects of dietary fiber and vitamin A metabolism (23).
Microbiota-mediated modulation of intestinal barrier function
It’s remarkable to think that a single layer of epithelial cells is all that separates the enormous number and variety of food antigens and microbes within the intestinal lumen from the underlying immune cell rich lamina propria. Specialized epithelial subpopulations have unique properties and functions that are important for enforcing barrier protection to prevent uncontrolled access of antigens to the lamina propria (53). These include the production of antimicrobial peptides (AMPs) by Paneth cells and of mucus by goblet cells which together act to form a physical barrier that limits the access of bacteria to the epithelial surface (71). Additionally, intestinal epithelial cells express tight junction proteins that act to seal off the underlying immune rich lamina propria from excessive exposure to luminal antigens (72). We have found that increased intestinal permeability contributes to food antigen sensitization (22). Stefka et al observed that sensitization of mice to peanuts is associated with an increased concentration of the major peanut allergens Ara h 2 and Ara h 6 in the serum (22). Colonization with a consortium of mucosa associated Firmicutes of the Clostridia class reduced serum Ara h 2 and Ara h 6 and ameliorated allergic sensitization to peanut, suggesting that commensal microbe-driven reinforcement of the epithelial barrier is important for protection against food allergy (22). The improved barrier function observed after colonization by this Clostridia consortium was associated with an increase in goblet cell numbers and mucus production in the intestine as well as upregulated expression of the AMPSs Reg3β and Reg3γ suggesting a broad effect on multiple intestinal epithelial cells types (22). Both intestinal mucus and AMP production are regulated by the barrier protective cytokine IL-22 (73). By utilizing treatment with a neutralizing antibody for IL-22 or depletion of innate lymphoid cells (ILCs), which produce IL-22, Stefka et al demonstrated that Clostridia-induced IL-22 production by type 3 ILCs (ILC3s) was both necessary and sufficient to reduce intestinal permeability and prevent food antigen sensitization in this model (22). This finding highlights the complex role of the microbiota in modulating innate immune responses that, in turn, influence intestinal epithelial cell function and regulate its barrier protective properties (Figure 2).
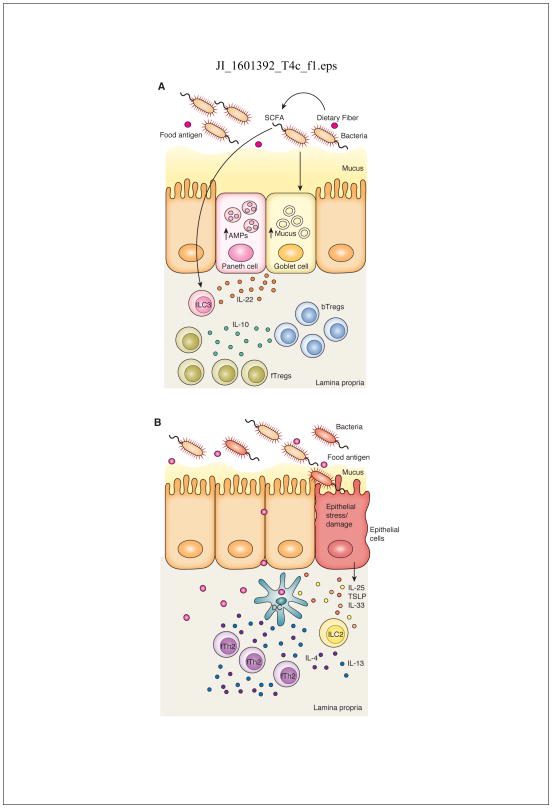
Figure 2. The microbiota regulates both protective and pathogenic barrier responses in the intestine.
A) A healthy microbiota will induce a barrier protective response in the intestine, in part, through production of SCFAs that are most likely to act on ILC3s to produce IL-22. IL-22 induces antimicrobial peptide (AMP) production by Paneth cells and mucus production by goblet cells to reinforce barrier function, controlling the location and composition of the microbiota. This barrier protective function prevents uncontrolled access to the lamina propria by food antigens to prevent allergic sensitization. B) Dysbiosis fails to induce these protective pathways. Dysregulated epithelial barrier function and a compromised mucus layer allow increased permeability to food antigens. Damaged or stressed epithelial cells release the alarmins IL-25, IL-33 and TSLP that activate ILC2s to produce IL-4 and IL-13 which promotes the development of allergic sensitization to food antigens through the generation of food antigen specific Th2 cells (fTh2).
