Abstract
Previous work with EM immunolocalization examined the intracuticular placement of several antibodies directed against cuticular proteins (CPs) in various structures of Anopheles gambiae. Those structures had long stretches of fairly uniform cuticle. We have now used 19 antibodies directed against members of five CP families on two adult structures with considerable complexity, Johnston's organ and the corneal lens of the compound eye. We also localized chitin with colloidal-gold labeled wheat germ agglutinin. Twelve of these antibodies recognized structures in Johnston's organ. Only 6 were detected in the outer pedicel wall, but the internal structures were more complex with distinct distributions of members of the five CP families in six different structures. The corneal lens had four distinct regions of laminar cuticle. Thirteen of the 15 members of the CPR family were detected, none from the other CP families. Specific antibodies were localized to different regions and in different laminae within a region. The specificity of deployment of cuticular proteins revealed in this study is helping to explain why An. gambiae allocates about 2% of its protein coding genes to structural CPs.
Keywords: cuticular laminae, antennae, compound eye, EM Immunolocalization, mosquito
Graphical abstract
1. Introduction
Anopheles gambiae allocates almost 2% (295) of its protein coding genes to structural cuticular proteins (CPs). In attempts to learn why there are so many CPs, we have examined the precise location of a small number of these proteins within the cuticle with EM immunolocalization (Vannini et al., 2014b, 2015; Vannini and Willis, 2016). The structures we examined had long stretches of seemingly identical cuticle and the epitopes detected for individual CPs covered large areas of these structures. We thought that we might gain additional information by using our available antibodies on two adult structures with special features. Johnston's Organ (JO), in the second antennal segment, also known as the pedicel of the antenna (Harbach and Knight, 1980), transmits information about wing beat frequency from nearby mosquitoes and contains many distinct cuticular structures (Clements, 1999; Hart et al., 2011). The corneal lens of the compound eye has laminae that are far better defined than what we saw in the surface cuticle.
The CPs of An. gambiae have been classified into 13 families based on sequence characteristics. (For review see Willis, 2010; Willis et al., 2012.). Chitin binding has been demonstrated for members of six of these families, CPR, CPAP1, CPAP3, CPCFC, CPFL and TWDL (Rebers and Willis, 2001; Jasrapuria et al., 2010; Tang et al., 2010; Dong et al., 2016).
The CPR family with 164 members in An. gambiae is the largest CP family in every other arthropod studied and has been divided into two major groups, RR-1 and RR-2. A convenient tool for recognizing these groups is the Web Site CutProtFam-Pred (http://aias.biol.uoa.gr/CutProtFam-Pred/home.php; Ioannidou et al., 2014). We recently used 13 antibodies against members of the CPR family and learned that the two hypotheses about the utilization of these groups, separation by type of cuticle or by exclusive location in exo- or endo-cuticle, were each somewhat incorrect. Rather some RR-2s while restricted to hard cuticle were found in both exo- and endo-cuticle, and one RR-1 protein was detected in hard cuticle (Vannini and Willis, 2016).
We had also published results of EM immunolocalization using antibodies against individual members of the CPF, CPLCG, and CPCFC families (Vannini et al., 2014b, 2015). Thus we had a collection of 19 antibodies that we thought might give us fresh insights if used against the cuticle in the two specialized structures that are the focus of this report, JO and corneal lens.
Results using antibodies against CPR130, CPR152 and CPAP3-C are reported here for the first time. CPR130 failed to give significant results in our earlier analysis. An analysis of transcript levels in whole animals across development revealed that transcripts from CPR152 were present at much higher levels in male than in female pharate and newly eclosed adults (Togawa et al., 2008). In situ hybridization revealed that its mRNA was restricted to JO (unpublished observations), so its precise location in JO was of special interest. CPAP3-C, is an ortholog of gasp (also known as obstr-C) in Drosophila melanogaster where it participates in the formation of the tracheal lumen (Behr and Hoch, 2005; Tiklova et al., 2013). We wondered if it might play a role in the lumen of JO.
Our analysis of CPs in different adult structures by tandem mass spectrometry identified 79 CPs in JO and 90 in the corneal lens, both preparations including surrounding cuticle (Zhou et al., 2016). These data too revealed the need to learn more about how individual CPs contribute to cuticle.
The CPs of mosquitoes present a challenge to EM immunolocalization and to MS analyses because many proteins come from sequence clusters, groups of almost identical genes (Cornman et al., 2008; Cornman and Willis, 2008, 2009). We have used three antibodies that recognize from 6 to 10 proteins within a sequence cluster because no suitable unique peptide could be obtained. Two other antibodies recognize proteins from two distinct genes that code for identical proteins. Another peptide is found in two otherwise unrelated proteins. Problems with such similar genes have been discussed in some detail in prior publications (Vannini and Willis, 2016; Zhou et al., 2016).
The results demonstrate that we were correct in anticipating that the two new structures we have examined would provide new information about the utilization of CPs.
2. Materials and methods
2.1. Mosquito rearing
Eggs were obtained from the colony of An. gambiae (G3 strain) maintained in the insectary of the University of Georgia Entomology Department. Larvae were reared in a 12/12 L/D photoperiod at 27° C, fed ground Koi food (Foster and Smith Aquatics, Rhinelander, WI USA), and adults had access to water and an 8% fructose solution. To obtain developmentally synchronized pharate adults, pupae were collected at hourly intervals, separated by sex and maintained in small groups until they reached the desired age. Adults were collected on the morning after emergence (d 0) and prepared for fixation.
2.2. Antibody production
Sixteen of the 19 primary antibodies tested in this study were used in previous studies by Vannini et al. (2014b; 2015) and Vannini and Willis (2016). All the antibodies, except for the anti-3RC Cluster and anti-CPCFC1, were raised against synthetic peptides at Genscript Inc. (Piscataway, NJ) and affinity purified. Peptides were selected to avoid similarity to other An. gambiae CP sequences using blastp at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). See Supplementary File 1 for further information.
Anti-CPR130, anti-CPR152 and anti-CPAP3-C were used here for the first time. Two isoforms are known for CPAP3-C and the antibody was designed to recognize both of them. (See sequences in Supplementary File 2).
The colloidal-gold conjugated secondary antibodies (Sigma-Aldrich Co., St. Louis, MO) were 10 nm goat-anti-mouse and 5 or 10 nm goat-anti-rabbit. The presence of chitin was monitored with colloidal-gold conjugated Wheat Germ Agglutinin (WGA) (15 nm particle size) obtained from EY Laboratories Inc. (San Matteo, CA).
2.3. Western blots
Crude protein extracts came from 5-day-old adults using the second antennal segment for CPR152 and CPAP3-C and legs for CPR130. These were reacted with antibodies following methods previously described (Vannini and Willis, 2016).
2.4. Immunogold localization
Part of the head including the eyes and the second antennal segment (pedicel) with Johnston's organ and a few segments of the more distal flagellum from less than 12h old males were dissected and prepared for TEM. Tissues were fixed in 4% formaldehyde, 0.3% glutaraldehyde + 4% sucrose in phosphate buffer 1X (PBS) (pH 7.4) overnight at 4° C, and embedded in LR White resin (Electron Microscopy Sciences). The last step, carried out under nitrogen at 55° C for two days. A more detailed description is in Vannini et al. (2014b).
Primary antibodies were diluted in 0.5 M NaCl, 0.1% BSA, 0.05% TWEEN 20 and 5% FBS. The dilutions used are given in Table 1 as well the list of tissues tested. All treatments were done as described in Vannini et al. (2014b). The grids with sections were incubated face down on drops of the appropriate solutions. All steps were performed at room temperature except the incubation of the primary antibody that was performed at 4°C. As a negative control, sections were treated in the same way without the primary antibody incubation step.
