Abstract
BACKGROUND
Prostate cancer is a heterogeneous disease, but current treatments are not based on molecular stratification. We hypothesized that metastatic, castration-resistant prostate cancers with DNA-repair defects would respond to poly(adenosine diphosphate [ADP]–ribose) polymerase (PARP) inhibition with olaparib.
METHODS
We conducted a phase 2 trial in which patients with metastatic, castration-resistant prostate cancer were treated with olaparib tablets at a dose of 400 mg twice a day. The primary end point was the response rate, defined either as an objective response according to Response Evaluation Criteria in Solid Tumors, version 1.1, or as a reduction of at least 50% in the prostate-specific antigen level or a confirmed reduction in the circulating tumor-cell count from 5 or more cells per 7.5 ml of blood to less than 5 cells per 7.5 ml. Targeted next-generation sequencing, exome and transcriptome analysis, and digital polymerase-chain-reaction testing were performed on samples from mandated tumor biopsies.
RESULTS
Overall, 50 patients were enrolled; all had received prior treatment with docetaxel, 49 (98%) had received abiraterone or enzalutamide, and 29 (58%) had received cabazitaxel. Sixteen of 49 patients who could be evaluated had a response (33%; 95% confidence interval, 20 to 48), with 12 patients receiving the study treatment for more than 6 months. Next-generation sequencing identified homozygous deletions, deleterious mutations, or both in DNA-repair genes — including BRCA1/2, ATM, Fanconi’s anemia genes, and CHEK2 — in 16 of 49 patients who could be evaluated (33%). Of these 16 patients, 14 (88%) had a response to olaparib, including all 7 patients with BRCA2 loss (4 with biallelic somatic loss, and 3 with germline mutations) and 4 of 5 with ATM aberrations. The specificity of the biomarker suite was 94%. Anemia (in 10 of the 50 patients [20%]) and fatigue (in 6 [12%]) were the most common grade 3 or 4 adverse events, findings that are consistent with previous studies of olaparib.
CONCLUSIONS
Treatment with the PARP inhibitor olaparib in patients whose prostate cancers were no longer responding to standard treatments and who had defects in DNA-repair genes led to a high response rate.
Prostate cancer is the most common cancer in men and the sixth leading cause of death from cancer among men throughout the world.1 The interpatient molecular heterogeneity of this disease is well recognized; however, treatment to date has not been molecularly stratified.2,3 It would be useful to identify predictive biomarkers in order to provide more precise treatment for this disease.4
Metastatic, castration-resistant prostate cancer can have genomic aberrations that interfere with DNA repair.3,5 Some of these aberrations have been associated with sensitivity to platinum and poly(adenosine diphosphate [ADP]–ribose) polymerase (PARP) inhibitors, suggesting that treatment with a PARP inhibitor may exploit a synthetic lethal interaction.6–9 PARP is involved in multiple aspects of DNA repair, and the PARP inhibitor olaparib (Lynparza, AstraZeneca) has recently been approved for treating ovarian cancers with BRCA1/2 mutations.10,11
PARP inhibition has durable antitumor activity in men with metastatic, castration-resistant prostate cancer and deleterious germline BRCA2 mutations, a disease subset associated with a poor prognosis.8,12–14 We hypothesized that olaparib would have antitumor activity in sporadic cases of metastatic, castration-resistant prostate cancer with DNA-repair defects. In this clinical trial (TOPARP-A, Trial of PARP Inhibition in Prostate Cancer), we treated men with metastatic, castration-resistant prostate cancer with olaparib, obtaining fresh tumor-biopsy samples from all patients to conduct biomarker studies from both germline and somatic DNA, including exome and transcriptome sequencing, in order to elucidate the genomic aberrations, if any, associated with sensitivity to PARP inhibition in this disease.
METHODS
STUDY OVERSIGHT
This investigator-initiated study was designed by the TOPARP Protocol Development Group (see the Supplementary Appendix, available with the full text of this article at NEJM.org) and was cosponsored by the Institute of Cancer Research and the Royal Marsden NHS Foundation Trust. AstraZeneca provided olaparib free of cost and some funding in collaboration with the National Institute for Health Research Cancer Research Network but had no other role in the study. The study was approved by the research ethics committee at each participating site and by the Medicines and Healthcare Products Regulatory Agency. It was overseen by an independent steering committee and an independent data monitoring committee. Data were collated and analyzed by the Clinical Trials and Statistical Unit at the Institute of Cancer Research and were interpreted by all the authors. The manuscript was written by the first and last authors. All the authors reviewed the manuscript and vouch for the accuracy and completeness of the data and for the fidelity of the study to the protocol, which is available at NEJM.org.