Indeed, the microbiota has a profound influence on mucus production and goblet cell homeostasis (74). Germ free mice have altered mucus production compared to colonized mice, with differences observed in both the small and large intestine (74). These mucus layers have unique features that limit access of bacteria to the intestinal epithelium (74). In the large intestine the mucus forms two layers, an inner sterile layer and a diffuse outer layer that is heavily colonized by bacteria which are thought to feed on mucins and other proteins within the mucus layer (75). The permeability of the mucus layer differs along the length of the intestine. In the distal colon the inner mucus layer is impenetrable to bacteria sized beads whereas in the proximal colon some penetrability is observed (76). In the small intestine the mucus layer is diffuse and allows limited penetration by bacteria, however secretion of AMPs and high gut motility (which removes this easily detached mucus) helps to limit access to the epithelium (76). Within five weeks post colonization of germ free mice mucus thickness, impenetrability and detachment properties in the colon and small intestine resemble that seen in conventional mice (74). Alteration in mucus properties post colonization is associated with changes in the microbiota (74). Since treatment with some antibiotics also alters the mucus layer (77) this suggests that changes in the microbiota affect mucus associated barrier function in the intestine. Stefka et al demonstrated that one mechanism by which Clostridia protect against allergic sensitization is by eliciting increased mucus production (22). Additionally, both germ free and antibiotic treated mice have increased susceptibility to oral antigen sensitization (22, 29). Given the profound defect in mucus production in these mice it is likely that the altered mucus production and subsequent defects in barrier function contribute to a breakdown in oral tolerance. However, goblet cells and mucus play a role in the maintenance of intestine homeostasis beyond their well-known contributions to barrier function. It has been reported that culturing DCs in vitro with the mucus protein Muc2 induces a tolerogenic DC phenotype that induces greater Treg differentiation from naïve T cells (78). Tolerance to OVA is impaired in Muc2 deficient mice; both mucosal and systemic tolerance is restored when OVA is co-administered in the presence of Muc2 protein (78). Other work identified that goblet cells form goblet cell-associated antigen passages (GAPs) by taking up luminal antigen and delivering it to intestinal DCs (79–81). GAP formation is a regulated process, induced by acetylcholine acting on goblet cells. GAPs form in the steady state in the small intestine, but goblet cell responsiveness to acetylcholine is inhibited by MyD88 dependent, goblet cell intrinsic, sensing of the gut microbiota in the colon (80). Deletion of MyD88 in goblet cells, or disruption of the microbiota by antibiotics, overrides the normal suppression by the microbiota and allows the formation of colonic GAPs, potentiating inflammation due to uncontrolled exposure to luminal contents (80, 81). Moreover, altering GAP formation in early life results in persistent Th2 responses (R. Newberry, personal communication), providing an additional link between alterations in the gut microbiota in early life and a predisposition to food allergy.
Intestinal epithelial cells also monitor the luminal environment and produce cytokines that direct immune responses in the underlying lamina propria (82). Stressed or damaged intestinal epithelial cells secrete the cytokines IL-33, TSLP and IL-25, also collectively referred to as alarmins, to induce protective immunity and promote repair (82). These cytokines are associated with the initiation of the protective type 2 immune response that is responsible for the clearance of enteric helminths and repair of epithelial damage induced by infection (83–85). Epithelial alarmins activate type 2 ILCs to produce IL-13 and prime intestinal DCs to promote the differentiation of naïve T cells to Th2 cells that mediate worm clearance (82, 86). Recently the population of epithelial cells that produces IL-25 was revealed to be a specialized lineage known as tuft cells (83–85, 87). Howitt et al showed that tuft cells release IL-25 in response to activation by parasites via chemosensory receptors (82). As some chemosensory receptors are GPCRs, this raises the interesting possibility that in addition to helminths, these receptors may also respond to microbe-derived signals, such as SCFAs, skewing the immune environment in the intestine. Indeed, there is evidence that the microbiota can directly regulate the expression of epithelial alarmins in the intestine. Intestinal expression of IL-25, IL-33 and TSLP is reduced in germ free mice and increases following colonization (88–90). Moreover, administration of IL-25 can alter the expression of AMPs, changing the composition of the microbiota and demonstrating a role for IL-25 in the host-microbe cross-talk that is essential for homeostasis (91). In the context of allergic inflammation, allergen exposure can induce the release of alarmins and drive the pathogenic type 2 immune response associated with disease (71). Overexpression of IL25 or IL-33 drives allergic responses to dietary antigens, suggesting that these epithelial-derived cytokines may contribute to the development of dietary allergies by skewing the immune environment in the intestine from tolerogenic to pro-allergic (92, 93). A dysbiotic microbiome might also elicit epithelial alarmins and prime for allergic sensitization to food (94). In this setting, characterized by IL-4 production by mast cells and Th2 T cells, oral allergen exposure results in reprogramming of Tregs from a tolerogenic to a Th2-like phenotype, further propagating the allergic response (95). IL-33 has been shown to drive the expansion of GATA3+ Tregs in the colon (96), which are similar in phenotype to reprogrammed Tregs implicated in the loss of tolerance to food antigens (95), supporting the idea that epithelial alarmin production may alter the immunological environment in the intestine and contribute to allergic sensitization. In further support of this hypothesis, consumption of a low fiber diet that exacerbates food allergen sensitization is associated with increased expression of both IL-33 and TSLP in the intestine (23). Conversely protection against allergic sensitization following high fiber diet administration alters the microbiota and is associated with reduced expression of IL-33 and TSLP. Taken together these data suggest that a “healthy” microbiota may protect against allergic sensitization by reducing expression of alarmins by intestinal epithelial cells. However, in conditions of dysbiosis the microbiota may induce elevated levels of these alarmins resulting in aberrant Th2 responses towards dietary antigens by skewing the immune environment in the intestine towards a type 2 rather than tolerogenic response (Figure 2). Identifying how epithelial cell alarmin production is regulated may help to identify new targets to prevent allergic sensitization to food.
Clinical considerations and potential therapeutics
The findings described above outline the complex interplay between host immunity and the microbiota. Translational studies are beginning to explore a role for microbe modulating therapeutics for diseases such as food allergy (97). Both murine and human studies have suggested that dysbiosis early in life contributes to the development of allergic disease and that therapeutic interventions that alter the microbial composition during this time period may be most effective to prevent allergic sensitization (22, 29, 98–106). Recent analysis of one cohort of 319 children emphasized that changes in the microbiota in the first 100 days of life were most likely to be associated with allergic disease (99). During this critical time period children at risk of developing allergic asthma exhibited marked reductions in the abundance of four key microbial genera (Faecalibacterium, Lachnospira, Rothia and Veillonella) and in SCFA metabolites. Moreover, colonization of germ free mice with these genera protected from the development of allergic airway inflammation (99). This protection was also associated with increased levels of fecal butyrate, suggesting that one mechanism of protection was via SCFA signaling (99). Most studies have concluded that the efficacy of conventional Lactobacillus-based probiotics is limited to infancy. Berni Canani et al showed that treatment with a Lactobacillus GG supplemented formula accelerated the acquisition of tolerance in infants with cow’s milk allergy (CMA) (107). Fecal samples collected from a small subset of infants from this study revealed stark differences in the microbiota of CMA patients when compared to healthy age-matched controls (108). Twelve months of treatment with the tolerance inducing formula was associated with an increase in butyrate producing bacteria and fecal butyrate levels, suggesting that one therapeutic effect of probiotic administration was the alteration of intestinal microbial community structure (108). Pre-clinical mouse data also suggests that dietary manipulation of the microbiota may be effective for the treatment of allergic disease (23, 70). These studies are providing exciting early evidence that the microbiota provides protective and inductive signals to shape the intestinal immune response and tip the balance in favor of tolerance over allergic sensitization.
Collectively, then, there is provide compelling evidence that allergic phenotypes are associated with alterations in the intestinal microbiome and that this dysbiosis may drive the allergic response. It is also possible that allergic inflammation itself induces changes in the microbiota. Both scenarios may contribute to allergic sensitization at different times and stages of disease, highlighting the complex and dynamic nature of host-microbial interactions. As we learn more about the specific immune modulating effects of particular members of the microbiota we may be able to identify microbial signatures that are associated with pro-allergic responses such as release of alarmins by epithelial cells that contribute to allergic sensitization to food. Conversely, identification of a healthy microbiota that reinforces tolerance and barrier function in the intestine will allow for better targeted treatments to harness the microbiota to restore health.