Table 1.
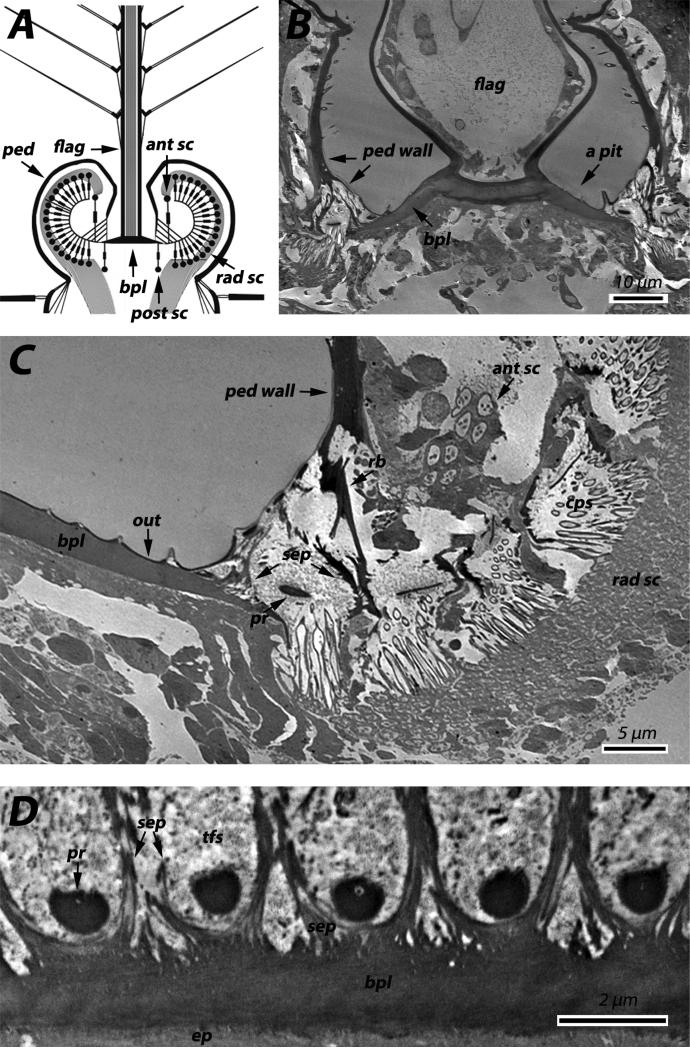
Details about structure tested, antibody dilution ([ ]) and size of gold particles used for immunolocalization. Antibodies against names of members of the CPR family are in color, green for RR-1group, brown for RR-2 and blue for unassigned. AR=antigen retrieval process used. Gray boxes indicate where positive label was found. Numbers refer to figure numbers and letters to figure panel where results are shown.
| Primary antibodies | [1°Ab] | 2°Ab [1”50] | Antenna | Corneal lens | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Antennal flagellum | Second antennal segment | ||||||||||
| Pedicel wall | Johnston's Organ | ||||||||||
| Basal plate | Prongs | Septa | Terminal filaments | Cap | Scolopidia lumen | ||||||
| Anti-CPR12=13 | [1:100] | anti-rabbit 10nm gold | 11C | ||||||||
| Anti-CPR22 | [1:100] | anti-rabbit 10nm gold | 13C | ||||||||
| Anti-CPR59/70 | [1:100] | anti-rabbit 5nm gold | 2B | 3A | 4C | 5A | 6A | 12A | |||
| Anti-CPR60 | [1:100] AR | anti-rabbit 10nm gold | 2A | 3C | 4B | 5B | 11B | ||||
| Anti-CPR61 | [1:100] | anti-rabbit 10nm gold | 11D | ||||||||
| Anti-CPR75 | [1:100] | anti-rabbit 10nm gold | 13A | ||||||||
| Anti-CPR125 | [1:100] | anti-rabbit 5nm gold | 2G,H | 3D | 13D | ||||||
| Anti-CPR130 | [1:100] AR | anti-rabbit 10nm gold | 5C | 12D | |||||||
| Anti-CPR133=153 | [1:100] | anti-rabbit 10nm gold | 6B | 8B | |||||||
| Anti-CPR140 | [1:100] | anti-rabbit 5nm gold | 4F | 13E | |||||||
| Anti-CPR151 | [1:100] | anti-rabbit 10nm gold | 8C | 13B | |||||||
| Anti-CPR152 | [1:100] | anti-rabbit 5nm gold | 4E | 6C | 7C | 8D | 9C,D | ||||
| Anti-2RA Cluster | [1:100] | anti-rabbit 10nm gold | 2C | 6D | 12C | ||||||
| Anti-2RB Cluster | [1:100] | anti-rabbit 5nm gold | 13F | ||||||||
| Anti-3RC Cluster | [1:10K] | anti-rabbit 10nm gold | 2D | 3B | 4D | 5D | 6E | 12B | |||
| Anti-CPAP3-C | [1:100] | anti-rabbit 5nm gold | 7B | 8E | 9E,F | ||||||
| Anti-CPCFC1 | [1:5K] | anti-rabbit 5nm gold | 2E | 3E | 6F | ||||||
| Anti-CPF3 | [1:100] | anti-mouse 10nm gold | 8F | ||||||||
| Anti-CPLCG3/4/5 | [1:20K] | anti-rabbit 5nm gold | 2F | 3F | |||||||
| WGA | [1:10] 15 nm gold | 4A | 7A | 8A | 9A,B | 11A | |||||
For CPR130 and CPR60, a post-embedding antigen retrieval (AR) procedure was necessary and performed following Saito et al. (2003), described in more detail in Vannini and Willis (2016). The ultrathin sections mounted on a nickel grid were placed on 100% EtOH followed by 0.05M TRIS pH 10 for 40 min at 99°C. After a brief step in the blocking solution, treatment with the primary antibody and the protocol following were done as usual.
The presence of chitin in the different regions of the Johnston's organ and in the corneal lens was revealed by labelling with WGA-gold that binds chitin, performed following Lee et al. (1990) and described in detail in Vannini and Willis (2016).
Sections were examined in a JEOL JEM-1011 transmission electron microscope (JEOL USA Inc. Peabody, MA) at 80 kV. The images were captured with a XR80M Wide-Angle Mid-Mount CCD Camera from AMT (Advanced Microscopy Techniques, Woburn, MA).
The secondary antibody used to detect the primary antibody had been conjugated with colloidal gold and did not react with the cuticular parts of the antenna or corneal lens when tested on sections that had been incubated with the binding buffer without the primary antibody (Supplementary File 3). Additional controls for many of these antibodies are in earlier papers (Vannini et al., 2014b, 2015; Vannini and Willis, 2016).
3. Results
3.1. Western blot
Western blot results for all but three of the primary antibodies used in this study were presented in previous work (Vannini et al., 2014b, 2015; Vannini and Willis, 2016).
Strong bands were detected for all three new proteins, with molecular masses larger than what was calculated for CPR151 and CPAP3-C, and lower for CPR130 (Supplementary File 4). The different molecular masses most likely reflect the already described abnormal electrophoretic mobility of many CPs (Cox and Willis, 1987).
