Abstract
Chronic wounds represent a major healthcare and economic problem worldwide. Advanced wound dressings that incorporate bioactive compounds have great potential for improving outcomes in patients with chronic wounds but significant challenges in designing treatments that are effective in long-standing, non-healing wounds. Here, we developed an optimized wound healing gel that delivers syndecan-4 proteoliposomes (“syndesomes”) with FGF-2 to enhance diabetic wound healing. In vitro studies demonstrated that syndesomes markedly increased migration of keratinocytes and fibroblasts isolated from both non-diabetic and diabetic donors. In addition, syndesome treatment led to increased endocytic processing of FGF-2 that included enhanced recycling of FGF-2 to the cell surface after uptake. The optimized syndesome formulation was incorporated into an alginate wound dressing and tested in a splinted wound model in diabetic, ob/ob mice. We found that wounds treated with syndesomes and FGF-2 had markedly enhanced wound closure in comparison to wounds treated with only FGF-2. Moreover, we show that syndesomes have an immunomodulatory effect on wound macrophages, leading to a shift towards the M2 macrophage phenotype and alterations in the wound cytokine profile. Together, these studies showed that delivery of exogenous syndecan-4 is an effective method for enhancing wound healing in the long-term diabetic diseased state.
Keywords: wound healing, diabetic ulcers, syndecan-4, immunomodulation, fibroblast growth factor-2
INTRODUCTION
Type 2 diabetes is a highly prevalent disorder that impacts 347 million people worldwide. Neuropathy and microvascular angiopathy are common complications of diabetes and contribute to a 12–25% lifetime risk of developing diabetic ulcers.[1] These diabetic ulcers are responsible for 25–50% of the total cost of diabetes treatment and are the most common cause of limb amputations in the United States.[2] Non-healing, diabetic ulcers are a complex clinical problem requiring a multifaceted treatment plan with standard therapeutic components including removal of necrotic tissue from wound (debridement), reduction of pressure in the wound (offloading), infection control, surgical revascularization, and limb elevation or compression. However, in many cases these treatments are ineffective, leaving patients with chronic ulcers and enhanced risk for limb amputation.
A number of advanced wound dressings have been used to enhance healing of chronic ulcers. The most prevalent approaches to bioactive dressings can be broadly classified into the categories of local delivery of growth factors,[3] delivery of therapeutic genes[4] or delivery of stem cells.[5] Of these strategies, only growth factors have been tested in large clinical trials, perhaps due to the safety and logistical challenges accompanying gene or stem cell therapies. However, the vast majority growth factor therapies have limited success in clinical trials for wound healing.[6] The only approved clinical growth factor treatment for chronic wounds is recombinant PDGF-BB (Becaplermin), and, while approved by the FDA, it has shown mixed results in clinical trials on chronic ulcers.[7] Other growth factors including FGF-2 and EGF have either shown no improvement or only moderate benefits in small clinical trials.[8] Thus, while clinical studies have shown that growth factor therapies are well tolerated by patients, there is a pronounced need to improve the efficacy of these treatments to maximize the benefit of these therapies and make them cost effective for our healthcare system.
Here, we hypothesized that the diabetic state prevents the effectiveness of growth factor therapies through alterations in expression and proteolytic degradation of receptors and co-receptors. This concept is supported by reduced efficacy of growth factors in many clinical trials for enhancing healing in chronic wounds[8] compared to healthy animals.[9] In addition, our group recently examined the expression of growth factor receptors and co-receptors in the heart and skeletal muscle of diabetic mice and found a significant loss in syndecan-4 and other cell surface proteoglycans that serve as co-receptors for growth factors including FGF-2, VEGF-A and PDGF-CC.[10] We have also shown that diabetes and other disease states increase expression of heparanase, an enzyme that cuts the heparan sulfate chains and increases shedding of cell surface proteoglycans.[11, 12] In this study, we demonstrate that there is a reduction in syndecan-4 in the skin of human patients with type 2 diabetes. We examined whether delivery of syndecan-4 proteoliposomes (“syndesomes”) could overcome the inherent resistance to growth factor signaling to enhance the healing of wounds in diabetic mice with severe disease. Our studies show that syndesomes delivered locally from alginate wound dressings markedly enhanced the efficacy of FGF-2 therapy for wound healing in the diabetic disease state through multiple mechanisms.
MATERIALS AND METHODS
Human samples
Human skin samples were obtained from the Glasgow Caledonian University Skin Research Tissue Bank, Glasgow, UK. The tissue bank has NHS research ethics to supply human skin for research (REC REF: 11/S1402/2). The samples were already paraffin embedded before being shipped to us. The samples were sectioned using a microtome to obtain 6 µm thick sections. The slides were for syndecan-4 (Abcam) using the Envision+ Dual Link Kit (Dako). The details of the staining procedure are described in a later section. We used 9 samples in both diabetic and non-diabetic groups.
Recombinant syndecan-4 protein production
HEK293-T cells were transduced with a custom made plasmid with full-length syndecan-4 gene using a lentiviral transduction system. The syndecan-4 overexpressing stable cells were grown in high glucose DMEM with 10% FBS, 5% Penicillin-Streptomycin and 5% L-glutamine. The cells were lysed using a lysis buffer containing 1% Triton X-100 and protease inhibitors (Roche). The cell lysate was sonicated, vortexed and then centrifuged (25000 × g). The supernatant was used for protein purification by a HiTrap Q HP column (GE Healthcare) on a FPLC (Amersham Biosciences). Pure protein was confirmed by silver stain and a western blot probing for the syndecan-4 protein, and the protein concentration was quantified using a BCA assay (Thermo Scientific).
Preparation of syndesomes
The lipids used for the preparation were 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), cholesterol, and sphingomyelin (Avanti Polar Lipids). Briefly, a solution mixture of the four lipids at 10 mg/ml concentration was made in a volumetric ratio of 2:1:1:1 chloroform. The mixture was prepared in a round bottom glass flask and the chloroform was removed using a rotatory evaporator for 1 hour followed by treatment with stream of argon gas for 15 minutes. Once all the chloroform was removed, the lipid film was resuspended in a HEPES-buffered salt solution by vortexing, sonicating, and freeze thawing three times each in order. This lipid solution was then extruded through a 400-nm polycarbonate membrane filter (Avestin) to generate liposomes. Syndecan-4 protein was added to the liposome suspension to a final concentration of 50 µg/ml. To this solution we added 1% n-octyl-β-D-glucopyranoside to permeablize the liposomes and incorporate the protein. The detergent was then removed through serial dilution, extensive dialysis and treatment with BioBeads (SM-2, Bio-Rad). The amount of protein incorporated into the liposomes was measured using a BCA assay (Thermo Scientific). FGF-2 (Peprotech) was mixed with the syndesomes when they needed to be delivered together.
Liposome characterization
The size and dispersion of the syndesomes and isolated syndecan-4 was characterized by dynamic light scattering (Malvern Zetasizer Nano ZS). The instrument was calibrated using 54-nm diameter polystyrene particles. The syndesomes were diluted 1:1000 to fit the detection region of the instrument and then aliquoted into a polystyrene cuvette to run in the machine. The results were an average of 50 size measurements. For imaging with cryoelectron microscopy, the liposomes were plunge-frozen in liquid ethane on carbon holey film grids as previously described (R2×2 Quantifoil®; Micro Tools GmbH, Jena, Germany)[13]. The grids were transferred to a cryo-specimen holder (Gatan 626) under liquid nitrogen and put in a microscope (JEOL 2100 LaB6) operating at 200 keV. Grids were maintained at close to liquid nitrogen temperatures during EM session (−172°C to −180°C). Liposomes were imaged at 20,000× EM magnification with a 4kx4k slow-scan CCD camera (UltraScan 895, GATAN, Inc.) using low-dose imaging procedure. Images were acquired with less than 20 electrons/Å2 electron dose.
Preparation of syndesome-releasing alginate gels
We created a 6.35 mm diameter alginate disk using a custom-designed mold to implant in the wound. Equal volumes of 4% sodium alginate (Sigma) solution and 0.85% NaCl solution were mixed and the syndesomes and/or FGF-2 were added to this solution. The alginate solution was pipetted into the mold and then cross-linked with a solution of 1.1% CaCl2 for 1 hour at 4°C. We used 5 µg of FGF-2 and/or 0.5 µg of syndecan-4 protein according to the sample (control, FGF-2, S4PL or S4PL with FGF-2) in each disk implanted. For the release studies, 2 alginate disks with different treatments were placed in a scintillation vial containing 10ml of 1X PBS with Ca2+ and Mg2+. At every time point, 200ul of buffer was aliquoted from the scintillation vial and frozen while replacing 200ul of fresh buffer into the vial. The buffer samples at each time point were analyzed using FGF-2 ELISA for the amount of FGF-2 released cumulatively over the course of a week. The alginate disks were flash frozen in liquid nitrogen (−195C) and lyophilized overnight (−110C, 0.0005mbar) in scintillation vials. The final freeze-dried gels were sputter coated with gold discharge for 30 seconds and then imaged using the scanning electron microscope (FEI Quanta 650 ESEM) at 10kV.
Electric cell-substrate impedance sensing (ECIS) assay for cell migration
The cells used for the assay were adult dermal fibroblasts and adult epidermal keratinocytes, both from either healthy or type 2 diabetic donors (Lonza). Fibroblasts were grown in high glucose DMEM (Gibco) with Pen-Strep, 10% FBS, L-glutamine and fibroblast supplements (Lonza) while the keratinocytes were grown in MCDB-131 (Gibco) with Pen-Strep, 10% FBS, L-glutamine and keratinocyte supplements (Lonza). However when the cells were used for the experiment, the media was serum free. The 96W1E+ plates (96 wells) were first coated with 100 µl of 2mM Cysteine (Sigma) per well for 30 minutes followed by a quick wash with 1X PBS. The wells were then coated with 40 µl/well of fibronectin (Sigma) at 8 µg/ml overnight. After a quick 1X PBS wash, to remove unbound fibronectin, the cells were plated at 10,000 cells per well and allowed to attach, while the plate was placed on the Z-Theta instrument (ECIS), which created an electric fence around the electrode. The cells were allowed to settle and attach for 4 hours. Finally, the electric fence was turned off and the cells were allowed to migrate over the electrode. The substrate impedance and resistance were measured every 48 seconds at a frequency of 40,000Hz. Six wells per treatment group in the migration assay were used.