PATIENTS
Eligible patients had histologically confirmed, metastatic, castration-resistant prostate cancer with progression after one or two regimens of chemotherapy. Additional eligibility criteria included an Eastern Cooperative Oncology Group (ECOG) performance-status score of 0 to 2 (on a scale of 0 to 5, with 0 indicating no symptoms and higher scores indicating increasing disability); no prior exposure to platinum, cyclophosphamide, mitoxantrone, or PARP inhibitors; documentation of cancer progression on the basis of Prostate Cancer Working Group 2 (PCWG2) criteria (see the Supplementary Appendix),15 Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1, or both sets of criteria16; adequate organ function; and 5 or more circulating tumor cells per 7.5 ml of blood. Complete eligibility criteria are available in the Supplementary Appendix. All patients provided written informed consent.
STUDY DESIGN, TREATMENT, AND END POINTS
TOPARP-A was an open-label, single-group, two-stage, phase 2, multisite study (Fig. S1 in the Supplementary Appendix). All the patients were treated with olaparib tablets at a dose of 400 mg twice a day until the occurrence of radiologic progression, unequivocal clinical progression, unacceptable side effects, withdrawal of consent, or death. Dose-modification guidelines for managing toxic effects were implemented.
The primary end point was the response rate. A response was defined as any of the following: a response according to RECIST, version 1.1; a reduction in the prostate-specific antigen (PSA) level of 50% or more; or a conversion in the circulating tumor-cell count, which is indicated by a reduction in the number of circulating tumor cells from 5 or more per 7.5 ml of blood at baseline to less than 5 per 7.5 ml during treatment, with a confirmatory assessment at least 4 weeks later. Secondary end points included radiologic progression–free survival (defined as the time to the first evidence of two new lesions on a bone scan plus two additional lesions on a confirmatory scan, as specified by PCWG2 criteria; progression according to RECIST, version 1.1; or death), progression-free survival, overall survival, time to PSA progression (a 25% increase in the PSA level), proportion of patients with conversion of the circulating tumor-cell count, and safety of olaparib and adverse events.
ASSESSMENTS
Disease assessments performed at baseline and every 12 weeks included computed tomographic (CT) studies or magnetic resonance imaging (MRI) and whole-body bone scanning, PSA measurements, and circulating tumor-cell counts. Additional circulating tumor-cell counts were performed after 1, 2, 4, and 8 weeks of treatment. Radiologic evaluations were subject to retrospective central review by radiologists who were unaware of the outcome data. Safety was assessed by monitoring adverse events, graded according to Common Terminology Criteria for Adverse Events (CTCAE), version 4.0 (see the Supplementary Appendix). Samples from protocol-specified biopsies were obtained from all patients during screening and in the first 4 weeks of treatment.
BIOMARKER STUDIES
All biomarker studies were prospectively planned. Whole-exome sequencing and transcriptome studies were performed on DNA from fresh-frozen tumor-biopsy samples obtained before treatment; germline whole-exome sequencing was performed on DNA from saliva samples. These studies were conducted at the University of Michigan as previously described.3,17 Targeted next-generation sequencing studies were conducted at the Institute of Cancer Research18,19; libraries were constructed with the use of the GeneRead DNAseq Panel (Qiagen) and run on a MiSeq sequencer (Illumina). Copy-number data were validated by means of droplet digital polymerase-chain-reaction (PCR) testing with the use of the QX100 Droplet Digital PCR System (Bio-Rad).19 Circulating tumor-cell counts were performed with the use of CellSearch (Janssen Diagnostics).20 For the purpose of correlating the results of next-generation sequencing with the response to treatment, patients were classified as positive or negative for genomic defects in DNA-repair genes. A patient was considered to be biomarker-positive if a homozygous deletion or deleterious mutation was identified in a gene reported to be involved either in DNA damage repair or sensitivity to PARP inhibition.21–23 PTEN and ERG protein expression was determined by means of immunohistochemical assessment24 (for details, see the Supplementary Appendix).