3.2 Cuticular protein localization in antennal structures
3.2.1 CP localization in antennal flagellum and pedicel wall of second antennal segment
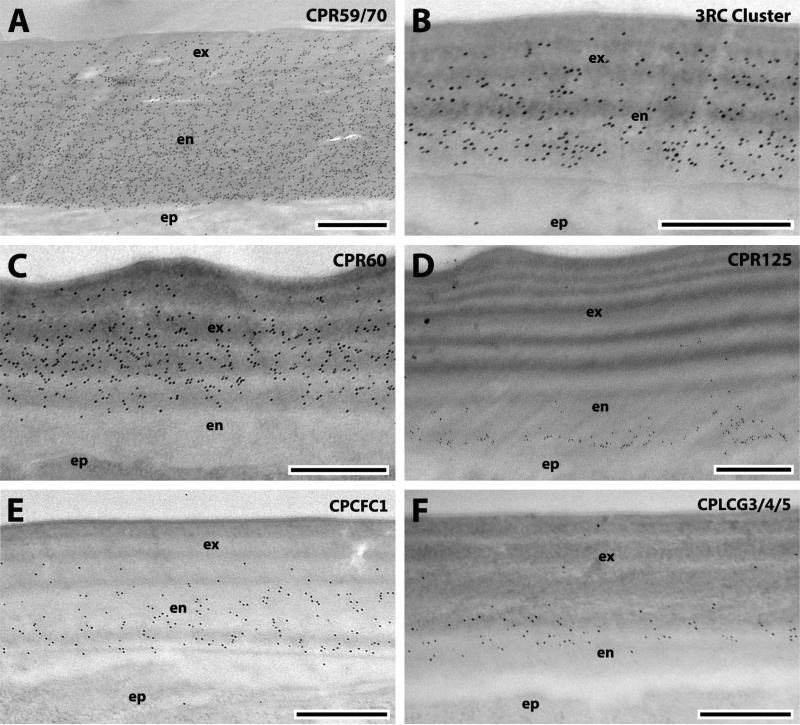
JO fills the enlarged pedicel of the second antennal segment in dipteran males (Fig. 1A) and females have a fully develop JO but it is far smaller (Supplementary File 5F) and has a reduced sensibility compared to males (Göpfert et al., 1999). The second antennal segment is covered with a cuticular pedicel wall that appears similar to the surface cuticle of the more distal flagellum that extends distally from the apical pit of the pedicel (Fig. 1A, B). The pedicel wall surrounded the JO up to where the flagellum extends and where it becomes thinner and organized in ribs (Fig. 1B). The results with the 19 antibodies that we tested were almost identical for the flagellum and pedicel wall (Table 1). In addition to the expected presence of chitin (data not shown), we detected binding of 7 antibodies, including those designed to recognize the RR-2 proteins: CPR60 (Figs. 2A, 3C); CPR59/70 (Figs. 2B, 3A); and the 3RC cluster (Figs. 2D, 3B). The only RR-1 protein detected was CPR125 (Figs. 2G, H, 3D). Also detected were members of two other CP families, CPCFC1 (Figs. 2E, 3E) and CPLCG3/4/5 (Figs. 2F, 3F). 2RA was found only in sections of the pedicel wall (Fig. 2C). Interestingly, the density of gold particles for 2RA was far lower in the outer regions of the exocuticle. Perhaps as sclerotization proceeds, some proteins become unavailable as antigens (Fig. 2C).
Fig. 1.
Overview of the antennal pedicel and Johnston's organ. A: Schematic representation of a transversal section of a pedicel (modified from Hart et al, 2011). B: TEM section of the junction between the flagellum and the pedicel. C: TEM transversal section of Johnston's organ. D: TEM section of the connections of the prongs and septa to the basal plate. Abbreviations: ant sc, anterior scolopidia; tfs, terminal filaments; flag, flagellum; bpl, basal plate; ped, pedicel; post sc, posterior scolopidia; pr, prong; rad sc, radial scolopidia; rb, rib; sep, septa; ped wall, pedicel wall; cps: caps; out, outer layer of the basal plate; ep, epidermis.
Fig. 2.
EM immunolocalization of CPR60, CPR59/70, 2RA Cluster, 3RC Cluster, CPCFC1, CPLCG3/4/5 and CPR125, in TEM sections of the flagellum of a young adult. Abbreviations as in Fig. 1. Scale bar 500nm.
Fig. 3.
EM immunolocalization of CPR59/70, 3RC Cluster, CPR60, CPR125, CPCFC1 and CPLCG3/4/5 in TEM sections of the pedicel wall of a young adult. Abbreviations as in Fig. 1. Scale bar 500nm.
The location of the antigens within the cuticle was also almost the same for both structures (Table 2). Three were present in both exo- and endo-cuticle (CPR59/70, 2RA, 3RC); CPR60 was restricted to the exocuticle, while CPCFC1 and CPLCG3/4/5 were predominantly detected in endocuticle. CPR125, was present throughout the endocuticle (Fig. 2G) of the flagellum closest to the pedicel. Distally its localization was restricted to a distinct row of gold particles at the junction between epidermis and endocuticle (Fig. 2H) as in the pedicel wall (Fig. 3D).
Table 2.
Comparison of intracuticular localization of peptides from this study with previously published work from our laboratory (Vannini et al., 2014b; Vannini et al., 2015; Vannini and Willis, submitted). Names of members of the CPR family are green for RR-1 group, brown for RR-2, and blue for unclassified. CPR152 falls just below the threshold for a RR-2 using CutProtFam-Pred for group assignment. Numbers in bottom row refer to figure numbers in current paper.
| Primary antibodies | ANTENNA | Previous work | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antennal flagellum | Second antennal segment | ||||||||||||
| Pedicel wall | Johnston's Organ | ||||||||||||
| exo | endo | exo | endo | Basal plate outer | Basal plate inner | Prongs | Septa | Caps | Terminal filaments? | Scolopidia lumen | exo | endo | |
| Anti-CPR12=13 | No Ab detected | soft | |||||||||||
| Anti-CPR22 | No Ab detected | soft | |||||||||||
| Anti-CPR59/70 | + | +++ | ++ | +++ | +++ | − | +++ | +++ | − | − | − | ++ | ++ |
| Anti-CPR60 | + | − | ++ | − | ++ | ++ | + | − | − | − | − | ++ | − |
| Anti-CPR61 | No Ab detected | soft | |||||||||||
| Anti-CPR75 | No Ab detected | soft | |||||||||||
| Anti-CPR125 | − | + | − | + | No Ab detected | − | + | ||||||
| Anti-CPR130 | − | − | − | − | − | − | + | − | − | − | − | No prior data | |
| Anti-CPR133=153 | − | − | − | − | − | − | − | + | ++ | − | − | soft | |
| Anti-CPR140 | − | − | − | − | − | ++ | − | − | − | − | − | +++ | + |
| Anti-CPR151 | − | − | − | − | − | − | − | − | + | − | − | soft | |
| Anti-CPR152 | − | − | − | − | − | + | − | + | ++ | ++ | ++ | No prior data | |
| Anti-2RA Cluster | + | ++ | − | − | − | − | − | + | − | − | − | +++ | ++ |
| Anti-2RB Cluster | No Ab detected | +++ | − | ||||||||||
| Anti-3RC Cluster | + | ++ | ++ | ++ | ++ | − | ++ | ++ | − | − | − | ++ | + |
| Anti-CPAP3-C | − | − | − | − | − | − | − | − | + | ++ | ++ | No prior data | |
| Anti-CPCFC1 | +? | +++ | − | ++ | − | − | − | + | − | − | − | + | +++ |
| Anti-CPF3 | − | − | − | − | − | − | − | − | +++ | − | − | +++ | − |
| Anti-CPLCG3/4/5 | − | ++ | − | +? | No Ab detected | − | +++ | ||||||
| Figure | 2 | 3 | 4 | 5 | 6 | 8 | 7 | 9 | |||||
3.2.2. JO within the second antennal segment
JO is a complex structure with many distinct cuticular components in addition to its neural components (Boo and Richards, 1975a,b). More CPs were detected within Johnston's organ than in its surrounding pedicel. Indeed only 7 of the 19 antibodies we tested failed to detect proteins in one or more of its structures. Furthermore, different structures had from 2-6 CPs associated with them and all but the caps had chitin (Table 1). Antibodies against 7 CPs were only detected in components of JO and not the flagellum or pedicel wall (Table 1).