Fibroblast invasion assays
Human adult dermal fibroblasts (Lonza) were used in the Trevigen Inc. The fibroblasts were cultured in with high glucose DMEM with Pen-Strep, 10% FBS, L-glutamine and fibroblast supplements (Lonza). The cells were starved for 24 hours in media without serum before the assay in high glucose DMEM with Pen-Strep, L-glutamine (no FBS). The treatments were added to this starvation media. Collagen-I cell invasion assay to assess the invasion potential of the fibroblasts in the presence of various treatments. The top invasion chamber was coated with collagen I and kept to attach overnight. The cells were plated in each well at a concentration of 106 cells/ml in the top chamber and treatments were added to the bottom chamber. The cells were incubated with the treatments for 24 hours and then the top chambers were moved to the assay plate with Calcein-AM and cell dissociation solution. This solution detaches the cells that have invaded from the top chamber to the bottom side. Finally the assay plate was read without the top chamber at 485 nm excitation and 520 nm emission. The intensity is a measure of the amount of cell invasion through the collagen I layer. Six wells per treatment group in the invasion assay were used.
Intracellular trafficking of FGF-2 using co-localization with Rab adapter proteins
The Rab5-GFP, Rab7-GFP and Rab11-GFP plasmids were provided by Dr. Mukhopadhyay and have been previously described.[14] The Rab4-GFP and Rab9-GFP constructs were purchased from Addgene.[15] FGF-2 (Peprotech) was conjugated with Alexa Fluor 594 (Life Technologies) using the heparin column that binds the active site of FGF-2 preserving its biological activity.[10, 16] HEK293Ta cells plated on 8-well glass slides (ibidi) at 10000cells/well and were transfected with the above-mentioned plasmids using DNA HTS jetPEI transfection reagent (Polyplus transfection™) using standard protocol. Twenty four hours post transfection, the cells were treated with AF594 tagged FGF-2 (1 µl) and/or syndesomes (1 µl). The cells were fixed at 15, 30, 60 and 120 minutes using 4% paraformaldehyde (Electron Microscopy Sciences) and washed three times with PBS for 10 minutes each. The cell nuclei were stained with DAPI (Vector Labs) and mounting media was added in the wells. The slides were then imaged using the laser scanning confocal microscope (Zeiss LSM710). The percentage of Rab-GFP labeled endosomes containing AF594 tagged FGF-2 was calculated as follows: each cell was divided into four roughly equal quadrants and one of the quadrants was chosen for quantification randomly (using RAND function of Excel). The total number of Rab-GFP labeled endosomes and endosomes that co-localize with AF594 tagged FGF-2 in the chosen quadrant were counted to calculate the percentage of co-localization. For each time point and treatment group, 10 cells were analyzed using Metamorph (Molecular Devices).
Animal studies
All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of University of Texas at Austin, and in accordance with NIH guidelines “Guide for Care and Use of Laboratory Animals” for animal care. All the animal experiments were performed on a diabetic, obese and hyperlipidemic mouse model (ob/ob). All the ob/ob mice (B6.Cg-Lepob/J) were purchased from the Jackson Laboratory. All animals were fed a high fat diet (Research Diets - D12331) for 10 weeks before performing wound healing surgeries.
Excisional wound healing model
To examine wound healing in the diabetic and obese mice we used a full-thickness excisional model with a splint to prevent wound contraction.[17] A sterile 5-mm biopsy punch was used to outline a pattern of four wounds, two on either side of midline on the dorsum of the mouse. A splint was fashioned using 0.5-mm thick silicone sheet and was placed so that the wound was centered within the splint. The splint was immobilized in place using 6-0 nylon sutures and cyanoacrylate glue to prevent wound contraction. Alginate gel disks encapsulating syndesomes and/or FGF-2 were then applied directly to the region of the open wound. A single sheet of Tegaderm was used to cover all the wounds. Photographs of the wounds were taken on days 0, 7 and 14. The animals were euthanized at 2, 6 and 14 days, and the wounds were biopsied with a 10-mm biopsy punch. The tissues were snap frozen in liquid N2-chilled isopentane and used for further analysis.
Laser speckle contrast imaging of tissue perfusion
A custom Laser Speckle Contrast Imager (LSCI) was used to image the tissue blood flow as previously described.[18] Briefly, a near infrared (785nm, 50mW) laser diode (Thor Labs) was used to illuminate the wounds on the back, and the speckle was captured using a Zoom-7000 lens (Navitar) linked to a Bassler CCD camera (Graftek). The wounds were imaged right after surgery (day 0), and at day 7. All wounds were imaged simultaneously within the laser field to allow the quantification of relative perfusion.
Histological analysis and immunostaining
Tissues from the in vivo experiments were embedded in paraffin and 6 µm thick sections were produced using a microtome. The slides were stained with H&E or Movat’s pentachrome stains. The wound healing samples were also immunostained using the Envision+ Dual Link Kit (Dako North America, Inc.) for cytokeratin (Abcam), CD86 (Bioss), CD163 (Bioss) or von Willebrand factor (Dako). The details about the antibodies are mentioned in the Supplemental Table S2. Briefly, the slides were de-paraffinized and placed in a bucket with Antigen Retrieval Solution (Dako), and placed in the microwave (1250 W) for 2 minutes and 40 seconds. Then the bucket was placed in a water bath maintained at 80°C for 3 hours. This reduces the background staining significantly. The slides were cooled in solution for 20 minutes and washed in PBS twice for 5 minutes each. Then they were blocked in 20% fetal bovine serum in PBS for 45 minutes at room temperature. The slides were then washed two times for 5 minutes in PBS and a circle was drawn around the section with a hydrophobic pen. The sections were peroxide blocked with dual enzyme block solution (Dako) and incubated for 30 minutes. This was followed by 3 washes in PBS for 5 minutes each. After that, the primary antibody in antibody diluent (Dako) was applied to the sections and the slides were incubated at 4°C for overnight. On the following day, the sections were washed in PBS thrice and then the peroxidase labeled polymer (HRP) was added and the slides were incubated for 30 minutes at room temperature. Following the incubation, nine washes with PBS were done after with a wait of 5 minutes after every 3 washes. In the meantime, the DAB+ solution was prepared and added to the sections once washing had been completed. The incubation period was optimized according to the intensity of staining. After 3 washes in PBS, the slides were stained in Mayer’s Hematoxylin for 3 minutes. Finally they were washed in distilled water three times, mounted with an aqueous mounting media and covered with a cover glass. For each treatment group we analyzed 5 slides per sample (total 40 slides) each with 2–3 sections.
Wound digestion and flow cytometry analysis
The wounds (day 2 and 6) were excised out using a 10 mm sterile biopsy punch and cut from the center into two disc shaped pieces. Half of the tissue was used for cryosectioning and histology. The other half was digested[19] in an enzyme cocktail and used for the flow cytometry experiments. The single cell suspension from the wound tissue was maintained at 106 cells/ml in the FACS staining buffer (BD). The samples were blocked with 1 µg/ml (final concentration) of Rat IgG2b for 20 minutes on ice. The cells were then stained with the following antibodies for 30 minutes on ice: anti-mouse F4/80 (Pe-Cy7), anti-human CD206 (FITC) and anti-mouse CD86 (Biotin; refer Supplemental Table S2 for additional antibody details). Two washes were performed with FACS staining buffer. The PerCP streptavidin antibody was used to stain the samples for another 20 minutes on ice. The samples were finally washed twice with the FACS staining buffer and resuspended in 1 ml buffer. The samples were fixed with 500 µl of cytofix buffer (BD Biosciences) and stored at 4°C. The samples were run together on the BD LSRFortessa cell analyzer recording at least 10,000 events in every sample. The final data was analyzed using FlowJo software (FlowJo, LLC).
Quantification of wound closure and immunohistochemical staining
The macroscopic pictures of the wounds taken during surgery at day 0, 7 and 14 were used for the wound closure analysis. The Meiji stereo-zoom surgical dissection microscope with a Nikon D70 camera was used to take the pictures of the wounds. The camera was height was fixed throughout the experiment and the inner diameter of the silicone splints (6 mm) served as the parameter to normalize the wound area. The images were quantified using Metamorph (Molecular Devices) and were compared to the day 0 area (100% open). The immunostained slides were imaged using a Meiji brightfield microscope with CCD camera at 10×, 20× and 40× magnifications. The images from each treatment group with five sections were used for the quantification. The number of cells that were positively stained was quantified in comparison to the total cells.
Measurement of cytokines in wound lysates
The wound tissues were frozen in liquid N2 cooled isopentane. The samples were sectioned into one-micron thick sections using a cryostat. The sections were solubilized using the lysis buffer with 1% Triton-X 100 and protease inhibitors. The lysate was centrifuged at 25,000g in a refrigerated centrifuge and the supernatant was applied to a Ray Biotech Mouse Inflammation Antibody Array 1 (G-series). The arrays were processed according to the manufacturer’s instructions. The images were scanned using a laser scanner with the Cy3 channel. The intensity data was background subtracted with the negative controls and normalized with respect to the positive controls.
Statistical Analysis
Comparisons between two groups were performed using a two-tailed Student’s t-test. Multiple comparisons between groups were analyzed by two-way ANOVA followed by a Two-sided Dunnett post-hoc testing. For the wound closure data, the test groups compared with a two-tailed p-value of < 0.05 were considered as statistically significant.