STATISTICAL ANALYSIS
A cohort of 45 patients was planned for TOPARP-A, which had a two-stage, phase 2 design (30 patients in stage 1, and 15 in stage 2), with a response rate of 5% or less for the null hypothesis and a response rate of more than 20% for the alternative hypothesis (at an alpha level of 0.02 and a beta level of 0.10). Safety analyses were descriptive and included all patients receiving at least one dose of olaparib. Analyses of response end points included all patients who could be evaluated for a response, with correlative biomarker-response analyses performed for all such patients. Statistical analyses were conducted with the use of Stata software, version 13 (StataCorp), on a snapshot of the data taken on April 24, 2015, as preplanned, when all patients had either discontinued treatment or received treatment for more than 6 months.
A two-sided exact binomial 95% confidence interval was calculated for the response rate. Percentage changes in PSA levels and circulating tumor-cell counts were represented in waterfall plots and longitudinal time plots. Time-to-event end points (radiologic progression–free, progression-free, and overall survival), measured from the time of study entry, were assessed by means of Kaplan–Meier methods. The association of biomarkers with treatment response was tested by means of the chi-square test or Fisher’s exact test, as appropriate. Univariable and multivariable logistic-regression models were used to calculate the unadjusted and adjusted odds ratios for a response. For the association of time-to-event end points with biomarker status, a log-rank test was used, and hazard ratios were calculated with the use of a Cox proportional-hazards model (with a hazard ratio of less than 1.00 indicating a reduced risk of events in the biomarker-positive subgroup). The proportional-hazards assumption was tested with the use of Schoenfeld residuals.
RESULTS
STUDY PATIENTS
From July 2012 through September 2014, a total of 50 patients (30 in stage 1 and 20 in stage 2 of the study) were enrolled at seven centers. All patients received at least one dose of olaparib. One patient was lost to follow-up after the first week; the data monitoring committee decided that this patient could not be evaluated for a response (Fig. S2 in the Supplementary Appendix). At the time of data cutoff, 35 of the 50 patients (70%) had died, with 4 patients remaining in the study after at least 40 weeks of therapy. The median overall survival was 10.1 months (interquartile range, 5.1 to 15.6), with a median follow-up of 14.4 months (range, 1.4 to 21.9). Table 1 summarizes the baseline characteristics of the patients.
Table 1.
Baseline Characteristics of the 50 Study Patients.*
| Characteristic | Value |
|---|---|
| Age — yr | |
| Median | 67.5 |
| Range | 40.8–79.3 |
| Time since initial diagnosis of prostate adenocarcinoma — yr |
|
| Median | 5.0 |
| Interquartile range | 3.4–7.9 |
| Metastatic disease at initial diagnosis — no. (%) | 23 (46) |
| Time since confirmation of CRPC — yr | |
| Median | 2.2 |
| Interquartile range | 1.7–3.9 |
| PSA — ng/ml | |
| Median | 349.5 |
| Interquartile range | 153–806 |
| CTC count — no. of cells/7.5 ml of blood | |
| Median | 37 |
| Interquartile range | 14–110 |
| ECOG performance-status score — no. (%)† | |
| 0 | 9 (18) |
| 1 | 35 (70) |
| 2 | 6 (12) |
| Received prior regimens for CRPC — no. (%) | |
| 2 | 3 (6) |
| 3 | 7 (14) |
| ≥4 | 40 (80) |
| Received prior treatments — no. (%) | |
| Radical prostatectomy or radiotherapy | 25 (50) |
| Castration (chemical or surgical) | 50 (100) |
| Abiraterone acetate | 48 (96) |
| Enzalutamide | 14 (28) |
| Docetaxel | 50 (100) |
| Cabazitaxel | 29 (58) |
| Radium-223 | 1 (2) |
CRPC denotes castration-resistant prostate cancer, CTC circulating tumor-cell, and PSA prostate-specific antigen.
The Eastern Cooperative Oncology Group (ECOG) performance-status score ranges from 0 to 5, with 0 indicating no symptoms and higher scores indicating increasing disability.
ANTITUMOR ACTIVITY
Sixteen of the 49 patients who could be evaluated had a response to olaparib on the basis of the composite definition of response specified in the study protocol (response rate, 33%; 95% confidence interval, 20 to 48). The median duration of treatment for the 16 patients who had a response was 40 weeks, with 12 patients receiving olaparib for more than 6 months and 4 patients receiving it for more than 12 months. Overall, 11 of the 49 patients (22%) had reductions in the PSA level of 50% or more. The median circulating tumor-cell count at baseline was 37 cells per 7.5 ml of blood (interquartile range, 14 to 110); 14 of the 49 patients (29%) had a confirmed reduction in the circulating tumor-cell count to less than 5 cells per 7.5 ml. Of the 49 patients who could be evaluated, 32 (65%) had measurable disease at baseline according to RECIST, version 1.1; 6 of these patients (19%) had a confirmed radiologic partial response (Table 2).