Conclusions
The marked increase in both incidence and severity of dietary allergies that has occurred in parallel with profound environmental and lifestyle changes suggests a link between alterations in the microbiota and the rising prevalence of allergic disease. Increasing knowledge of how the immune response is influenced by the microbiota is revealing new approaches to treat diseases such as food allergy. While there is already promising evidence in support of manipulating the microbiota during early life to prevent allergic sensitization, it is not yet clear if a stably established gut microbiota can be effectively manipulated to treat food allergy (109). We do know that, in adults, the microbiota can be readily altered, on even daily timescales, by changes in components of the diet, particularly fiber (4, 15, 110). Moreover, the utilization of microbial metabolites to treat complex immune mediated-diseases is starting to generate results (111). These observations lend promise to the vision that microbiome-modulating therapeutics will have efficacy later in life, particularly as an adjunctive strategy to potentiate antigen specific desensitization protocols, and promote long lasting tolerance (112).
Acknowledgments
This work was supported by National Institutes of Health Grant R01 AI 106302, the Sunshine Foundation, UChicago Center for Translational Medicine, UChicago Digestive Diseases Research Core Center (DK42086) and F.A.R.E. (Food Allergy Research and Education).
The authors would like to thank P. Belda Ferre and E. Beltran for critical review of this manuscript.
Abbreviations used in this manuscript
- AMP
antimicrobial peptide
- bTreg
bacterial specific regulatory T cell
- CMA
cow’s milk allergy
- DC
dendritic cell
- fTh2
food antigen specific T helper 2 cell
- fTreg
food antigen specific regulatory T cell
- GAP
goblet cell-associated antigen passage
- GPCR
G-protein coupled receptor
- HDAC
histone deacetylase
- IgE
immunoglobulin E
- ILC
innate lymphoid cell
- ILC3
type 3 innate lymphoid cell
- ILC2
type 2 innate lymphoid cell
- LPS
lipopolysaccharide
- mLN
mesenteric lymph node
- OVA
ovalbumin
- PBMC
peripheral blood mononuclear cell
- PRR
pattern recognition receptor
- RA
retinoic acid
- SCFA
short chain fatty acid
- TCR
T-cell receptor
- TLR
toll-like receptor
- TLR4
toll-like receptor 4
- Treg
regulatory T cell
References
- 1.Feehley T, Stefka AT, Cao S, Nagler CR. Microbial regulation of allergic responses to food. Semin Immunopathol. 2012;34:671–688. doi: 10.1007/s00281-012-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 3.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strachan DP. Hay fever, hygiene, and household size. Bmj. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 7.Riedler J, Eder W, Oberfeld G, Schreuer M. Austrian children living on a farm have less hay fever, asthma and allergic sensitization. Clin Exp Allergy. 2000;30:194–200. doi: 10.1046/j.1365-2222.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- 8.Braun-Fahrlander C, Gassner M, Grize L, Neu U, Sennhauser FH, Varonier HS, Vuille JC, Wuthrich B. Prevalence of hay fever and allergic sensitization in farmer’s children and their peers living in the same rural community. SCARPOL team. Swiss Study on Childhood Allergy and Respiratory Symptoms with Respect to Air Pollution. Clin Exp Allergy. 1999;29:28–34. doi: 10.1046/j.1365-2222.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- 9.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, Maisch S, Carr D, Gerlach F, Bufe A, Lauener RP, Schierl R, Renz H, Nowak D, von Mutius E. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347:869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 10.Waser M, von Mutius E, Riedler J, Nowak D, Maisch S, Carr D, Eder W, Tebow G, Schierl R, Schreuer M, Braun-Fahrlander C. Exposure to pets, and the association with hay fever, asthma, and atopic sensitization in rural children. Allergy. 2005;60:177–184. doi: 10.1111/j.1398-9995.2004.00645.x. [DOI] [PubMed] [Google Scholar]
- 11.Bloomfield SF, Rook GA, Scott EA, Shanahan F, Stanwell-Smith R, Turner P. Time to abandon the hygiene hypothesis: new perspectives on allergic disease, the human microbiome, infectious disease prevention and the role of targeted hygiene. Perspectives in public health. 2016;136:213–224. doi: 10.1177/1757913916650225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Frontiers in cellular and infection microbiology. 2012;2:94. doi: 10.3389/fcimb.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parfrey LW, Walters WA, Knight R. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front Microbiol. 2011;2:153. doi: 10.3389/fmicb.2011.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 22.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, Antonopoulos DA, Zhou L, Chang EB, Fu YX, Nagler CR. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE, Macia L, Mackay CR. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell reports. 2016;15:2809–2824. doi: 10.1016/j.celrep.2016.05.047. [DOI] [PubMed] [Google Scholar]