RESULTS
Syndecan-4 is reduced in the skin of patients with type 2 diabetes
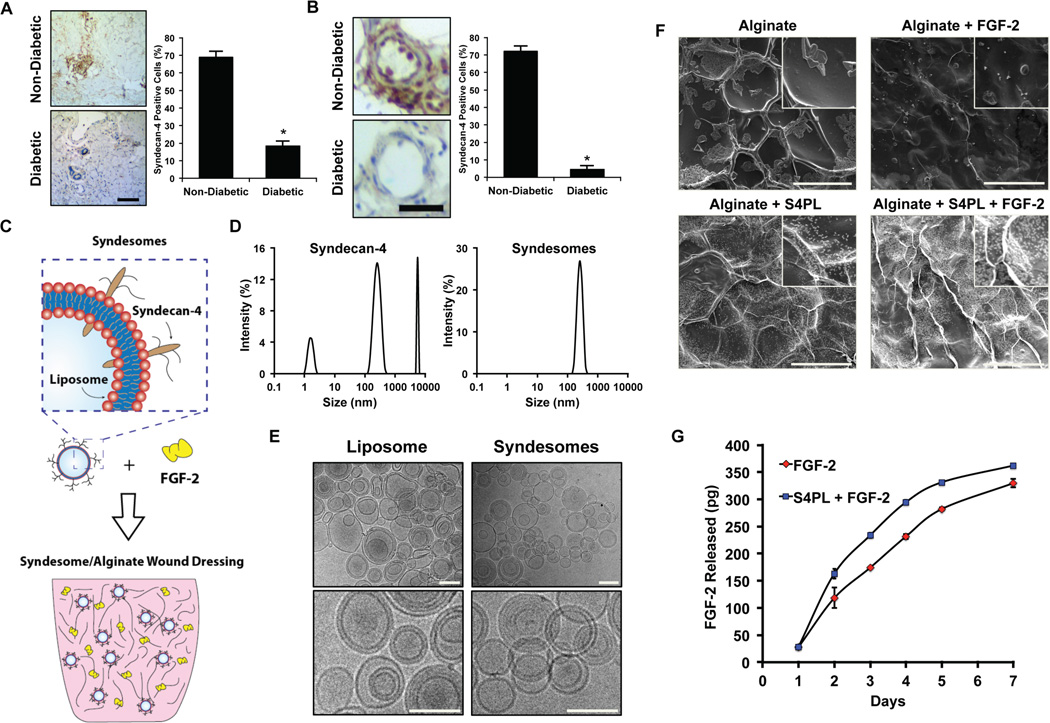
We hypothesized that the long-term disease state of diabetic patients may reduce the levels of syndecan-4 and thus reduce the effectiveness of growth factor therapies in this patient population. To examine whether there was loss of syndecan-4 in diabetic humans, we collected skin samples from patients with type 2 diabetes and non-diabetic patients (Supplemental Table S1), and then performed immunostaining of syndecan-4 in these tissues. We found a significant reduction in the staining of syndecan-4 in both the overall tissue samples (Fig. 1A) and in the blood vessels (Fig. 1B). We next aimed to examine whether syndesomes would be able to overcome enhance FGF-2 activity in the context of the diabetic disease state in which there is a loss of syndecan-4 and reduced responsiveness to growth factor therapy. The overall concept was to deliver FGF-2 with liposomes incorporating syndecan-4 (syndesomes) from a non-adhesive alginate wound dressing that could be applied to non-healing wounds in diabetic patients for enhancing wound healing (Fig. 2).
Figure 1. Measurement of syndecan-4 levels in the skin of diabetic and healthy patients; synthesis and characterization of syndesomes.
(A) Expression of syndecan-4 in human skin from diabetic and non-diabetic patients for the overall skin histology. Bar = 25 µm. (B) Expression of syndecan-4 in the blood vessels in the human skin. Bar = 25 µm. (C) Diagram of incorporating syndesomes and FGF-2 in an alginate wound dressing. (D) Dynamic light scattering analysis for syndecan-4 protein and liposomes incorporated with syndecan-4 (syndesomes). (E) Cryo-electron microscopy images liposomes and syndesomes. Bar = 400 nm. (F) Scanning electron microscopy images of desiccated alginate disks with various treatments. Bar = 100 µm. (G) Release kinetics of the FGF-2 from alginate beads containing syndesomes with FGF-2 or FGF-2 alone. *Statistically different from the non-diabetic patient group (p < 0.05, n = 9).
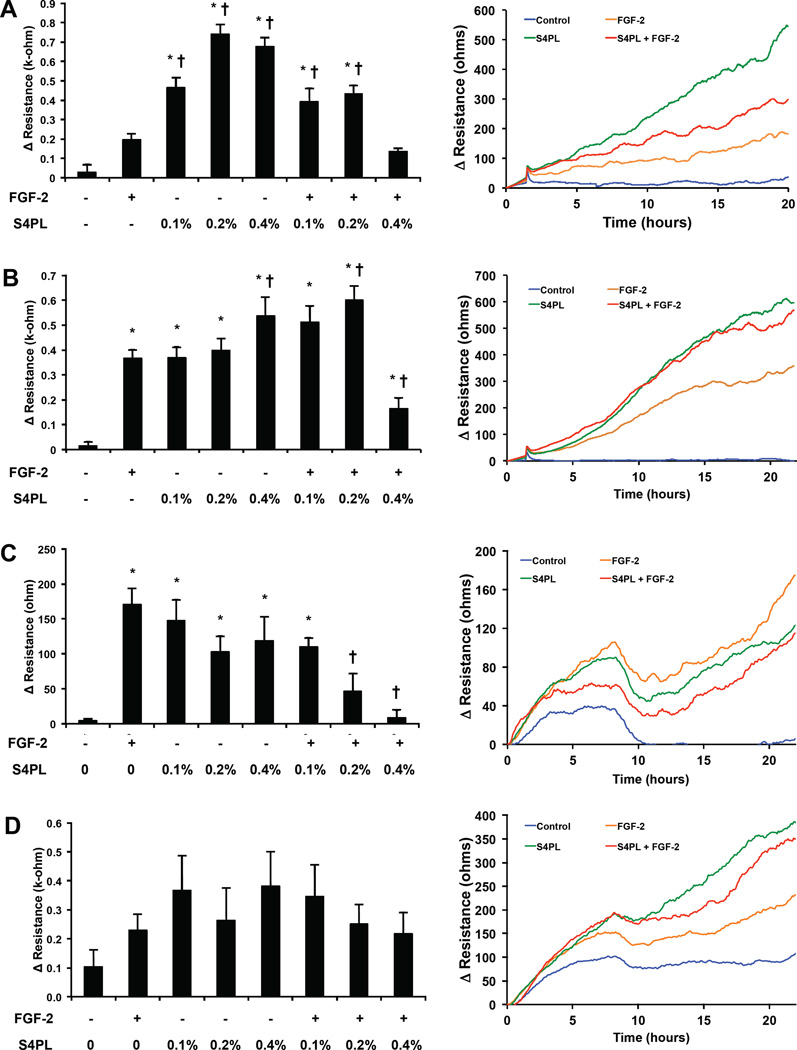
Figure 2. Effect of syndesomes on migration of keratinocytes and fibroblasts.
(A) Migration of keratinocytes from a non-diabetic donor at the 20 hours, with various treatments shown as bar graphs (left). Line graphs showing the time course of the change of resistance for the four treatment groups (right). Note that S4PL concentration is 0.4%. (A) Migration of keratinocytes from a diabetic donor at the 20 hours, with various treatments shown as bar graphs (left). Line graphs showing the time course of the change of resistance for the four treatment groups (right). Note that S4PL concentration is 0.4%. (C) Change in resistance due to migration at 20 hours in dermal fibroblasts from a non-diabetic donor, shown as bar graphs (left). Line graphs show the time course of the change of resistance for all the treatments with 0.4% S4PL concentration (right). (D) Change in resistance due to migration at 20 hours in dermal fibroblasts from a diabetic donor, shown as bar graphs (left). Line graphs show the time course of the change of resistance for all the treatments with 0.4% S4PL concentration (right). *p < 0.05 compared with no treatment group and †p < 0.05 compared with the FGF-2 group.
Synthesis and characterization of syndesomes and alginate gels
We created syndesomes by isolating recombinant syndecan-4 proteins and fusing them into the membrane of liposomes using a detergent extraction method. We confirmed the purity of the syndecan-4 protein using SDS-PAGE with silver staining as well as western blotting for syndecan-4 (Supplemental Fig. S1). A high molecular weight smear was observed in the blots implying that majority of the syndecan-4 was glycosylated. We also measured the size distribution of the isolated recombinant protein and the syndesomes using dynamic light scattering. This analysis demonstrated that the recombinant protein in isolation had significant self-association and separated into three distinct peaks, most likely representing protein aggregate formation and varying degrees of glycosylation (Fig. 1D). In contrast, the syndesomes had a single distinct peak corresponding to the approximate liposome diameter of 400 nm. In addition, we confirmed the integrity of the liposomes by performing cryo-electron microscopy (cryo-EM) analysis of the liposomes and syndesomes (Fig. 1E). To further confirm the incorporation of the protein into the lipid membrane, we examined the change in size of the liposome with varying amounts of protein using transmission electron microscopy (TEM). We measured the amount of syndecan-4 before and after incorporation into liposomes and found the majority of the protein was associated with the liposomes after detergent removal (98%±2% protein encapsulation; n = 3). We found that the liposome size increased with increasing amounts of protein incorporation (Supplemental Fig. S2A, B). In addition, the zeta potential of the liposomes was altered by syndecan-4 incorporation (Supplemental Fig. S2C). To create a local delivery platform for release of the syndesomes into the wound, we encapsulated the compounds into alginate disks (Fig. 1F). The release kinetics of FGF-2 from the alginate was similar over the implantation time of 7 days (Fig. 1G). We confirmed that the proteoliposomes were released intact by measuring the size of the released liposomes before and after release with DLS (Supplemental Fig. S3).