Table 2.
Changes in PSA Levels and CTC Counts, Best Radiologic Response, Duration of Treatment, and Biomarker Status for the 16 Patients with a Treatment Response.
| Patient No. |
Maximum PSA Decline |
Measurable Disease at Baseline |
Best Radiologic Response* |
Confirmed CTC Conversion |
CTC Count | Duration of Treatment |
Biomarker Status |
|
|---|---|---|---|---|---|---|---|---|
| Baseline | Maximum Decline |
|||||||
| % |
no./7.5 ml of blood |
% | wk | |||||
| 1 | 85 | No | No | 87 | 100 | 73 | Positive | |
| 5 | 51 | No | Yes | 24 | 100 | 58 | Positive | |
| 6 | 29 | Yes | SD | Yes | 105 | 97.1 | 16† | Positive |
| 8 | 47 | No | Yes | 38 | 94.7 | 62 | Positive | |
| 11 | No decline | Yes | PD | Yes | 6 | 83.3 | 12 | Negative |
| 14 | 83 | No | Yes | 102 | 100 | 36 | Positive | |
| 15 | 80 | Yes | PR | Yes | 18 | 100 | 36 | Positive |
| 16 | 88 | Yes | PR | Yes | 5 | 100 | 40‡ | Negative |
| 17 | 95 | Yes | PR | Yes | 8 | 100 | 24 | Positive |
| 20 | 88 | Yes | PR | NE | <5 | 100 | 48 | Positive |
| 26 | No decline | No | Yes | 12 | 100 | 19§ | Positive | |
| 30 | 70 | No | Yes | 100 | 100 | 44‡ | Positive | |
| 35 | 96 | Yes | PR | Yes | 513 | 100 | 40‡ | Positive |
| 36 | 59 | No | Yes | 22 | 100 | 57 | Positive | |
| 39 | 68 | Yes | PR | Yes | 24 | 100 | 44‡ | Positive |
| 48 | No decline | Yes | SD | Yes | 9 | 100 | 39 | Positive |
Best radiologic response was assessed in patients with measurable disease at baseline, according to Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1. NE denotes not evaluated, PD progressive disease, PR partial response, and SD stable disease.
The patient required repeated reductions in the dose of olaparib, to 200 mg twice a day.
The patient was still receiving treatment at the time of data cutoff.
Treatment was discontinued because of an adverse event.
DEFECTS IN DNA DAMAGE-REPAIR GENES
Paired samples from tumor biopsies performed before treatment and during treatment were available for all patients participating in the study; 28 patients underwent bone marrow biopsies, and 22 underwent imaging-guided biopsies of nodal or visceral metastases. Of the 49 patients who could be evaluated for a response, 43 had tumor-biopsy material that was suitable for next-generation sequencing. For the other 6 patients, archival tumor samples obtained at diagnosis were analyzed.