- 24.Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133:291–307. doi: 10.1016/j.jaci.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Iweala OI, Nagler CR. Immune privilege in the gut: the establishment and maintenance of non-responsiveness to dietary antigens and commensal flora. Immunol Rev. 2006;213:82–100. doi: 10.1111/j.1600-065X.2006.00431.x. [DOI] [PubMed] [Google Scholar]
- 26.Iweala OI, Burks AW. Food Allergy: Our Evolving Understanding of Its Pathogenesis, Prevention, and Treatment. Curr Allergy Asthma Rep. 2016;16:37. doi: 10.1007/s11882-016-0616-7. [DOI] [PubMed] [Google Scholar]
- 27.Brandt EB, Strait RT, Hershko D, Wang Q, Muntel EE, Scribner TA, Zimmermann N, Finkelman FD, Rothenberg ME. Mast cells are required for experimental oral allergen-induced diarrhea. J Clin Invest. 2003;112:1666–1677. doi: 10.1172/JCI19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoy KD, Harris NL, Diener P, Hatak S, Odermatt B, Hangartner L, Senn BM, Marsland BJ, Geuking MB, Hengartner H, Macpherson AJ, Zinkernagel RM. Natural IgE production in the absence of MHC Class II cognate help. Immunity. 2006;24:329–339. doi: 10.1016/j.immuni.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wannemuehler MJ, Kiyono H, Babb JL, Michalek SM, McGhee JR. Lipopolysaccharide (LPS) regulation of the immune response: LPS converts germfree mice to sensitivity to oral tolerance induction. J Immunol. 1982;129:959–965. [PubMed] [Google Scholar]
- 31.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 32.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, Peet A, Tillmann V, Uibo R, Mokurov S, Dorshakova N, Ilonen J, Virtanen SM, Szabo SJ, Porter JA, Lahdesmaki H, Huttenhower C, Gevers D, Cullen TW, Knip M, Xavier RJ. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson DW, Kim YB. Modification of Host Responses to Bacterial Endotoxins. I. Secificity of Pyrogenic Tolerance and the Role of Hypersensitivity in Pyrogenicity, Lethality, and Skin Reactivity. J Exp Med. 1963;118:425–446. doi: 10.1084/jem.118.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Feehley T, Belda-Ferre P, Nagler CR. What’s LPS Got to Do with It? A Role for Gut LPS Variants in Driving Autoimmune and Allergic Disease. Cell Host Microbe. 2016;19:572–574. doi: 10.1016/j.chom.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 37.Faria AM, Weiner HL. Oral tolerance: mechanisms and therapeutic applications. Adv Immunol. 1999;73:153–264. doi: 10.1016/s0065-2776(08)60787-7. [DOI] [PubMed] [Google Scholar]
- 38.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007:1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, Muller W, Sparwasser T, Forster R, Pabst O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokota-Nakatsuma A, Takeuchi H, Ohoka Y, Kato C, Song SY, Hoshino T, Yagita H, Ohteki T, Iwata M. Retinoic acid prevents mesenteric lymph node dendritic cells from inducing IL-13-producing inflammatory Th2 cells. Mucosal Immunol. 2014;7:786–801. doi: 10.1038/mi.2013.96. [DOI] [PubMed] [Google Scholar]
- 44.Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, Lee JY, Lee M, Surh CD. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016;351:858–863. doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- 45.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 47.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 48.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanoue T, Atarashi K, Honda K. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol. 2016;16:295–309. doi: 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- 51.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, Busslinger M, Cerf-Bensussan N, Boneca IG, Voehringer D, Hase K, Honda K, Sakaguchi S, Eberl G. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 52.Houston SA, Cerovic V, Thomson C, Brewer J, Mowat AM, Milling S. The lymph nodes draining the small intestine and colon are anatomically separate and immunologically distinct. Mucosal Immunol. 2016;9:468–478. doi: 10.1038/mi.2015.77. [DOI] [PubMed] [Google Scholar]