Syndesomes enhance keratinocyte migration, and reduce both fibroblast invasion and migration
We next assessed whether exogenous delivery of syndecan-4 could enhance the migration and invasion of dermal fibroblasts and keratinocytes, two key cellular effectors of wound healing. Keratinocytes from non-diabetic donors showed a nearly four-fold increased in migration with treatment with the syndecan-4 proteoliposomes at the optimal concentration (0.2% S4PL; Fig. 2A). Surprisingly, the syndesomes alone were more effective at inducing migration than in combination with FGF-2 but migration was increased under both conditions. Keratinocytes from diabetic patients showed only a moderate increase in migration in the groups treated with both syndecan-4 and FGF-2 (Fig. 2B). Higher doses of syndecan-4 with FGF-2 demonstrated a reduction in migration for both cell lines. In contrast, treatment with the syndesomes appeared to decrease migration of normal dermal fibroblasts (Fig. 2C) and did not alter the migration of fibroblasts from diabetic patients (Fig. 2D). We also measured the invasion of fibroblasts through a collagen gel under various treatment conditions and found a moderate reduction in migration with syndesome treatment in diabetic fibroblasts and no significant difference between the treatment groups in the normal fibroblasts (Supplemental Fig. S4).
Syndesomes increase endosomal processing and recycling of FGF-2 to the cell surface
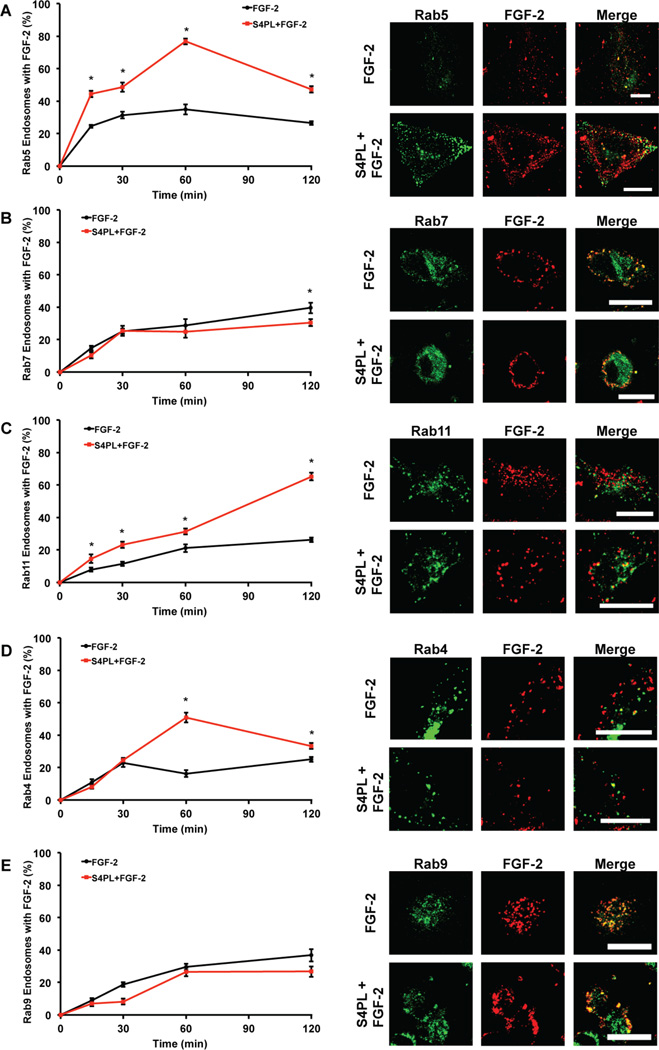
FGF-2 can be internalized by receptor-mediated and heparan sulfate proteoglycan-mediated mechanisms. The FGF receptor-1 (FGFR1) is endocytosed into early endosomes in a both a caveolin and clathrin-dependent mechanisms, from which it can be recycled through both the slow and fast pathways, or shuttled to the lysosomal compartment for degradation.[20] We next investigated how syndesomes altered the endosomal processing of FGF-2 to better understand the mechanism of action. Cells were transfected with plasmids containing GFP conjugated Rab proteins to label specific endosome subsets and then treated with fluorescently labeled FGF-2. We quantified the percentage of endosomes that co-localized with labeled FGF-2 for each time point. We found a significant increase in percentage of Rab5 (early endosome marker) labeled endosomes with FGF-2 in the syndesome with FGF-2 group at all the time points indicating higher FGF-2 uptake (Fig. 3A). The level of FGF-2 that co-localized with the late endosomal marker Rab7 (late endosomal marker) remained low throughout the experiment, showing significant differences between the treatment groups only at 120 minutes (Fig. 3B). There was an increase in the percentage of FGF-2 positive Rab11 endosomes (late recycling endosomes) at all time points when we compare the FGF-2 only treatment with the syndesomes with FGF-2 treatment (Fig. 3C). Thus, the majority of the FGF-2 that was processed through endosomal pathways was likely getting recycled to the plasma membrane surface through the Rab11 pathway rather than getting degraded by the Rab7 pathway. This hypothesis was supported by a significant increase in the Rab4 endosomes (early recycling endosomes) co-localizing with FGF-2 in the syndesome with FGF-2 group (Fig. 3D). We did not see any differences in the colocalization of Rab9 endosomes (late endosomes that transport to trans-golgi network) with FGF-2 (Fig. 3E).
Figure 3. Syndesomes alter endosomal processing of FGF-2.
HEK cells were transfected with GFP-Rab5, GFP-Rab7 and GFP-Rab11 separately. These cells were treated with AF594 labeled FGF-2 and syndesomes. (A) Percentage of Rab5 endosomes that co-localize with labeled FGF-2 with or without syndesomes. The image panel shows GFP-Rab5, AF594 FGF-2 and merge channels for the two treatments. (B) Percentage of Rab7 endosomes that co-localize with labeled FGF-2 with or without syndesomes. The image panel shows GFP-Rab7, AF594 FGF-2 and merge channels for the two treatments. (C) Percentage of Rab11 endosomes that co-localize with labeled FGF-2 with or without syndesomes. The image panel shows GFP-Rab11, AF594 FGF-2 and merge channels for the two treatments. (D) Percentage of Rab4 endosomes that co-localize with labeled FGF-2 with or without syndesomes. The image panel shows GFP-Rab4, AF594 FGF-2 and merge channels for the two treatments. (E) Percentage of Rab9 endosomes that colocalize with labeled FGF-2 with or without syndesomes. The image panel shows GFP-Rab9, AF594 FGF-2 and merge channels for the two treatments. *Statistically different from FGF-2 group at the same time point (p < 0.05; n =10).
Syndesomes improve wound healing in obese, diabetic mice
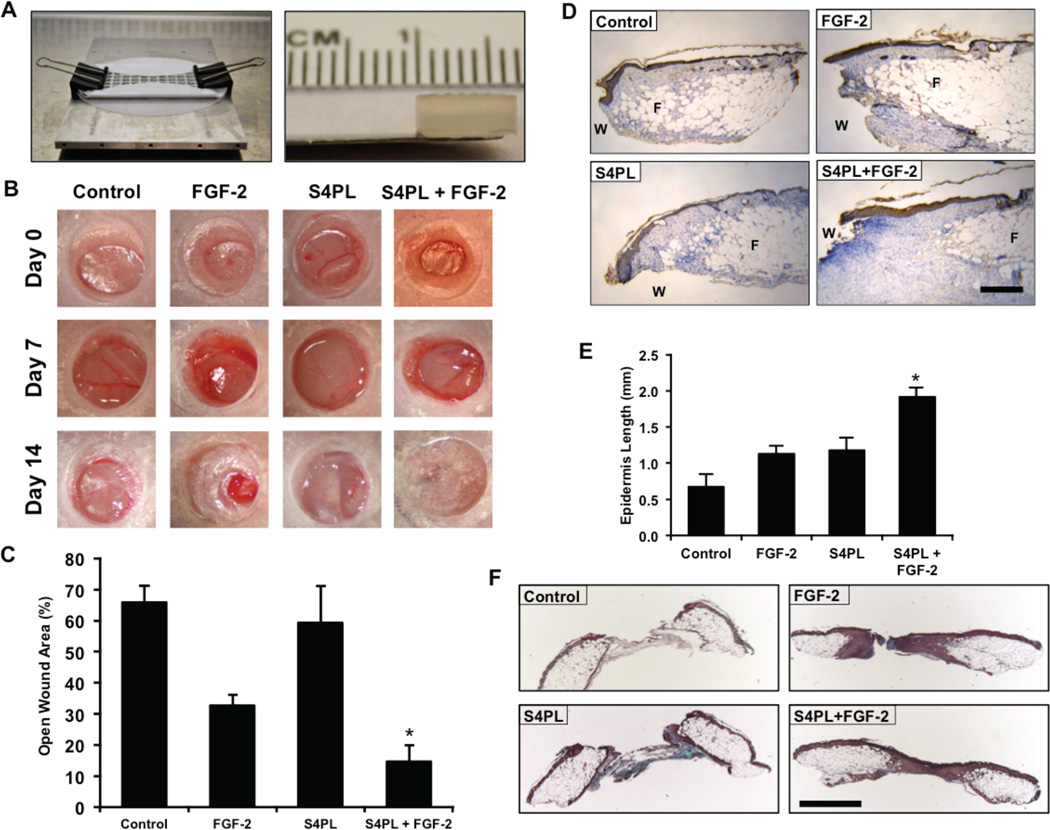
We next tested the effectiveness of syndesome therapy for enhancing wound healing in the diabetic ob/ob mouse model. Previous studies have shown that these mice have reduced wound closure[21] and have reduced responsiveness to FGF-2[22], when placed on a high fat diet. To examine whether syndesomes could enhance wound healing in a diabetic and obese animal model we created full-thickness wounds on the dorsal surface of these mice and attached a silicone splint around the wound using glue and sutures to prevent contraction. We created alginate wound dressings that matched the size of the wounds using a custom-designed mold (Fig. 4A). The gels were replaced seven days after the initial wounding and the mice were allowed to heal for an additional seven days. A macroscopic analysis of wound closure revealed a two-fold decrease in wound size after 14 days in the syndesomes with FGF-2 treatment compared to FGF-2 alone (Fig. 4B, 4C and Supplemental Fig. 5A, 5B). We performed immunostaining for cytokeratin and measured the regeneration of the epidermis beyond the initial wound defect. A morphometric quantification showed increase re-epithelization in the syndesome with FGF-2 group over the other treatment groups (Fig. 4D, 4E). Analysis of the granulation tissue area revealed similar levels of granulation tissue in all wounds with a slight increase in the FGF-2 treated samples (Supplemental Fig. S5C, S5D). A histological analysis of the wound beds demonstrated increased cellular infiltration in the syndesome with FGF-2 group in comparison to the other groups including the syndesomes alone (Fig. 4F).