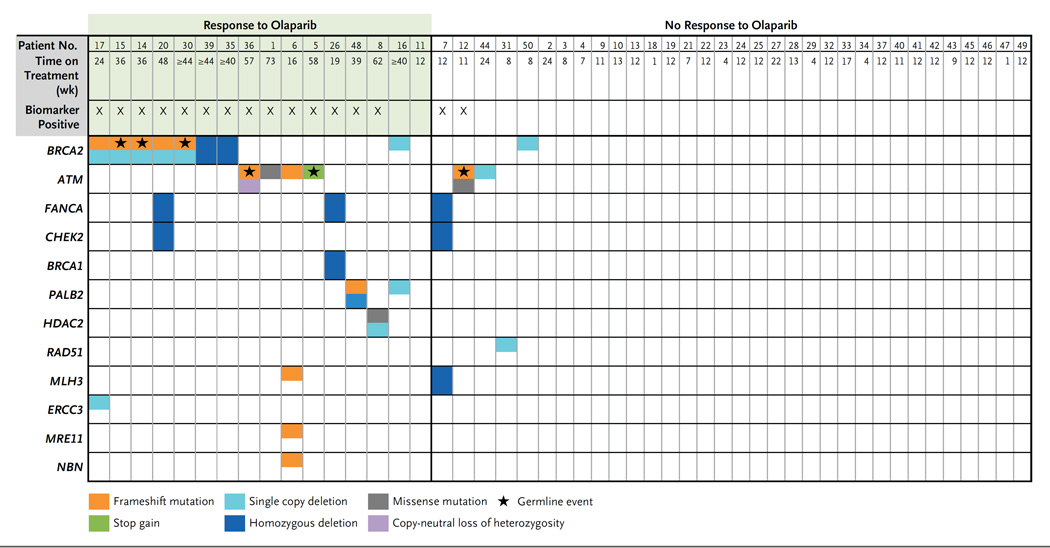
Overall, 16 patients (33%) had tumor aberrations in DNA-repair genes (Fig. 1). BRCA2 aberrations were detected in 7 patients: 2 had somatic homozygous deletions, 2 had a combination of somatic mutations and loss of heterozygosity, and 3 had a previously unidentified germline pathogenic mutation with somatic loss of the second allele. Five other patients had tumors with ATM aberrations. Three of these patients had germline mutations predicted to cause truncation of the ATM protein, and 2 of the 3 also had aberrant alleles in somatic DNA (1 with loss of heterozygosity and 1 with a missense mutation in the kinase domain–coding region of ATM). The other 2 patients had ATM mutations with no germline events: 1 had a frameshift mutation (p.V2288fs*1) predicted to cause truncation before the C-terminal phosphoinositide 3-kinase (PI3K) catalytic domain (normally formed by amino acid residues 2712 through 2962), as well as ATM domains required for p53 activation and the response to DNA damage,25,26 and 1 had a missense mutation (p.N2875H) within the PI3K catalytic domain of ATM.27 Homozygous somatic deletions of BRCA1 or CHEK2 occurred with FANCA deletion in 3 patients. A somatic frameshift mutation in PALB2 (partner and localizer of BRCA2) was also detected in a patient with a heterozygous PALB2 deletion. Finally, biallelic somatic aberrations in histone deacetylase 2 (HDAC2), which has a role in ATM function and the response to DNA damage,28–30 were identified in 1 patient.
Figure 1. Genomic Aberrations in DNA Repair in Patients with Metastatic, Castration-Resistant Prostate Cancer.
Data are shown for the 49 patients who could be evaluated for a response. Mutations and deletions in DNA-repair genes were identified through next-generation sequencing studies. Green shading indicates patients who were classified as having a response to olaparib in the clinical trial. Patients were considered to be biomarker-positive if homozygous deletions, deleterious mutations, or both were detected in DNA-repair genes (but not single copy deletions without events detected in the second allele). A star indicates that a particular genomic event was detected in germline DNA. Archival tumor samples were used for the sequencing studies in Patients 13, 18, 21, 40, 41, and 49 because the biopsy samples obtained during the trial were negative for tumor content.