In situ hybridization, revealed the presence of transcripts for CPR152, CPAP3-C, CPCFC1, CPLCG3 and CPLCG4 in Johnston's organ of pharate adults (Supplementary File 5).
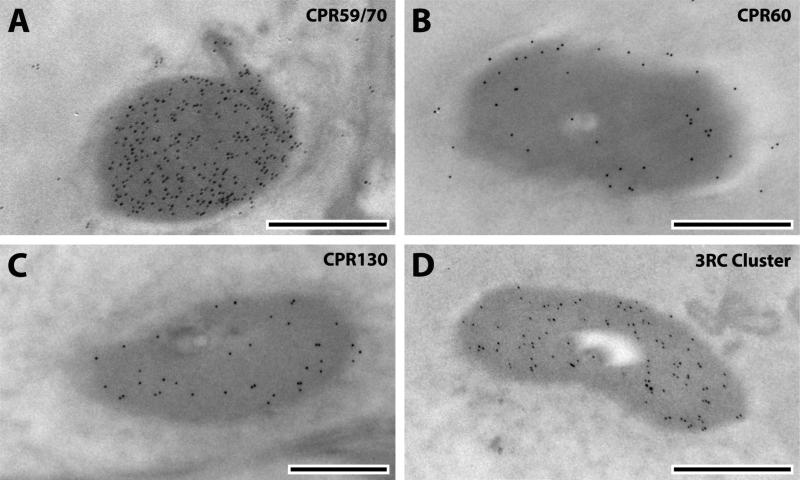
The bottom of the apical pit is occupied by the basal plate, a circular disc of unsclerotized cuticle (Clements, 1999) that lacks laminae and any morphological distinction between exo- and endo-cuticle (Figs.1, 4). Cuticular projections, such as prongs and septa, spread from it (Fig. 1C, D). The septa lie between the prongs that curve out and up from the basal plate and are attached to the ribs by fragile strips of cuticle that form a network of cuticle. Thanks to these connections the basal plate is extremely flexibly suspended and capable of responding to each flagellum vibration. The basal plate shows an outer layer (Fig. 1C) interrupted by dorsal crests that Clemens (1999) called “possibly epicuticle.” Treatment with WGA showed not only that this layer had chitin but the gold particles were more densely arrayed there than in the inner regions of the basal plate below (Fig 4A). The presence of chitin eliminates the possibility that this is epicuticle, because epicuticle is chitin-free. (See Willis et al., 2012 for review and mention of one possible exception to this generalization.) CPR60, CPR59/70 and the 3RC Cluster were also localized in the outer layer (Fig. 4B-D). CPR59/70 and the 3RC cluster were found in both exoand endo-cuticle of the flagellum and pedicel, while CPR60 was restricted to the exocuticle (Table 2). Thus it is of interest that of these three, only CPR60 was also detected in the inner regions of the basal plate (Fig. 4B, Table 2). Interestingly, the other CPs localized exclusively in the endocuticle of the pedicel wall and flagellum (CPR125, CPCFC1 and CPLCG3/4/5) were not found in the outer layer of the basal plate (Table 2). This suggests that the outer layer of the basal plate is an extension of the pedicel wall exocuticle. CPR152 and CPR140 were the only two CPs restricted to the inner region of the basal plate (Fig. 4E, F). CPR152 was found in several structures of JO, whereas CPR140 was only detected in JO's basal plate in this study (Tables 1, 2). This is the first record of proteins localized in the basal plate, a key structure in mosquito reproduction and environmental sensing.
Fig. 4.
EM immunolocalization of WGA, CPR60, CPR59/70, 3RC Cluster, CPR152 and CPR140 in TEM sections of the basal plate of a young adult. Abbreviations as in Fig. 1. Scale bar 500nm.
Four CPs were detected in prongs, 6 in septa (Figs. 5-6). Treatment with WGA showed that these structures contain chitin (data not shown). The prongs were the only antennal structure that had evidence for CPR130 (Fig. 5C). CPR152 also was detected in the region surrounding the prongs (Fig. 7C) that we believe to show transverse sections of terminal filaments connecting the scolopidia to the prongs (Hart et al., 2011).
Fig. 5.
EM immunolocalization of CPR59/70, CPR60, CPR130, and 3RC Cluster in TEM sections of the prongs of a young adult. Abbreviations as in Fig. 1. Scale bar 500nm.
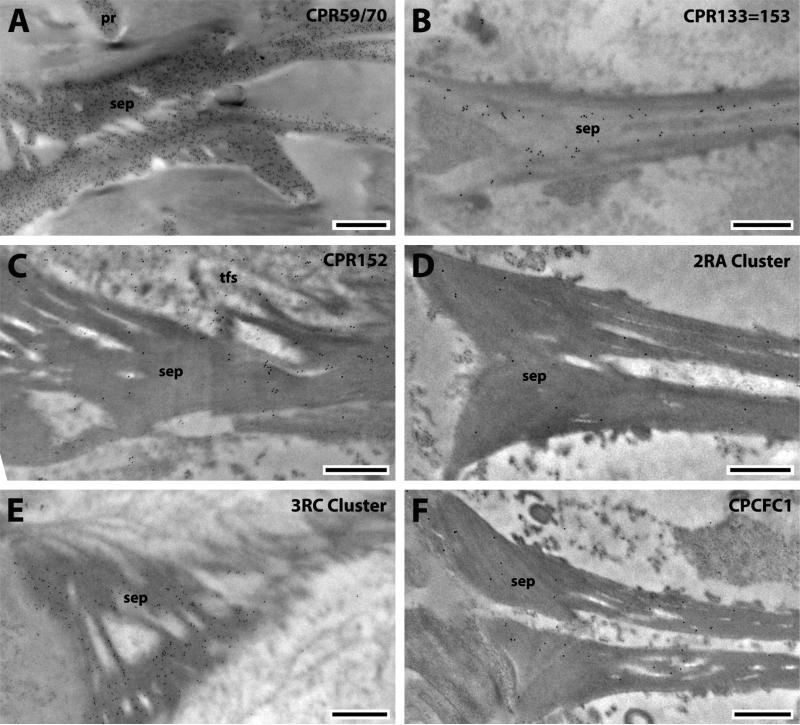
Fig. 6.
EM immunolocalization of CPR59/70, CPR133=153, CPR152, 2RA Cluster, 3RC Cluster and CPCFC1 in TEM sections of the septa of a young adult. Abbreviations as in Fig. 1. Scale bar 500nm.
Fig. 7.
EM immunolocalization of WGA, CPAP3-C and CPR152 in TEM sections of terminal filaments of a young adult. Abbreviations as in Fig. 1. Scale bar 500nm.