Figure 4. Syndesomes enhance cutaneous wound healing in ob/ob mice on high fat diet.
(A) Custom-made mold for fabricating alginate disks and the alginate disk. (B) Macroscopic images of wound closure over 14 days with various treatments. (C) Quantification of the open wound area (%) over 14 days using the macroscopic wound images. (D) Immunostaining of the wound sections for cytokeratin to visualize the epidermal regrowth following wounding. “W” refers to the initial wound and “F” refers to the subcutaneous fat area. The edge of the fat layer marks the wound edge. Bar = 250 µm. (E) Quantification of the regrowth of the epidermis beyond the wound edge in various treatment groups. (F) Histological sections from the wounds in ob/ob mice after 14 days with treatment with syndesomes (S4PL) and FGF-2. The sections were stained with Movat’s pentachrome stain. Bar = 1 mm. *Statistically different from all treatment groups (p < 0.05; n = 8).
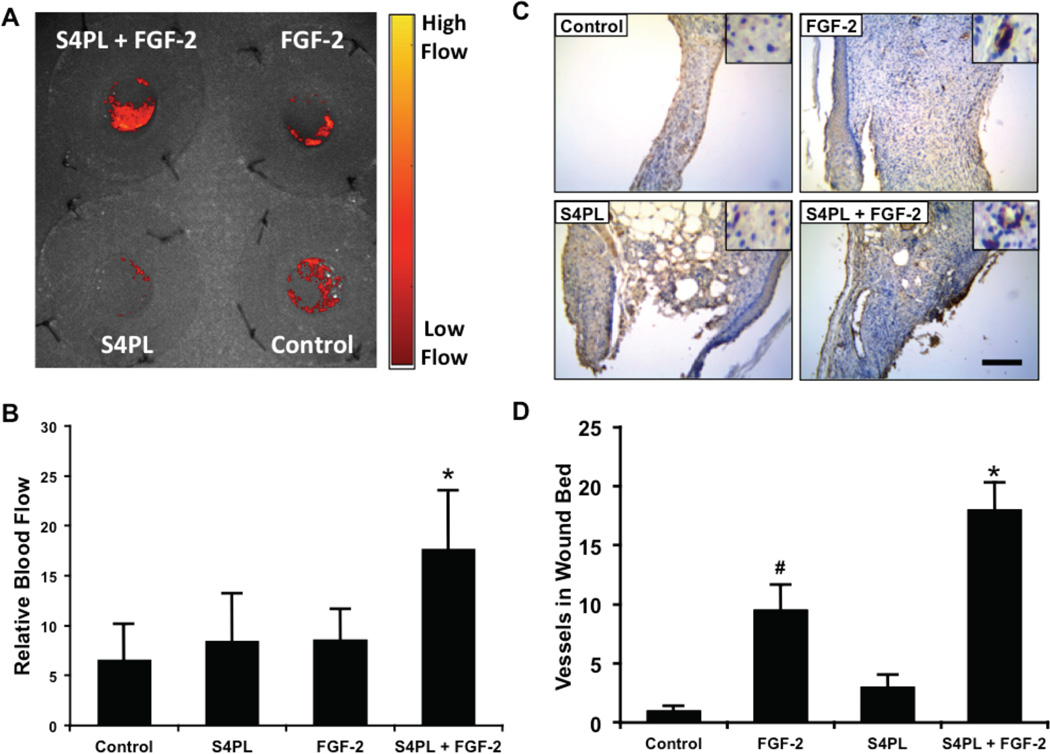
Syndesomes increase perfusion in the healing wound bed
We measured the blood perfusion in the wounds immediately after wounding and seven days later. Due to the variation in healing between the groups we did not measure blood perfusion in the wounds at day 14 following wounding. We found that there was significant increase of blood perfusion in the syndesomes with FGF-2 compared to all other groups at seven days (Fig. 5A, B). After 14 days, we harvested the wound beds and performed immunostaining for endothelial cells. This analysis showed increased blood vessels in the wound bed of the FGF-2 with syndesome treated group in comparison to FGF-2 alone and other groups (Fig. 5C, D).
Figure 5. Syndesomes increase perfusion in the developing wound beds.
(A) Laser speckle contrast image of the dorsal surface of the mice with the four wounds 7 days post surgery and a heat map showing relative blood flow. (B) Quantification of blood flow in the wounds at day 7, relative to perfusion on the day of surgery. (C) Histological sections of wound bed after 14 days after wounding immunostained for an endothelial marker (von Willebrand factor). Bar = 125 µm and insets are magnified 1.5 times. (D) Quantification of the number of vessels per field of view in the wound bed for different treatment groups. *Statistically different from all other groups (p < 0.05; n = 8). #Statistically different from the control and S4PL groups (p < 0.05; n = 8).
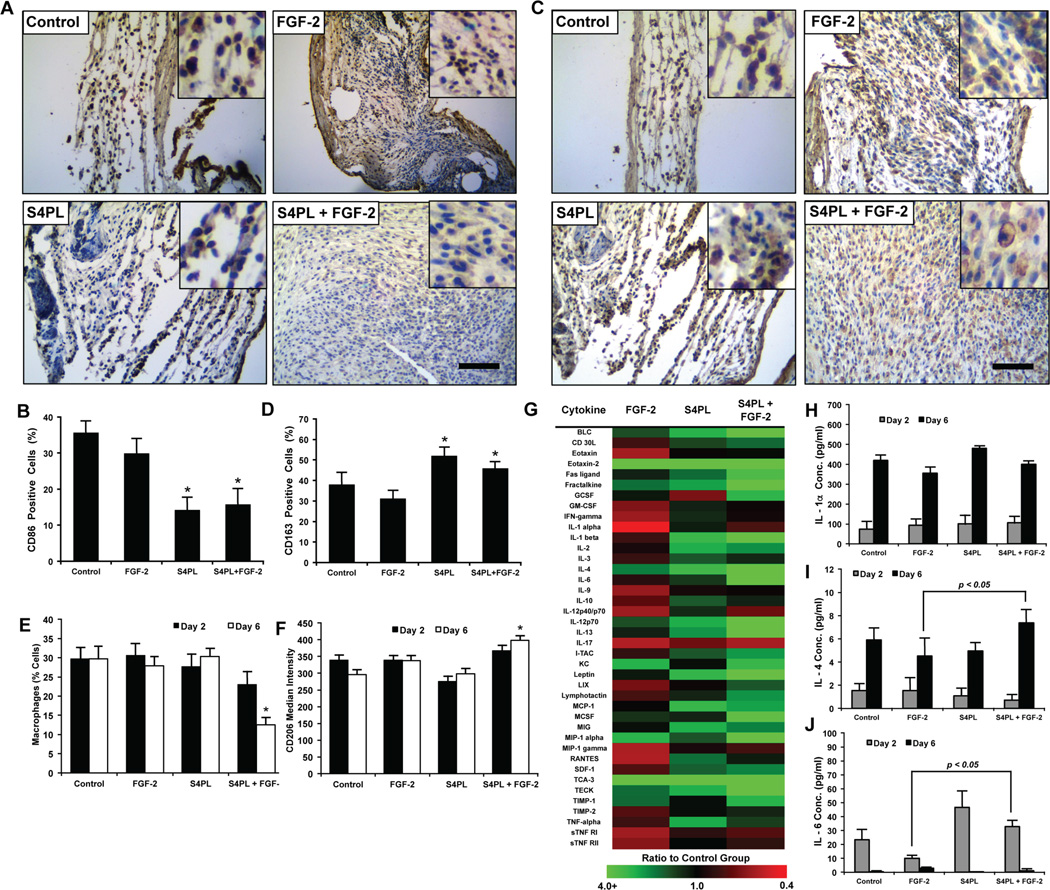
Syndesomes enhance wound healing phenotype in macrophages
Macrophages are key players in the wound healing cascade through the regulation of inflammation and healing responses. Macrophages can express a continuum of phenotypes that are often broadly classified into M1 macrophages with pro-inflammatory activities or M2 macrophages that orchestrate matrix deposition and wound healing. We examined the expression of CD86 (M1 marker) and CD163 (M2 marker) using immunostaining of histological sections from the mice treated with syndesome-incorporating alginate wound dressings after 14 days. This analysis demonstrated a decrease in the expression of CD86 with syndesome treatment (Fig. 6A, B). In addition, the levels of the M2 marker CD163 were increased in the wound beds (Fig. 6C, D). Interestingly, the modulation of the marker expression was present in both the syndesomes with FGF-2 and in the S4PL alone groups, suggesting that the syndesomes were directly inducing immunomodulation in the wounds.