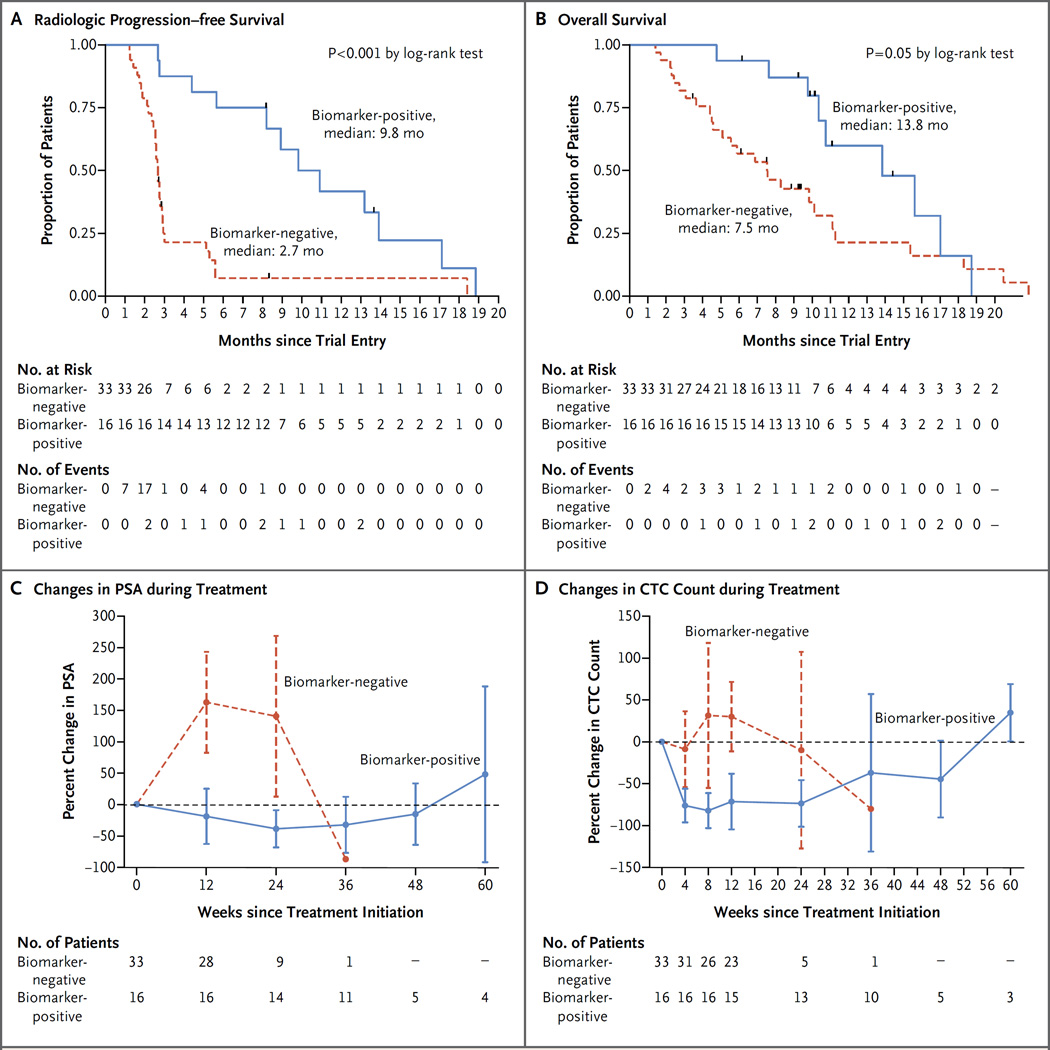
Overall, patients with aberrations in DNA-repair genes had a significantly higher response rate in unadjusted and adjusted analyses (P<0.001 in a multivariable logistic-regression model for response) (Tables S2 through S5 in the Supplementary Appendix); 14 of 16 biomarker-positive patients (88%) had a response to olaparib. Conversely, only 2 of 33 biomarker-negative patients (6%) were classified as having a response (sensitivity, 88%; specificity, 94%). Radiologic progression– free survival was significantly longer in the biomarker-positive group than in the biomarker-negative group (median, 9.8 vs. 2.7 months; P<0.001 by the log-rank test) (Fig. 2). Overall survival was also prolonged in the biomarker-positive group (median, 13.8 months, vs. 7.5 months in the biomarker-negative group; P = 0.05 by the log-rank test), even though established prognostic factors were balanced between the two groups (Table S6 in the Supplementary Appendix).
Figure 2. Antitumor Activity of Olaparib and Association with Defects in DNA-Repair Genes, According to Biomarker Status.
Panels A and B show radiologic progression–free survival and overall survival curves, respectively, for patients with genomic defects in DNA-repair genes (biomarker-positive group) and patients without such defects (biomarker-negative group). The hazard ratio for radiologic progression in the biomarker-positive group as compared with the biomarker-negative group was 0.24 (95% confidence interval [CI], 0.11 to 0.50), and the hazard ratio for death was 0.47 (95% CI, 0.22 to 1.02). Panels C and D show mean percentage changes in prostate-specific antigen (PSA) levels and circulating tumor-cell (CTC) counts, respectively, over the course of treatment in the biomarker-positive and biomarker-negative groups.
All seven patients with BRCA2 loss had PSA levels that fell by 50% or more from baseline; of these seven patients, all five with measurable disease also had a radiologic partial response (Fig. 3). Four of the five patients with deleterious ATM mutations had a response to olaparib, including all three patients with DNA mutations predicted to cause loss of the kinase catalytic domain of ATM but with no defects in the second ATM allele.