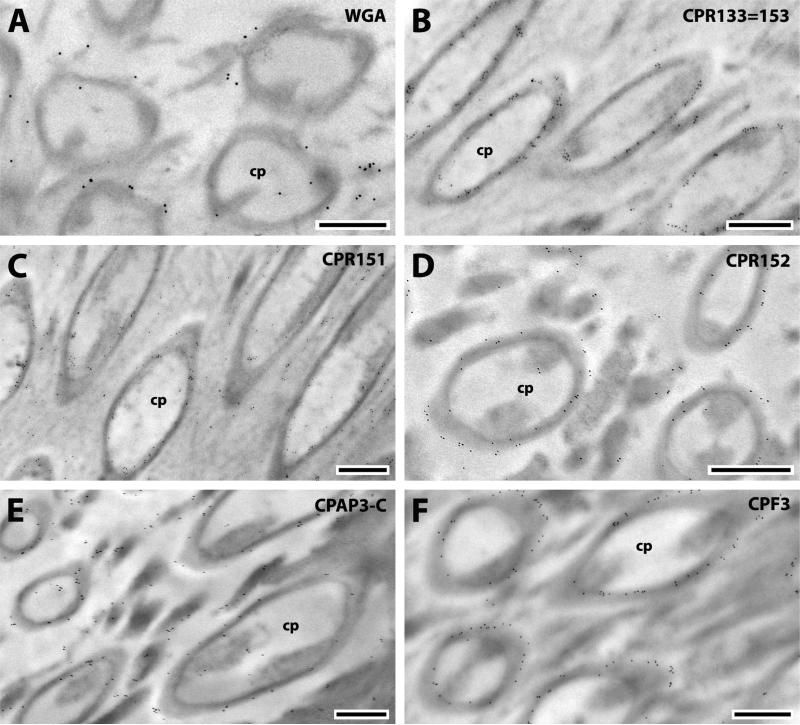
The scolopidia are the fundamental sensory units of Johnston's organ (Clements, 1999; Hart et al., 2011). Each scolopidium is covered by a conical cuticular cap (Hart et al., 2011) where CPR133=153, CPR151, CPR152, CPAP3-C and CPF3 were found (Fig. 8). WGA failed to react with caps itself but sporadic gold particles were found on the surrounding rods (Fig. 8A). The absence of chitin in the caps themselves suggests a different nature for these structures even though all of the CPs detected there except for CPF3 have chitin-binding domains.
Fig. 8.
EM immunolocalization of CPR133=153, CPR151, CPR152, CPAP3-C and CPF3 in TEM sections of the caps of a young adult and WGA surrounding caps. Abbreviations as in Fig. 1. Scale bar 500nm.
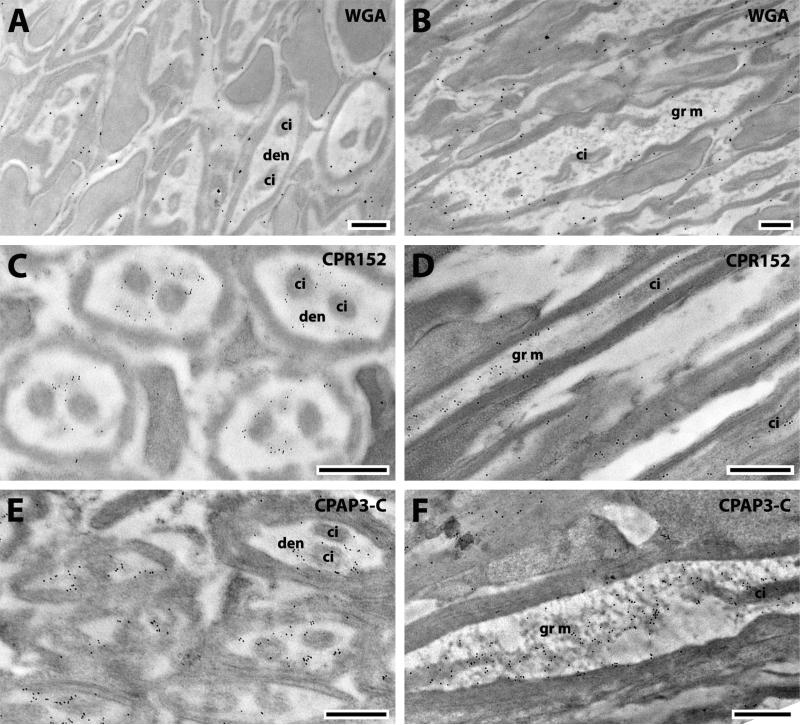
Radial scolopidia represent 97% of all the sensitive units; they have two similar neurons with a dendrite consisting of two ciliary structures (Hart et al., 2011). CPR152 and CPAP3-C were found in the dendrite lumen (Fig. 9 C, E). Longitudinal sections of the dendrites revealed the localization of the two proteins in the granular material that fills the lumen (Fig. 9 D, F). This material of unknown composition deposited between the ciliary structures does not contain chitin (Fig. 9 A, B), yet both of these proteins have chitin-binding domains.
Fig. 9.
EM immunolocalization of WGA, CPR152 and CPAP3-C in TEM sections of radial scolopidia of a young adult. Abbreviations: den, dendrite; ci, ciliary structure; gr m, granular material. Scale bar 500nm.
3.3. CP localization in the corneal lens
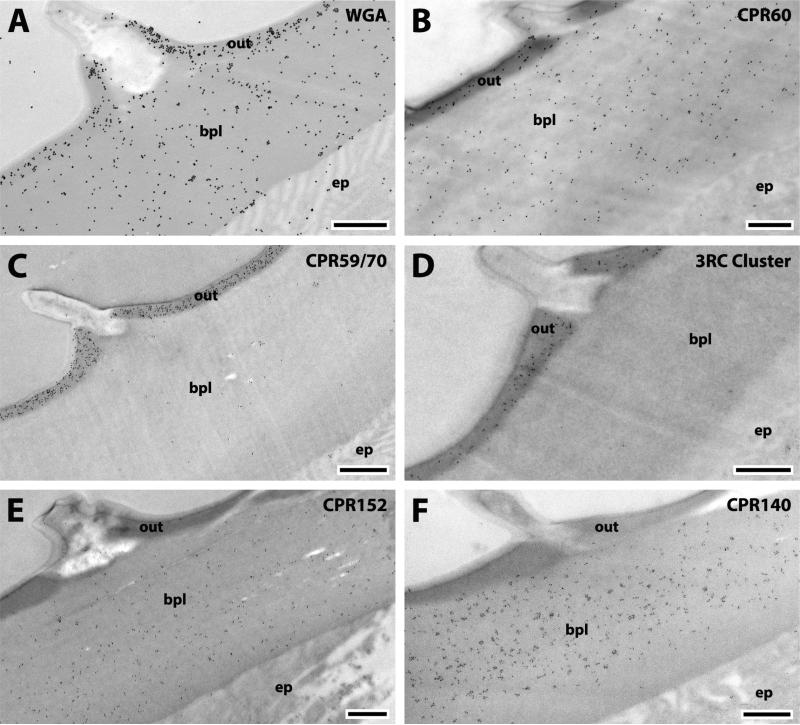
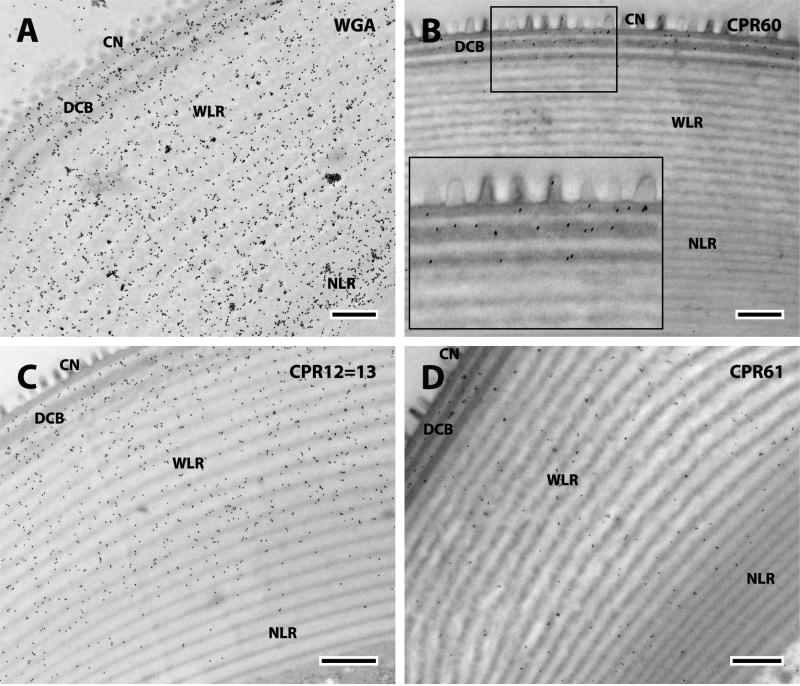
The corneal lens of the compound eye (Fig.10) is a modified transparent portion of the chitinous exoskeleton made of concentric layers covered externally by small conical protuberances known as the corneal nipples partially embedded in the outer layer of the corneal lens in Diptera (Trujillo-Cenóz, 1972). Immediately below the nipples are three dark bands with two clear bands in-between that we have called the DCB (dense and clear band region). The largest inner portion of the cornea can be classified in two regions: wide laminae (WLR) and narrow laminae (NLR) connected to the crystalline cone and darker (Fig.10). The outer alternating bands are the substratum of a system of interference filters tinting the light reaching the photoreceptors (Trujillo-Cenóz, 1972). WGA was localized throughout the corneal lens except for the corneal nipples (Fig. 11A).