Figure 6. Syndesomes temporally modulate the macrophage response to wound healing phenotype.
(A) Histological sections of the wound beds 14 days post-surgery with immunostaining for an M1 macrophage marker (CD86). Bar = 125 µm and insets are magnified threefold. (B) Quantification of CD86 positive cells within the wound beds. *p < 0.05 versus FGF-2 group (n = 8). (C) Histological sections of the wound beds 14 days post-surgery with immunostained for an M2 macrophage marker (CD163). Bar = 125 µm and insets are magnified threefold. (D) Quantification of the number of CD163 positive cells in the wound beds. *p < 0.05 versus FGF-2 group (n = 8). (E) Analysis of cells harvested from wounds in ob/ob mice after 2 or 6 days post surgery using flow cytometry. Cells were stained for macrophage marker (F4/80) and compared to the total cells measured in the wound. *p < 0.05 versus all other groups at day 6 (n = 5). (F) Median intensity of staining for M2 macrophage marker (CD206) in macrophages harvested from wounds 2 or 6 days post surgery using flow cytometry. *p < 0.05 versus all other groups at day 6 (n = 5). (G) Heat map image of the fold change (compared to control group) in the concentration of various cytokines in the wounds 6 days after surgery. (H, I, J) Concentrations (pg/ml) of IL-1α, IL-4 and IL-6, respectively, in the wound bed at days 2 and 6 measured using ELISA normalized to the total protein concentration. *p < 0.05 versus the FGF-2 group at the same day (n = 5).
To further examine whether syndesomes could modulate the immune response during healing, we treated ob/ob mice with the various wound dressings and harvested the wound tissues at two and six days following wounding. We digested half of the harvested wound beds into a single cell suspension and used flow cytometry to quantify the expression of macrophage markers. Six days after wounding, we found a decreased number of macrophages (F4/80+ cells) and increased expression of the M2 marker CD206 in the syndesomes with FGF-2 treated wounds in comparison to FGF-2 only treated wounds (Fig. 6E, 6F). We took the other half of the wound bed from day two and six after wounding, lysed the tissue, and performed an analysis of the cytokines in the wound using ELISA and cytokine antibody arrays. The cytokine antibody array at day 6 revealed many changes in the cytokine concentrations compared to the control group (Fig. 6G; Supplemental Fig. S6). The ELISA revealed similar levels of IL-1α, an inflammatory cytokine released by many cell types including neutrophils and macrophages, between the four treatment groups (Fig. 6H). We also observed an increase in IL-4 and IL-6 in the syndesomes with FGF-2 treated wounds over FGF-2 alone treated wounds (Fig. 6I, 6J). Both of these cytokines have been linked to alternative activation of macrophages and this finding is consistent with the increased CD163 expression and decreased CD86 expression in the syndesomes with FGF-2 treatment group. In addition, there were increased levels of stromal cell-derived factor-1 (SDF-1), IL-1β, monokine induced by gamma interferon (MIG/CXCL9) and IL-2 in the syndesomes with FGF-2 group versus wounds treated with FGF-2 alone (Supplemental Fig. S7; Supplemental Table S3). Previous studies have shown that exogenously applied SDF-1 or IL-2 enhances wound healing[23]. A proteomic analysis of chronic pressure ulcers in human patients found that MIG increases in chronic ulcers that heal but remains constant in those that do not heal.[24]
DISCUSSION
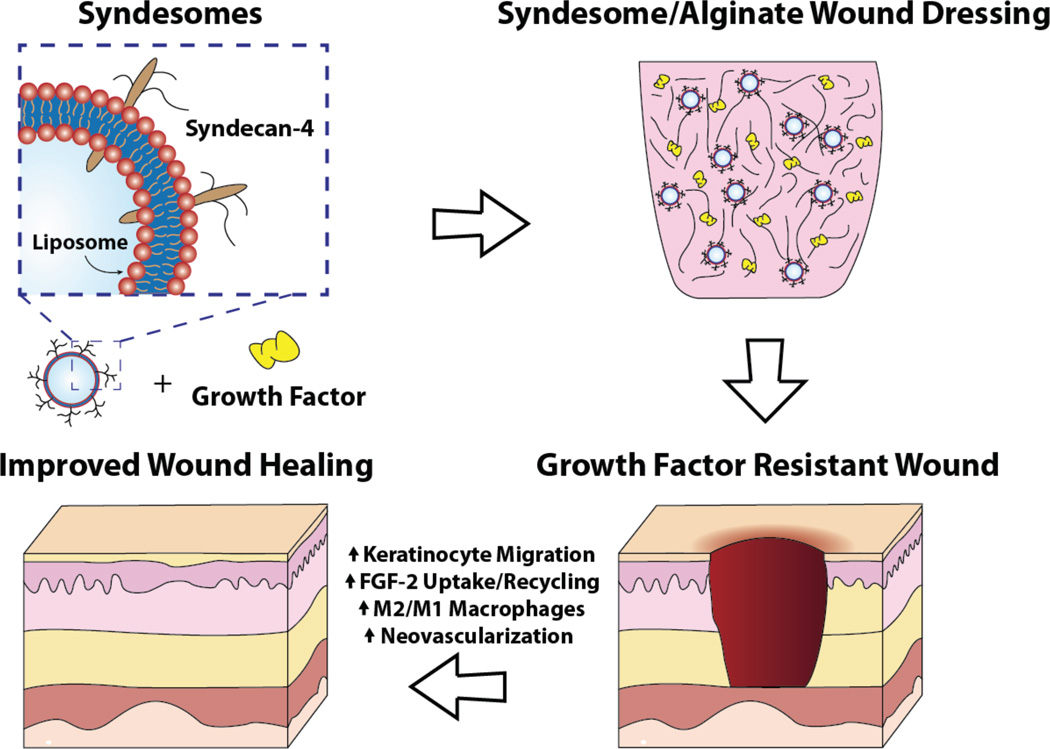
Clinical trials using growth factor therapies to enhance diabetic wound healing have produced poor or equivocal results.[8] Here, we hypothesized that the current approach of delivering growth factors is not effective because it does not account for the changes in tissue responsiveness due to disease. We examined skin samples from patients with type 2 diabetes and found that the levels of syndecan-4 were reduced in both the overall skin and blood vessels relative to non-diabetic patients. We then tested the ability of syndecan-4 liposomes in controlling wound healing in in vitro and in vivo studies. Overall, the delivery of syndecan-4 protein with FGF-2 markedly improves many indices of wound healing including the migration of keratinocytes, wound closure and shifting the macrophages towards the wound healing M2 phenotype (Fig. 7). In addition, our results support the validity of the concept that by delivering co-receptors downregulated by a disease state, one can markedly improve the efficacy of a delivered therapeutic ligand.
Figure 7. Summary diagram of the findings of the study and the enhancing activity of the syndesomes.
A key point of our study is that the diabetic disease state must be considered when developing protein therapeutics for wound healing. Many clinical trials have been performed for therapies to improve the healing of chronic wounds based on the findings in large animal models in the absence of diabetics. In fact, the most commonly used preclinical wound model is performed in the non-diabetic pig, a model that has relatively rapid wound healing and does not have healing resistant wounds. Our findings that diabetic patients have reduced syndecan-4 in blood vessels of the skin illustrates the profound difference that exists between healthy patients and those that are prone to developing chronic wounds. We hypothesize that much of this loss of syndecan-4 can be attributed to the shedding and degradation of the protein by diabetes-induced changes in protease activity. This hypothesis is supported by the loss of the glycans observed in diabetes[25] and that syndecan-4 is shed by factors increased in diabetes including reactive oxygen species,[26] proteases[27] and inflammation.[28] This concept is also supported by our prior studies in diabetic mice in which we found a significant reduction in many of the growth factor receptors and co-receptors in the skeletal muscle and myocardium.[10] Therefore, we would argue that it is essential to perform preclinical studies for wound healing in animal models that represent the disease states that commonly accompany non-healing wounds in patients. As diabetic porcine models have been used extensively in the study of atherosclerosis,[11] it would seem that there is both an economic and ethical imperative to use these types of model in preclinical studies of wound healing to reduce the risk of failed clinical trials.
While previous studies have shown endogenous syndecan-4 plays a role wound healing, it is surprising that delivery of exogenous syndecan-4 protein is able to enhance keratinocyte migration and wound healing to the extent we observed in our studies. Syndecan-4 is induced in the skin following acute wounding in both mice and neonatal humans.[29] Mice lacking syndecan-4 have delayed wound healing and impaired angiogenesis.[30] Therefore, in the context of wound healing there are likely multiple benefits to increasing syndecan-4 in the wound bed. Several prior studies have also supported a role for syndecan-4 in wound healing independent of its activity as a co-receptor for FGF-2. Endogenous syndecan-4 expression promotes fibroblast migration and regulates integrin signaling and small GTPases during wound healing.[31] In addition, syndecan-4 also enhances keratinocyte migration[32] and is necessary for migration of fibroblasts in fibrin gels.[33] Our previous work has shown that delivery of syndecan-4 in a proteoliposome was more potent in inducing cell proliferation/migration, activation of ERK1/2 signaling pathway, in vitro endothelial tube formation, nuclear trafficking of growth factors and angiogenesis in comparison to the free syndecan-4 protein.[16] In our studies showed increased migration in keratinocytes and decreased invasion activity when the cells were treated with exogenous syndecan-4 protein in a proteoliposome. This would suggest that the developed treatments would be able to synergistically enhance cell therapies, including those delivered from electrospun materials.[34] With exogenous delivery of the protein there is the possibility that the syndecan-4 works as a competitive inhibitor to the binding of the endogenous syndecan-4. Whether this occurs is likely a function of concentration of the ligand bound by syndecan-4, the concentration of the relevant receptor and the concentration of endogenous syndecan-4. In this case, it would suggest there is additional capability in keratinocytes to have enhanced FGF-2 signaling through the addition of syndecan-4 but that additional syndecan-4 serves as a competitive inhibitor for fibroblasts undergoing invasion.