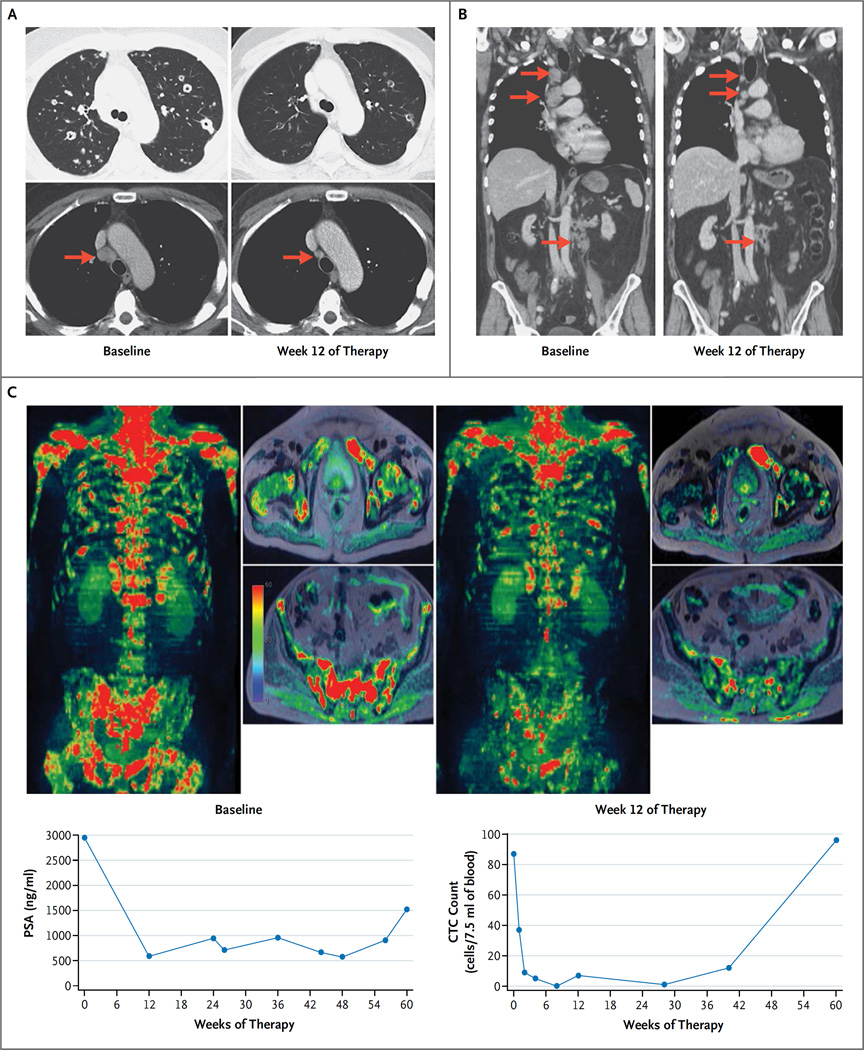
Figure 3. Radiologic Evidence of Tumor Responses to Olaparib at Week 12.
Panel A shows CT scans of the chest, obtained in the lung and soft-tissue window settings, from a 61-year-old man with metastatic, castration-resistant prostate cancer (Patient 39) who had a response to olaparib; there was shrinkage of the lung and nodal (arrows) metastatic deposits after 12 weeks of therapy (right), as compared with baseline (left). Whole-exome sequencing showed a somatic homozygous deletion of BRCA2. Panel B shows CT scans with coronal reconstruction in a 70-year-old man with a somatic BRCA2 frameshift insertion (p.Y2154fs*21) and somatic deletion of the second allele (Patient 20). The scans show the response in the mediastinal and abdominal lymph nodes (arrows). The patient received treatment for a total of 48 weeks. Panel C shows multiparametric whole-body MRI scans, including diffusion-weighted imaging, with coronal three-dimensional reconstruction and selected axial images in a 79-year-old man (Patient 1) who had a response to olaparib, with an 85% reduction in the PSA level. The patient received treatment for a total of 73 weeks. The images show reduction in the water content within the skeletal metastasis, which in conjunction with other findings on imaging would be consistent with tumor regression during therapy (right), as compared with baseline (left). Next-generation sequencing of the baseline bone marrow–biopsy specimen revealed a somatic missense mutation within the ATM phosphoinositide 3-kinase catalytic domain (p.N2875S), with no evidence of genomic loss of the second allele and with maintenance of ATM expression on immunohistochemical assessment.
An additional three patients who were classified as biomarker-positive had a response to olaparib. Patient 26, who had a somatic homozygous deletion of both BRCA1 and FANCA, had a response, with a reduction in the circulating tumor-cell count from 12 to 0 cells per 7.5 ml; olaparib was discontinued prematurely after 19 weeks because of myelosuppression with radiologic stable disease (Table 2). Patient 48, whose tumor had biallelic PALB2 aberrations, had a durable response that lasted for 39 weeks. The third patient with a response (Patient 8), who had biallelic somatic aberrations in HDAC2, received the study treatment for 62 weeks. His transcriptome revealed low HDAC1 and HDAC2 messenger RNA expression (Fig. S4 in the Supplementary Appendix). Conversely, a patient with defective mismatch DNA repair due to biallelic MLH3 loss with somatic homozygous deletion of FANCA and CHEK2 did not have a response to therapy, nor did the fifth patient with mutant ATM, who had radiologic progression of the liver metastases and an increase in the PSA level at the first response assessment.