Fig. 10.
Corneal lens. CN, corneal nipples; DCB, dense and clear bands; WLR, wide lamellar region; NLR, narrow lamellar region. Scale bar 500nm.
Fig. 11.
EM immunolocalization of WGA, CPR60, CPR12=13, and CPR61 in TEM sections of a single corneal lens of a young adult. Abbreviations as in Fig. 10. Scale bar 500nm.
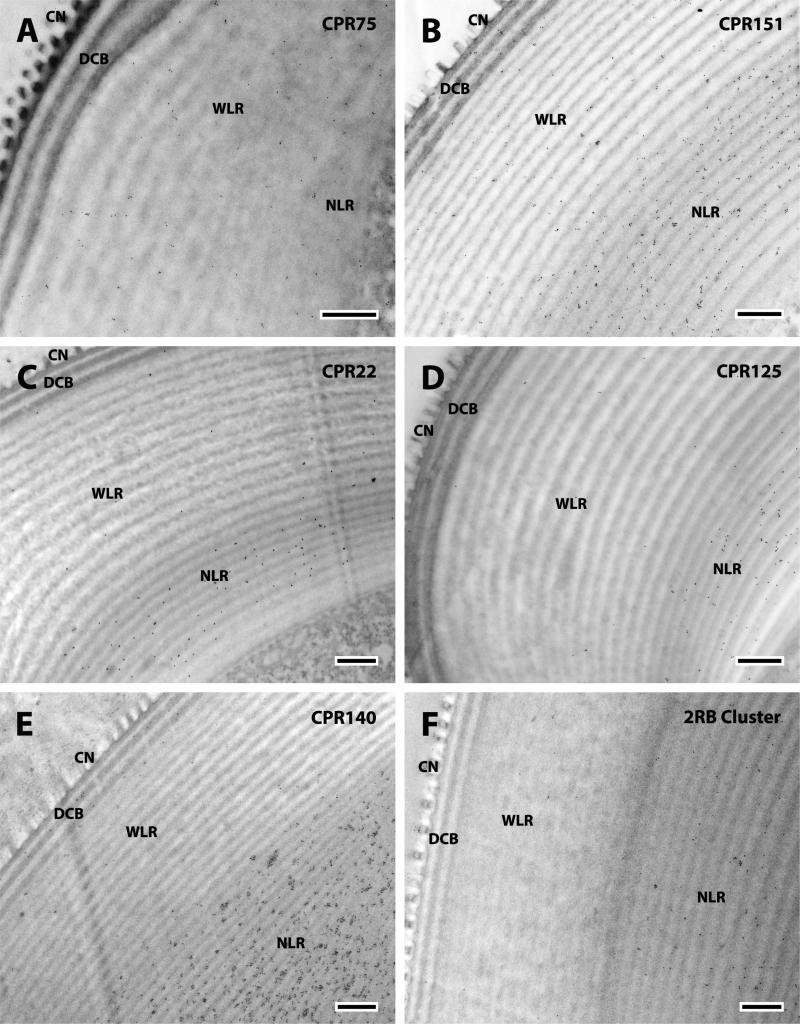
We detected even more (13) of the 19 CPs in the corneal lens than we had in JO. Importantly, they were all members of the CPR family (Tables 1, 3). The localization of the CPRs in the corneal lens fell into 4 patterns according to the region where proteins were detected: i) DCB only (Fig. 11 B); ii) the DCB and WLR, (Fig. 11 C, D); iii) the DCB and NLR (Fig. 12); iv) the NLR only (Fig. 13). These locations and their specific proteins are summarized in Table 3.
Table 3.
Location of antibodies in the corneal lens of a young adult showing the distribution of antibodies within each of the three main regions. The number of pluses was determined by eye to depict relative abundance of gold particles from a single antibody. For several antibodies, the distinction between dark and light laminae in the narrow laminar region was not obvious. The Figure numbers are on the right. Names of members of the CPR family are green for RR-1 group, brown for RR-2, and blue for unclassified.
| Primary antibodies | CORNEAL LENS | Figure | |||||
|---|---|---|---|---|---|---|---|
| Dense and clear band (DCB) | Wide laminar region (WLR) | Narrow laminar region (NLR) | |||||
| Dark laminae | Light laminae | Dark laminae | Light laminae | Dark laminae | Light laminae | ||
| Anti-CPR12=13 | + | +++ | ++ | ++ | + | + | 11C |
| Anti-CPR22 | − | − | − | − | + | + | 13C |
| Anti-CPR59/70 | ++++ | + | − | − | ++ | 12A | |
| Anti-CPR60 | + | − | − | − | − | − | 11B |
| Anti-CPR61 | + | + | + | + | + | − | 11D |
| Anti-CPR75 | − | − | + | + | + | 13A | |
| Anti-CPR125 | − | − | − | − | ++ | ++ | 13D |
| Anti-CPR130 | ++ | + | − | − | ++++ | 12D | |
| Anti-CPR140 | + | − | − | − | ++++ | 13E | |
| Anti-CPR151 | + | + | + | + | ++ | ++ | 13B |
| Anti-2RA Cluster | ++ | + | − | − | +++ | 12C | |
| Anti-2RB Cluster | − | − | − | − | + | + | 13F |
| Anti-3RC Cluster | ++ | − | + | + | + | + | 12B |
| WGA (chitin) | ++++ | ++ | +++ | +++ | ++++ | 11A | |
| Anti-CPR133=153 | NO ANTIBODY DETECTED | ||||||
| Anti-CPR152 | |||||||
| Anti-CPAP3-C | |||||||
| Anti-CPCFC1 | |||||||
| Anti-CPF3 | |||||||
| Anti-CPLCG3/4/5 | |||||||
Fig. 12.
EM immunolocalization of CPR59/70, 3RC Cluster, 2RA Cluster, and CPR130 in TEM sections of a single corneal lens of a young adult. Abbreviations as in Fig. 10. Scale bar 500nm.
Fig. 13.
EM immunolocalization of CPR75, CPR151, CPR22, CPR125, CPR140 and 2RB Cluster in TEM sections of a single corneal lens of a young adult. Abbreviations as in Fig. 10. Scale bar 500nm.
In situ hybridization had revealed transcripts in pharate adults for CPR59 and CPR75. They code for two of the proteins detected in this study and were found in cells beneath the corneal lens (Vannini et al., 2014a).
WGA detected chitin in all regions and in both the dark and light laminae (Fig. 11A). But the patterns of CP localization were different for the different proteins, for some were exclusively present or more abundant in the dark laminae where they could be recognized, for it was impossible to distinguish the two types of laminae in some sections of the NLR. Thus CPR60, CPR59/70, and the 3RC Cluster were found only in the darker bands of the DCB (Figs. 11B, 12A, B). The only protein that was most abundant in the lighter bands of the DCB was the RR-1 protein CPR12=13 (Fig. 11C). These distributions are summarized in Table 3.