Our current study suggests that a major activity of syndesomes is to enhance FGF-2 recycling through endosome-mediated mechanisms. Syndecan-4 is a key co-receptor in the FGF-2 and FGFR-1 signaling cascade[35] where the heparan sulfate chains bind to FGF-2, and syndecan-4 dimerizes to aid as a co-receptor to FGFR-1. Furthermore, FGF-2 and FGFR-1 reside in the same endosomal compartment as syndecan-syntenin-PIP2 complex.[36] Both FGF-2 and FGFR-1 are taken up through the clathrin-mediated pathway, followed by trafficking to the early and late endosomes.[37] Syndecan-4 also recycles to the surface through both early and late endosomes[38] In our studies, we found significantly higher amounts of FGF-2 in the early endosomes (Rab5 positive). This finding would be expected with overexpression of the syndecan-4 gene, as an increased amount of syndecan-4 would facilitate receptor binding and uptake through pinocytosis.[39] However, as the proteoliposomes used do not undergo direct membrane fusion it is less clear how these changes might occur with syndesome treatment. We hypothesize that syndesomes increase FGF-2 uptake by enhancing the binding of FGF-2 to its receptor through the heparan sulfate chains of syndecan-4 and then facilitating the uptake of additional FGF-2 bound to syndecan-4 in the liposomal membrane during internalization. This hypothesis is consistent with our previous finding that free syndecan-4 is not as effective in enhancing FGF-2 uptake, signaling and angiogenesis.[16] It would also be consistent with our finding of increased recycling of FGF-2 through the slow (Rab11) and fast (Rab4) mechanisms that we observed in our studies. This increased recycling may be the result of FGF-2 bound syndecan-4 being trafficked back to the surface after being internalized through the syndesome construct. This recycled FGF-2 would therefore be available for additional receptor interactions,[40] making the FGF-2 more effective for an equivalent dose and leading to a prolonged effect.
Our study supports that the delivery of exogenous syndecan-4 protein drives wound macrophages toward the M2 phenotype. Wound healing is a delicate balance between the necessary inflammation that facilitates wound closure and angiogenesis, and excessive inflammation that can impede the wound healing process. A recently proposed scheme classifies macrophages as ranging in a phenotypic continuum from inflammatory (M1) to “alternatively activated” (M2) phenotypes.[41] However, the exact nature of these phenotypic states remains unclear and multiple other sub-phenotypes have been hypothesized. Efficient wound healing requires both the necessary inflammatory state and the pro-fibrotic, scaffold generating state. Thus, one hypothesis is that chronic non-healing wounds can arise when too much or too little inflammation is present, and scar formation can occur when the “healing” response is overactive and leads to wound fibrosis and scarring.[42] Within the wound environment, various subsets of macrophages have been associated with inflammation or with fibrosis but the strict definitions of M1 and M2 phenotypes do not adequately characterize the full complexity of wound macrophage phenotypes.[43]
The detailed mechanistic role of syndecan-4 in macrophage phenotype is unknown but several studies support that it is functionally involved in LDL uptake,[44] signaling in response to RANTES and SDF-1,[45] and the response to endotoxic shock.[46] Our study demonstrated increased M2 macrophage phenotype at early time points in the wounds, suggesting that syndesomes can alter the initial immune response to the wound. The presence of increased M2 macrophages would have a number of benefits for healing in chronic wounds including enhancing angiogenesis and the production of pro-healing cytokines.[47] Consistent with M2 macrophage phenotype, we observed an increase in cytokines associated with M2 macrophages (IL-4 and IL-6) as well as a number of pro-healing cytokines including IL-2 and SDF-1. Topically applied IL-4 can increase wound healing in mice and facilitated macrophage differentiation into the M2 phenotype.[48] IL-6 is also essential for wound healing and facilitates keratinocyte migration, wound contraction and macrophage infiltration.[49] In addition, both IL-2 and SDF-1 have been shown to increase wound healing in small animal models.[23] Thus, our studies demonstrate that syndesomes act in the early stages of wound healing to enhance the production of pro-healing cytokines including those that would facilitate an M2 macrophage phenotype.
CONCLUSIONS
In summary, we have shown that there is a reduction of syndecan-4 protein in diabetic patients and that syndesomes enhance FGF-2 therapy in the context of wound healing in the diabetic disease state. Our studies support that the syndesomes increase keratinocyte migration, endosomal recycling of FGF-2, and enhance the M2 macrophage phenotype and production of pro-healing cytokines. While it is likely that there exist many other mechanisms of growth factor resistance diabetes it is encouraging that significant improvements in therapeutic potential can be garnered by targeting only the FGF-2/syndecan-4 signaling pathway. Further studies are needed to both understand the detailed mechanisms of growth factor resistance and to further develop therapeutics to be effective in disease states. In this context, our therapeutic paradigm of delivering co-receptors as enhancers of growth factor activity may be applicable to a number of other biological systems in which disease modifies the responsiveness of the tissue such has hyperlipidemia and metabolic syndrome.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge support through the American Heart Association (10SDG2630139), the Welch Foundation (F-1836) and the NIH Director’s New Innovator Grant (1DP2 OD008716-01) to A.B.B. This work was also supported through the NIH (EB-011556, NS-078791 and NS-082518), NSF (CBET-0644638), AHA (14EIA18970041), and the Coulter Foundation grants to A.K.D. as well as NIH R00-ES020844 to S.M. In addition, this work was supported by the Glasgow Caledonian University Skin Research Tissue Bank, Glasgow, UK (P.E.M. and C.S.W.). C.S.W. is currently funded by the Dr. Hadwen Trust (DHT) and did not participate in experiments involving animals, animal tissue, cells, cell lines or human embryonic stem cells. The authors acknowledge the Sealy Center for Structural Biology and Molecular Biophysics at the University of Texas Medical Branch at Galveston and the W. M. Keck foundation for providing research resources.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors have filed a patent application on the compounds described in this work.
REFERENCES
- 1.Singh N, Armstrong DG, Lipsky BA. JAMA. 2005;293:217. doi: 10.1001/jama.293.2.217. [DOI] [PubMed] [Google Scholar]
- 2.A. American Diabetes. Diabetes Care. 2013;36:1033. [Google Scholar]; Larsson J, Agardh CD, Apelqvist J, Stenstrom A. Clin Orthop Relat Res. 1998:149. [PubMed] [Google Scholar]
- 3.Goldman R. Adv Skin Wound Care. 2004;17:24. doi: 10.1097/00129334-200401000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Branski LK, Pereira CT, Herndon DN, Jeschke MG. Gene Ther. 2007;14:1. doi: 10.1038/sj.gt.3302837. [DOI] [PubMed] [Google Scholar]
- 5.Zahorec P, Koller J, Danisovic L, Bohac M. Cell Tissue Bank. 2014 doi: 10.1007/s10561-014-9440-2. [DOI] [PubMed] [Google Scholar]
- 6.Buchberger B, Follmann M, Freyer D, Huppertz H, Ehm A, Wasem J. Exp Clin Endocrinol Diabetes. 2011;119:472. doi: 10.1055/s-0031-1279713. [DOI] [PubMed] [Google Scholar]
- 7.Bhansali A, Venkatesh S, Dutta P, Dhillon MS, Das S, Agrawal A. Diabetes Res Clin Pract. 2009;83:e13. doi: 10.1016/j.diabres.2008.10.005. [DOI] [PubMed] [Google Scholar]; Fang RC, Galiano RD. Biologics. 2008;2:1. doi: 10.2147/btt.s1338. [DOI] [PMC free article] [PubMed] [Google Scholar]; Smiell JM, Wieman TJ, Steed DL, Perry BH, Sampson AR, Schwab BH. Wound Repair Regen. 1999;7:335. doi: 10.1046/j.1524-475x.1999.00335.x. [DOI] [PubMed] [Google Scholar]; Steed DL. J Vasc Surg. 1995;21:71. doi: 10.1016/s0741-5214(95)70245-8. [DOI] [PubMed] [Google Scholar]; Wieman TJ, Smiell JM, Su Y. Diabetes Care. 1998;21:822. doi: 10.2337/diacare.21.5.822. [DOI] [PubMed] [Google Scholar]