- 53.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 54.Cao S, Feehley TJ, Nagler CR. The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS letters. 2014;588:4258–4266. doi: 10.1016/j.febslet.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, Riethmacher D, Si-Tahar M, Di Santo JP, Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S, Charbonnier LM, Noval Rivas M, Georgiev P, Li N, Gerber G, Bry L, Chatila TA. MyD88 adaptor-dependent microbial sensing by regulatory T cells promotes mucosal tolerance and enforces commensalism. Immunity. 2015;43:289–303. doi: 10.1016/j.immuni.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, Du K, Singh S, Pike JW, Darnay BG, Choi Y, Wu H. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 58.Han D, Walsh MC, Cejas PJ, Dang NN, Kim YF, Kim J, Charrier-Hisamuddin L, Chau L, Zhang Q, Bittinger K, Bushman FD, Turka LA, Shen H, Reizis B, Defranco AL, Wu GD, Choi Y. Dendritic cell expression of the signaling molecule TRAF6 is critical for gut microbiota-dependent immune tolerance. Immunity. 2013;38:1211–1222. doi: 10.1016/j.immuni.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han D, Walsh MC, Kim KS, Hong SW, Lee J, Yi J, Rivas G, Surh CD, Choi Y. Microbiota-Independent Ameliorative Effects of Antibiotics on Spontaneous Th2-Associated Pathology of the Small Intestine. PLoS One. 2015;10:e0118795. doi: 10.1371/journal.pone.0118795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AS. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, Roberts LK, Wong CH, Shim R, Robert R, Chevalier N, Tan JK, Marino E, Moore RJ, Wong L, McConville MJ, Tull DL, Wood LG, Murphy VE, Mattes J, Gibson PG, Mackay CR. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nature communications. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 62.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 64.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 66.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of GPR109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, Prasad PD, Ganapathy V. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murukami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 69.Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 71.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 72.Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Annu Rev Pathol. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- 73.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nature reviews Drug discovery. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 74.Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, Arike L, Wising C, Svensson F, Backhed F, Hansson GC. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013;305:G341–347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M, Comerma L, Huang B, Blander JM, Xiong H, Mayer L, Berin C, Augenlicht LH, Velcich A, Cerutti A. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 2015;8:198–210. doi: 10.1038/mi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knoop KA, McDonald KG, Kulkarni DH, Newberry RD. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016;65:1100–1109. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hammad H, Lambrecht BN. Barrier Epithelial Cells and the Control of Type 2 Immunity. Immunity. 2015;43:29–40. doi: 10.1016/j.immuni.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 83.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529:226–230. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, Artis D, Garrett WS. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351:1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529:221–225. doi: 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hung LY, I, Lewkowich P, Dawson LA, Downey J, Yang Y, Smith DE, Herbert DR. IL-33 drives biphasic IL-13 production for noncanonical Type 2 immunity against hookworms. Proc Natl Acad Sci U S A. 2013;110:282–287. doi: 10.1073/pnas.1206587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gronke K, Diefenbach A. Tuft cell-derived IL-25 activates and maintains ILC2. Immunol Cell Biol. 2016;94:221–223. doi: 10.1038/icb.2016.10. [DOI] [PubMed] [Google Scholar]
- 88.Zaph C, Du Y, Saenz SA, Nair MG, Perrigoue JG, Taylor BC, Troy AE, Kobuley DE, Kastelein RA, Cua DJ, Yu Y, Artis D. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mosconi I, Geuking MB, Zaiss MM, Massacand JC, Aschwanden C, Kwong Chung CK, McCoy KD, Harris NL. Intestinal bacteria induce TSLP to promote mutualistic T-cell responses. Mucosal Immunol. 2013;6:1157–1167. doi: 10.1038/mi.2013.12. [DOI] [PubMed] [Google Scholar]