4. Discussion
4.1 Overview
The most striking finding of this study was the precise localization of 19 CPs in specific components of the structures studied. Most of the cuticular structures within JO were detected with only a subset of the antibodies tested but the total number of CPs detected in JO (12) was greater than in the structurally less specialized antenna (7) or pedicel of JO (6). The corneal lens also showed specific localization of individual CPs. We had 15 antibodies against members of the CPR family, 13 recognized structures in the corneal lens, including 6 that had not reacted with JO components. Interestingly, the 4 antibodies we had directed against members of other CP families, failed to recognize components of the corneal lens, while they did recognize antennal structures. This precise deployment of specific CPs to components of a single structure helps to explain why there are so many distinct genes for CPs in insect genomes and invites further investigation to correlate protein properties with localization.
4.2 Localization of CPs within the cuticle of the antennal flagellum and Johnston's organ
Many regions of cuticle have a laminar structure and other investigators have reported that certain antibodies preferentially recognize proteins in specific regions of the procuticle (reviewed in Willis et al., 2012; more recent findings in Vannini and Willis, 2016). This study extended these findings, for once again we found proteins restricted to the exo- or endo-cuticle of the antennal flagellum and pedicel wall. The distribution of proteins in the two regions of the procuticle of these structures was the same as we had previously found in legs and sclerites with four RR-2 proteins (CPR59/70, CPR140, 2RA and 3RC) present in both exo- and endo-cuticle, one, CPR60, exclusively in exocuticle. The only RR-1 protein, CPR125, that we had found present in hard cuticle where it was restricted to the endocuticle, also appeared in the endocuticle of the flagellum and pedicel wall (Table 2).
The antennae are sensitive hearing organs, and their auditory mechanism depends on the ability of the Johnston's organ to transmit information received by the flagellae to the central nervous system. Essential for this is the basal plate of Johnston's organ that vibrates after wing vibrations from another mosquito or other signals have induced deflection of the antennae. This gives the basal plate a crucial role in hearing and everything that depends on that, including frequency matching during courtship (Göpfert et al., 1999; Eberl and Boekhoff-Falk, 2007). The internal structures of JO did not show any laminar cuticle nor a distinction between exo- and endo-cuticle. The only region with two distinct areas was the basal plate. CPR60 was in both regions, while CPR59/70 was found in the outer plate and CPR140 was restricted to the inner region of the basal plate where it appeared in small clusters, as it did in other structures. So, we cannot equate the distinction between these two regions to exo- and endo-cuticle. We included CPR152 in our analysis because MS/MS data revealed that it is found only in antennae and predominantly in Johnston's organ (Zhou et al., 2016). It was also found in the inner region of the basal plate, but intriguingly also in most of the other components of JO.
We were particularly interested in CPAP3-C for its ortholog in D. melanogaster binds the intraluminal chitin fibrils during tracheal development (Barry et al., 1999). The extent of growth of the diameter of tracheae is determined by the amount of intraluminal protein present. The chitin cable is transient and cleared from the lumen before larval hatching to enable gas filing (Tiklová et al., 2013). We can speculate a similar function for the Anopheles CPAP3-C in the radial scolopidia, suggesting that the amount of the protein bond to or part of the granular material determines the size of the dendritic lumen and is not cleared out after development is complete, because there is no need to open airways or other spaces in this tissue. But the role of CPAP3-C in the terminal filaments and caps will need another explanation.
4.3 Localization of CPs in the corneal lens of the compound eye
It is well known that diverse species have built their lenses with proteins that also serve other roles, a phenomenon that has been named “gene sharing” (Piatigorsky, 2007). What was surprising was the large number of proteins we found in the corneal lens, first with in situ hybridization (Vannini et al., 2014a), next with MS/MS (Zhou et al., 2016) and now with EM immunolocalization.
In our previous work with EM immunolocalization, we failed to detect a distribution of antibodies biased against either dark or light laminae (Vannini et al., 2014b, 2015; Vannini and Willis, 2016). But in this study where we examined the corneal lens, we found evidence for CPs localized exclusively or primarily in the dark regions of cuticular laminae and one, CPR125, in the light laminae of the DCR (Table 3). We are not the first to observe differential localization of a protein within the laminae of the corneal lens. Komori et al. (1992) identified a 52 kDa protein found exclusively in the eye. They had only obtained N-terminal sequence data, but used an antibody raised against their 13 aa to localize the peptide in the corneal lens. With precise quantitation, they found 2.3 times as many gold particles from the secondary antibody in the dark layers as in the light layers of the corneal laminae. A subsequent study (Janssens and Gehring, 1999) obtained the complete sequence for this protein, named it crystalline (crys, CG16963) and recognized that it had the R&R Consensus chitin-binding domain. This protein scores as an RR-2 on CutProtFam-Pred (Ioannidou et al., 2014). There is no ortholog of crys in An. gambiae.
4.4 Comparison of localization of specific CPs between antennal structures and eye with other adult structures previously examined with LC MS/MS
We have compared our finding with the antibodies to data obtained from an LC MS/MS analysis carried out on the second antennal segment with its JO, the rest of the antenna and the corneal lens (Zhou et al., 2016). Data are summarized in Supplementary File 6 where we go beyond scoring presence and absence to include a rough quantitation of the MS/MS data obtained from spectral counts. We show the rank order of the proteins we studied compared to all possible CPs. The limitations of using spectral counts with data that include proteins found in sequence clusters and others that share peptides are discussed in Zhou et al. (2016), but spectral counts provide at least a rough view of relative abundance.
In general, the MS/MS data fit our current results. Three of the antibodies tested (anti-CPR59, -CPR70 and -CPR60) recognized proteins in all three structures and were among the most abundant proteins in all three structures. They were, however, absent from terminal filaments, caps and the scolopidia lumen. The MS/MS data indicate that although we had an antibody that potentially recognized CPLCG3, CPLCG4 and CPLCG5 (Supplementary File 2), it is unlikely that it was recognizing CPLCG3 since that protein had not been identified in our MS analysis,
A protein of low abundance in the second antennal segment (CPF3) was detected in only one of the two MS/MS samples where it scored #46. This fits nicely with our only finding it in the caps of JO. In contrast, CPR130 which scored well in the MS/MS analysis of all three structures, was only detected in the prongs and abundantly in the narrow laminar region of the corneal lens. Perhaps it becomes modified so that it is less accessible to the specific antibody that we used.
5. Conclusions
The data we obtained provide the first record of CPs localized in the cuticle of the different components that make up JO. With one exception, Dmelcrys, they are also, as far as we know, the first to localize specific proteins in distinct regions and layers of the corneal lens. Our previous immunolocalization studies examined structures with almost no differentiation of cuticle types, except soft (intersegmental membranes) and hard (sclerites, legs). Now that we have examined two structures with well demarcated components, we found that CP deployment can be complex. These data that revealed the precise localization of different CPs in different components of both JO and the eye lens, is helping us to understand why An. gambiae devotes so many genes to structural CPs.
Supplementary Material
Highlights.
EM immunolocalization used 19 antibodies vs. Anopheles gambiae cuticular proteins.
The second antennal segment with Johnston's organ (JO) and corneal lens were studied.
Internal structures of JO were recognized by 12 antibodies from 4 CP families.
Antibodies against 13 CPRs reacted with the corneal lens in a region specific manner.