- 8.Acosta JB, Savigne W, Valdez C, Franco N, Alba JS, del Rio A, Lopez-Saura P, Guillen G, Lopez E, Herrera L, Fernandez-Montequin J. Int Wound J. 2006;3:232. doi: 10.1111/j.1742-481X.2006.00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fernandez-Montequin JI, Betancourt BY, Leyva-Gonzalez G, Mola EL, Galan-Naranjo K, Ramirez-Navas M, Bermudez-Rojas S, Rosales F, Garcia-Iglesias E, Berlanga-Acosta J, Silva-Rodriguez R, Garcia-Siverio M, Martinez LH. Int Wound J. 2009;6:67. doi: 10.1111/j.1742-481X.2008.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mohan VK. Diabetes Res Clin Pract. 2007;78:405. doi: 10.1016/j.diabres.2007.06.004. [DOI] [PubMed] [Google Scholar]; Richard JL, Parer-Richard C, Daures JP, Clouet S, Vannereau D, Bringer J, Rodier M, Jacob C, Comte-Bardonnet M. Diabetes Care. 1995;18:64. doi: 10.2337/diacare.18.1.64. [DOI] [PubMed] [Google Scholar]
- 9.Fiddes JC, Hebda PA, Hayward P, Robson MC, Abraham JA, Klingbeil CK. Ann N Y Acad Sci. 1991;638:316. doi: 10.1111/j.1749-6632.1991.tb49042.x. [DOI] [PubMed] [Google Scholar]; Lynch SE, Colvin RB, Antoniades HN. J Clin Invest. 1989;84:640. doi: 10.1172/JCI114210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Singh G, Baker AB. Biomaterials. 2014;35:196. doi: 10.1016/j.biomaterials.2013.09.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker AB, Chatzizisis YS, Beigel R, Jonas M, Stone BV, Coskun AU, Maynard C, Rogers C, Koskinas KC, Feldman CL, Stone PH, Edelman ER. Atherosclerosis. 2010;213:436. doi: 10.1016/j.atherosclerosis.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker AB, Gibson WJ, Kolachalama VB, Golomb M, Indolfi L, Spruell C, Zcharia E, Vlodavsky I, Edelman ER. J Am Coll Cardiol. 2012;59:1551. doi: 10.1016/j.jacc.2011.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baker AB, Groothuis A, Jonas M, Ettenson DS, Shazly T, Zcharia E, Vlodavsky I, Seifert P, Edelman ER. Circ Res. 2009;104:380. doi: 10.1161/CIRCRESAHA.108.180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA. J Virol. 2006;80:10395. doi: 10.1128/JVI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay S, Linstedt AD. Proc Natl Acad Sci U S A. 2011;108:858. doi: 10.1073/pnas.1013642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury A, Dominguez M, Puri V, Sharma DK, Narita K, Wheatley CL, Marks DL, Pagano RE. J Clin Invest. 2002;109:1541. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. Infect Immun. 2003;71:5855. doi: 10.1128/IAI.71.10.5855-5870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang E, Albadawi H, Watkins MT, Edelman ER, Baker AB. Proc Natl Acad Sci U S A. 2012;109:1679. doi: 10.1073/pnas.1117885109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Ge J, Tredget EE, Wu Y. Nat Protoc. 2013;8:302. doi: 10.1038/nprot.2013.002. [DOI] [PubMed] [Google Scholar]
- 18.Parthasarathy AB, Tom WJ, Gopal A, Zhang X, Dunn AK. Opt Express. 2008;16:1975. doi: 10.1364/oe.16.001975. [DOI] [PubMed] [Google Scholar]
- 19.Brubaker AL, Schneider DF, Palmer JL, Faunce DE, Kovacs EJ. Journal of Immunological Methods. 2011;373:161. doi: 10.1016/j.jim.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irschick R, Trost T, Karp G, Hausott B, Auer M, Claus P, Klimaschewski L. Histochem Cell Biol. 2013;139:135. doi: 10.1007/s00418-012-1009-1. [DOI] [PubMed] [Google Scholar]
- 21.Seitz O, Schurmann C, Hermes N, Muller E, Pfeilschifter J, Frank S, Goren I. Exp Diabetes Res. 2010;2010:476969. doi: 10.1155/2010/476969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Weel V, de Vries M, Voshol PJ, Verloop RE, Eilers PH, van Hinsbergh VW, van Bockel JH, Quax PH. Arterioscler Thromb Vasc Biol. 2006;26:1383. doi: 10.1161/01.ATV.0000219234.78165.85. [DOI] [PubMed] [Google Scholar]
- 23.Barbul A, Knud-Hansen J, Wasserkrug HL, Efron G. J Surg Res. 1986;40:315. doi: 10.1016/0022-4804(86)90193-9. [DOI] [PubMed] [Google Scholar]; Xu X, Zhu F, Zhang M, Zeng D, Luo D, Liu G, Cui W, Wang S, Guo W, Xing W, Liang H, Li L, Fu X, Jiang J, Huang H. Cells Tissues Organs. 2013;197:103. doi: 10.1159/000342921. [DOI] [PubMed] [Google Scholar]
- 24.Edsberg LE, Wyffels JT, Brogan MS, Fries KM. Wound Repair Regen. 2012;20:378. doi: 10.1111/j.1524-475X.2012.00791.x. [DOI] [PubMed] [Google Scholar]
- 25.Salmon AH, Satchell SC. J Pathol. 2012;226:562. doi: 10.1002/path.3964. [DOI] [PubMed] [Google Scholar]
- 26.Singh A, Ramnath RD, Foster RR, Wylie EC, Friden V, Dasgupta I, Haraldsson B, Welsh GI, Mathieson PW, Satchell SC. PLoS One. 2013;8:e55852. doi: 10.1371/journal.pone.0055852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarty SM, Percival SL. Adv Wound Care (New Rochelle) 2013;2:438. doi: 10.1089/wound.2012.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramnath R, Foster RR, Qiu Y, Cope G, Butler MJ, Salmon AH, Mathieson PW, Coward RJ, Welsh GI, Satchell SC. FASEB J. 2014;28:4686. doi: 10.1096/fj.14-252221. [DOI] [PubMed] [Google Scholar]
- 29.Gallo R, Kim C, Kokenyesi R, Adzick NS, Bernfield M. J Invest Dermatol. 1996;107:676. doi: 10.1111/1523-1747.ep12365571. [DOI] [PubMed] [Google Scholar]
- 30.Echtermeyer F, Streit M, Wilcox-Adelman S, Saoncella S, Denhez F, Detmar M, Goetinck P. J Clin Invest. 2001;107:R9. doi: 10.1172/JCI10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bass MD, Williamson RC, Nunan RD, Humphries JD, Byron A, Morgan MR, Martin P, Humphries MJ. Dev Cell. 2011;21:681. doi: 10.1016/j.devcel.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brooks R, Williamson R, Bass M. Small GTPases. 2012;3:73. doi: 10.4161/sgtp.19301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Araki E, Momota Y, Togo T, Tanioka M, Hozumi K, Nomizu M, Miyachi Y, Utani A. Mol Biol Cell. 2009;20:3012. doi: 10.1091/mbc.E08-09-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin F, Ren XD, Doris G, Clark RA. J Invest Dermatol. 2005;124:906. doi: 10.1111/j.0022-202X.2005.23740.x. [DOI] [PubMed] [Google Scholar]
- 34.Jayasinghe SN. Analyst. 2013;138:2215. doi: 10.1039/c3an36599a. [DOI] [PubMed] [Google Scholar]; Townsend-Nicholson A, Jayasinghe SN. Biomacromolecules. 2006;7:3364. doi: 10.1021/bm060649h. [DOI] [PubMed] [Google Scholar]
- 35.Elfenbein A, Lanahan A, Zhou TX, Yamasaki A, Tkachenko E, Matsuda M, Simons M. Sci Signal. 2012;5:ra36. doi: 10.1126/scisignal.2002495. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elfenbein A, Simons M. J Cell Sci. 2013;126:3799. doi: 10.1242/jcs.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]; Horowitz A, Tkachenko E, Simons M. J Cell Biol. 2002;157:715. doi: 10.1083/jcb.200112145. [DOI] [PMC free article] [PubMed] [Google Scholar]; Simons M, Horowitz A. Cell Signal. 2001;13:855. doi: 10.1016/s0898-6568(01)00190-5. [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann P, Zhang Z, Degeest G, Mortier E, Leenaerts I, Coomans C, Schulz J, N'Kuli F, Courtoy PJ, David G. Dev Cell. 2005;9:377. doi: 10.1016/j.devcel.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Jean S, Mikryukov A, Tremblay MG, Baril J, Guillou F, Bellenfant S, Moss T. Dev Cell. 2010;19:426. doi: 10.1016/j.devcel.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Lambaerts K, Wilcox-Adelman SA, Zimmermann P. Curr Opin Cell Biol. 2009;21:662. doi: 10.1016/j.ceb.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tkachenko E, Lutgens E, Stan RV, Simons M. J Cell Sci. 2004;117:3189. doi: 10.1242/jcs.01190. [DOI] [PubMed] [Google Scholar]
- 40.Hausott B, Vallant N, Hochfilzer M, Mangger S, Irschick R, Haugsten EM, Klimaschewski L. Eur J Cell Biol. 2012;91:129. doi: 10.1016/j.ejcb.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Sica A, Mantovani A. J Clin Invest. 2012;122:787. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brancato SK, Albina JE. Am J Pathol. 2011;178:19. doi: 10.1016/j.ajpath.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daley JM, Brancato SK, Thomay AA, Reichner JS, Albina JE. J Leukoc Biol. 2010;87:59. doi: 10.1189/jlb.0409236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyanovsky BB, Shridas P, Simons M, van der Westhuyzen DR, Webb NR. J Lipid Res. 2009;50:641. doi: 10.1194/jlr.M800450-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slimani H, Charnaux N, Mbemba E, Saffar L, Vassy R, Vita C, Gattegno L. Biochim Biophys Acta. 2003;1617:80. doi: 10.1016/j.bbamem.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Ishiguro K, Kojima T, Muramatsu T. Glycoconj J. 2002;19:315. doi: 10.1023/A:1025308702966. [DOI] [PubMed] [Google Scholar]
- 47.Jetten N, Verbruggen S, Gijbels MJ, Post MJ, De Winther MP, Donners MM. Angiogenesis. 2014;17:109. doi: 10.1007/s10456-013-9381-6. [DOI] [PubMed] [Google Scholar]
- 48.Salmon-Ehr V, Ramont L, Godeau G, Birembaut P, Guenounou M, Bernard P, Maquart FX. Lab Invest. 2000;80:1337. doi: 10.1038/labinvest.3780141. [DOI] [PubMed] [Google Scholar]
- 49.Lin ZQ, Kondo T, Ishida Y, Takayasu T, Mukaida N. J Leukoc Biol. 2003;73:713. doi: 10.1189/jlb.0802397. [DOI] [PubMed] [Google Scholar]; McFarland-Mancini MM, Funk HM, Paluch AM, Zhou M, Giridhar PV, Mercer CA, Kozma SC, Drew AF. J Immunol. 2010;184:7219. doi: 10.4049/jimmunol.0901929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.