- 90.De Salvo C, Wang XM, Pastorelli L, Mattioli B, Omenetti S, Buela KA, Chowdhry S, Garg RR, Goodman WA, Rodriguez-Palacios A, Smith DE, Abbott DW, Cominelli F, Bamias G, Xin W, Lee JJ, Vecchi M, Pizarro TT. IL-33 Drives Eosinophil Infiltration and Pathogenic Type 2 Helper T-Cell Immune Responses Leading to Chronic Experimental Ileitis. Am J Pathol. 2016;186:885–898. doi: 10.1016/j.ajpath.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fricke WF, Song Y, Wang AJ, Smith A, Grinchuk V, Mongodin E, Pei C, Ma B, Lu N, Urban JF, Jr, Shea-Donohue T, Zhao A. Type 2 immunity-dependent reduction of segmented filamentous bacteria in mice infected with the helminthic parasite Nippostrongylus brasiliensis. Microbiome. 2015;3:40. doi: 10.1186/s40168-015-0103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee JB, Chen CY, Liu B, Mugge L, Angkasekwinai P, Facchinetti V, Dong C, Liu YJ, Rothenberg ME, Hogan SP, Finkelman FD, Wang YH. IL-25 and CD4(+) TH2 cells enhance type 2 innate lymphoid cell-derived IL-13 production, which promotes IgE-mediated experimental food allergy. J Allergy Clin Immunol. 2016;137:1216–1225. doi: 10.1016/j.jaci.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chu DK, Llop-Guevara A, Walker TD, Flader K, Goncharova S, Boudreau JE, Moore CL, Seunghyun In T, Waserman S, Coyle AJ, Kolbeck R, Humbles AA, Jordana M. IL-33, but not thymic stromal lymphopoietin or IL-25, is central to mite and peanut allergic sensitization. J Allergy Clin Immunol. 2013;131:187–200. doi: 10.1016/j.jaci.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 94.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, Chehoud C, Kuczynski J, DeSantis T, Warrington J, Hyde ER, Petrosino JF, Gerber GK, Bry L, Oettgen HC, Mazmanian SK, Chatila TA. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–212. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noval Rivas M, Burton OT, Wise P, Charbonnier LM, Georgiev P, Oettgen HC, Rachid R, Chatila TA. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity. 2015;42:512–523. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA, Pott J, Griseri T, Bollrath J, Hegazy AN, Harrison OJ, Owens BM, Lohning M, Belkaid Y, Fallon PG, Powrie F. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–568. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4:137rv135. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Mohn WW, Turvey SE, Brett Finlay B. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 100.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy. 1999;29:342–346. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 101.Thompson-Chagoyan OC, Fallani M, Maldonado J, Vieites JM, Khanna S, Edwards C, Dore J, Gil A. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow’s milk protein allergy. Int Arch Allergy Immunol. 2011;156:325–332. doi: 10.1159/000323893. [DOI] [PubMed] [Google Scholar]
- 102.Nakayama J, Kobayashi T, Tanaka S, Korenori Y, Tateyama A, Sakamoto N, Kiyohara C, Shirakawa T, Sonomoto K. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunol Med Microbiol. 2011;63:397–406. doi: 10.1111/j.1574-695X.2011.00872.x. [DOI] [PubMed] [Google Scholar]
- 103.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 104.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, Mandhane PJ, Turvey SE, Subbarao P, Becker AB, Scott JA, Kozyrskyj AL. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. 2015;45:632–643. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 105.Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X, Yuan L, Wang Y, Sun J, Li L, Xiang C. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22:713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 107.Berni Canani R, Nocerino R, Terrin G, Frediani T, Lucarelli S, Cosenza L, Passariello A, Leone L, Granata V, Di Costanzo M, Pezella V, Troncone R. Formula selection for managment of children with cow milk allergy influences the rate of acquisition of tolerance: a prospective multicenter study. J Pediatr. 2013;163:771–777. doi: 10.1016/j.jpeds.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 108.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, Calignano A, Khan AA, Gilbert JA, Nagler CR. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. Isme j. 2016;10:742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wesemann DR, Nagler CR. The Microbiome, Timing, and Barrier Function in the Context of Allergic Disease. Immunity. 2016;44:728–738. doi: 10.1016/j.immuni.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olle B. Medicines from microbiota. Nat Biotechnol. 2013;31:309–315. doi: 10.1038/nbt.2548. [DOI] [PubMed] [Google Scholar]
- 112.Vickery BP, Berglund JP, Burk CM, Fine JP, Kim EH, Kim JI, Keet CA, Kulis M, Orgel KG, Guo R, Steele PH, Virkud YV, Ye P, Wright BL, Wood RA, Burks AW. Early oral immunotherapy in peanut-allergic preschool children is safe and highly effective. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.05.027. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]