ACKNOWLEDGMENTS
We thank Mark R. Brown and Anne Elliot for maintaining the mosquito facility from which the animals were obtained and M.R. Brown for advice on immunolabeling and comments on the MS. We also thank Mary B. Ard and John P. Shield of the Center for Advanced Ultrastructural Research at the University of Georgia for technical support. This work was supported by a grant from the U.S. National Institutes of Health R01AI055624.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barry MK, Triplett AA, Christensen AC. A peritrophin-like protein expressed in the embryonic tracheae of Drosophila melanogaster. Insect Biochem Mol Biol. 1999;29:319–327. doi: 10.1016/s0965-1748(99)00004-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- Behr M, Hoch M. Identification of the novel evolutionary conserved obstructor multigene family in invertebrates. FEBS. 2005;579:6827–6833. doi: 10.1016/j.febslet.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Boo KS, Richards AG. Fine structure of the scolopidia in the Johnston’s organ of male Aedes aegypti (L.) (Diptera:Culicidae). Int.J. Insect Morph. & Embryol. 1975a;4:549–566. doi: 10.1016/0022-1910(75)90126-2. [DOI] [PubMed] [Google Scholar]
- Boo KS, Richards AG. Fine structure of scolopidia in Johnston’s organ of female Aedes aegypti compared with that of the male. J Insect Physiol. 1975b;21:1129–1139. doi: 10.1016/0022-1910(75)90126-2. [DOI] [PubMed] [Google Scholar]
- Clements AN. The biology of mosquitoes. Vol. 2. Johnston's organ, CABI Publishing; New York: 1999. The antennae and hearing. pp. 62–71. 1999. [Google Scholar]
- Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, Willis JH. Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles gambiae. BMC Genomics. 2008;9:22. doi: 10.1186/1471-2164-9-22. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH. Extensive gene amplification and concerted evolution within the CPR family of cuticular proteins in mosquitoes. Insect Biochem Mol Biol. 2008;38:661–676. doi: 10.1016/j.ibmb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH. Annotation and analysis of low-complexity protein families of Anopheles gambiae that are associated with cuticle. Insect Molec Biol. 2009;18:607–622. doi: 10.1111/j.1365-2583.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DL, Willis JH. Analysis of the cuticular proteins of Hyalophora cecropia with two dimensional electrophoresis. Insect Biochem. 1987;17:457–468. [Google Scholar]
- Dong Z, Zhang W, Zhang Y, Zhang X, Zhao P, Xia Q. Identification and characterization of novel chitin-binding proteins from the larval cuticle of silkworm, Bombyx mori. J Proteome Res. 2016;15:1435–1445. doi: 10.1021/acs.jproteome.5b00943. [DOI] [PubMed] [Google Scholar]
- Eberl DF, Boekhoff-Falk G. Development of Johnston's organ in Drosophila. Int J Dev Biol. 2007;51:679–687. doi: 10.1387/ijdb.072364de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göpfert MC, Briegel H, Robert D. Mosquito-hearing: sound-induced antennal vibrations in male and female Aedes aegypti. J Exp Biol. 1999;202:2727–2738. doi: 10.1242/jeb.202.20.2727. [DOI] [PubMed] [Google Scholar]
- Harbach RE, Knight KL. Taxonomist's glossary of mosquito anatomy. first ed. Plexus Publishing, Inc.; Marlton, New Jersey: 1980. [Google Scholar]
- Hart M, Belton P, Kuhn R. European Mosquito Bulletin. 2011;29:103–113. The Risler Manuscript. [Google Scholar]
- Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas S. CutProtFam-Pred: detection and classification of putative structural cuticular proteins from sequence alone, based on profile hidden Markov models. Insect Biochem. Mol. Biol. 2014;52:51–59. doi: 10.1016/j.ibmb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens H, Gehring WJ. Isolation and characterization of drosocrystallin, a lens crystallin gene of Drosophila melanogaster. Devel. Biol. 1999;207:204–214. doi: 10.1006/dbio.1998.9170. [DOI] [PubMed] [Google Scholar]
- Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem Mol Biol. 2010;40:214–227. doi: 10.1016/j.ibmb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Komori N, Usukura J, Matsumoto H. Drosocrystallin, a major 52 kDa glycoprotein of the Drosophila melanogaster corneal lens. Purification, biochemical characterization, and subcellular localization. J Cell Sci. 1992;102(Pt 2):191–201. doi: 10.1242/jcs.102.2.191. [DOI] [PubMed] [Google Scholar]
- Lee SA, Clarke BS, Jenner DW, Williamson FA. Cytochemical demonstration of the effects of the acylureas flufenoxuron and diflubenzuron on the incorporation of chitin into insect cuticle. Pestic Sci. 1990;28:367–375. [Google Scholar]
- Piatigorsky J. Gene Sharing and Evolution: the diversity of protein functions. fourth ed. Harvard University Press; Cambridge, Massachusetts: 2007. [Google Scholar]
- Rebers JE, Willis JH. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem Mol Biol. 2001;31:1083–1093. doi: 10.1016/s0965-1748(01)00056-x. [DOI] [PubMed] [Google Scholar]
- Saito N, Konishi K, Takeda H, Kato M, Sugiyama T, Asaka M. Antigen retrieval trial for post-embedding immunoelectron microscopy by heating with several unmasking solutions. J Histochem Cytochem. 2003;51:989–994. doi: 10.1177/002215540305100802. [DOI] [PubMed] [Google Scholar]
- Tang L, Liang J, Zhan Z, Xiang Z, He N. Identification of the chitin binding proteins from the larval proteins of silkworm, Bombyx mori. Insect Biochem Mol Biol. 2010;40:228–234. doi: 10.1016/j.ibmb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Tiklová K, Tsarouhas V, Samakovlis C. Control of airway tube diameter and integrity by secreted chitin-binding proteins in Drosophila. PLOS one. 2013;8:e67415. doi: 10.1371/journal.pone.0067415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Dunn WA, Emmons AC, Nagao J, Willis JH. Developmental expression patterns of cuticular protein genes with the R&R Consensus from Anopheles gambiae. Insect Biochem Mol Biol. 2008;38:508–519. doi: 10.1016/j.ibmb.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-Cenóz O. In: The structural organization of the compound eye in insects. In Handbook of sensory physiology. Fuortes MGF, editor. Vol. 7. Springer; Heidelberg, (NY): 1972. pp. 5–62. [Google Scholar]
- Vannini L, Bowen JH, Reed T, Willis JH. The CPCFC Cuticular Protein Family: anatomical and cuticular locations in Anopheles gambiae and distribution throughout Pancrustacea. Insect Biochem Mol Biol. 2015;65:57–67. doi: 10.1016/j.ibmb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Dunn AW, Reed TW, Willis JH. Changes in transcript abundance for cuticular proteins and other genes three hours after a blood meal in Anopheles gambiae. Insect Biochem Mol Biol. 2014a;44:33–43. doi: 10.1016/j.ibmb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Reed TW, Willis JH. Temporal and spatial expression of cuticular proteins of Anopheles gambiae implicated in insecticide resistance or differentiation of M/S incipient species. Parasites & Vectors. 2014b;7:24. doi: 10.1186/1756-3305-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Willis JH. Localization of RR-1 and RR-2 Cuticular Proteins within the cuticle of Anopheles gambiae. 2016 doi: 10.1016/j.asd.2016.10.002. (submitted to Arthropod Structure and Development) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem Mol Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Papandreou NC, Iconomidou VA, Hamodrakas SJ. In: Cuticular proteins. In Insect Molecular Biology and Biochemistry. Gilbert LI, editor. Academic; Chapel Hill (NC): 2012. pp. 134–166. [Google Scholar]
- Zhou Y, Badgett MJ, Bowen JH, Vannini L, Orlando R. Distribution of cuticular proteins in different structures of adult Anopheles gambiae. Insect Biochem Mol Biol. 2016;75:1–13. doi: 10.1016/j.ibmb.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.