Finally, in two other patients categorized as having a response according to trial criteria, tumor-exome sequencing identified no genomic defects fulfilling the study definition of defective DNA repair. However, one of the two patients (Patient 11) had little evidence of a true response, with a drop in circulating tumor-cell counts from 6 cells to 1 cell per 7.5 ml and radiologic progression at 12 weeks. The other patient (Patient 16) had a clear radiologic response according to RECIST, as well as an 86% reduction in the PSA level. This patient’s tumor had monoallelic deletions of both BRCA2 and PALB2, with no detectable loss of the other allele evident on whole-exome sequencing.
SAFETY
The median duration of olaparib treatment was 12 weeks (interquartile range, 11 to 24). The average delivered-dose intensity was 87%. The most commonly reported adverse events that developed during treatment are summarized in Table S7 in the Supplementary Appendix. Grade 3 or 4 drug-related events were primarily anemia (in 10 of the 50 patients [20%]), fatigue (in 6 patients [12%]), leukopenia (in 3 patients [6%]), thrombocytopenia (in 2 patients [4%]), and neutropenia (in 2 patients [4%]). Overall, 13 patients (26%) required a reduction in the dose of olaparib to 300 mg twice a day; anemia was the most common indication for the dose reduction (in 7 patients). These episodes of anemia were considered to be largely drug-related, although most of the patients with anemia had extensive bony disease involvement; the median baseline hemoglobin level was 11.2 g per deciliter. Three of the 13 patients required a second dose reduction, to 200 mg of olaparib twice a day. Olaparib was permanently discontinued in 3 patients (6%) because of adverse events.
DISCUSSION
The results of this clinical trial suggest that a common subset of metastatic prostate cancers can be molecularly stratified for treatment. This subset, characterized by defects in DNA repair, accounts for approximately 25 to 30% of all sporadic, castration-resistant prostate cancers.3,17 The protocol for our therapeutic trial mandated the acquisition of fresh tumor-biopsy samples from patients with metastatic, castration-resistant prostate cancer in order to identify predictive biomarkers. Our findings suggest that this approach is feasible and that next-generation sequencing analyses of tumor-biopsy samples can increase our understanding of treatment responses. Additional studies are needed to assess whether the frequency of DNA-repair defects in patients with untreated prostate cancer is identical to the frequency in patients with castration-resistant prostate cancer.
Evidence for the antitumor activity of olaparib in this study cohort includes concurrent falls in PSA levels and circulating tumor-cell counts, with radiologic responses on CT scans and regression of bone disease on serial whole-body MRI scans (Fig. 3). Responses to treatment commonly lasted for more than 6 months and were associated with prolonged radiologic progression–free survival and impressive falls in circulating tumor-cell counts; however, we cannot yet determine whether olaparib improves overall survival among patients with metastatic, castration-resistant prostate cancer and DNA-repair defects.20,31
Using a high-throughput, next-generation sequencing assay, we detected multiple types of genomic aberrations associated with PARP-inhibitor sensitivity. Alterations were observed in BRCA2, ATM, BRCA1, PALB2, CHEK2, FANCA, and HDAC2, all of which have been reported to have a synthetic lethal interaction with PARP inhibition.21,22
BRCA2 loss is well recognized in prostate cancer.3,5 Of the seven patients in our study who had BRCA2 genomic aberrations, three also had previously unknown deleterious germline mutations in BRCA2, for an overall prevalence of 6% in our cohort of unselected patients. This finding is in keeping with emerging data17 and suggests that the prevalence of germline BRCA2 mutations in castration-resistant prostate cancer has been underestimated.32 We also identified truncating ATM germline mutations in three patients (6%) in this cohort of patients with advanced prostate cancers. Further work is needed to determine the relevance of these findings.
A notable finding of this study is the response to olaparib in patients whose tumors had monoallelic ATM aberrations with mutations affecting the kinase catalytic domain, despite the continued presence of the second allele and ATM protein expression on immunohistochemical assessment. Multiple studies have described how the loss of one kinase domain can alter ATM protein function.33–35 A proposed explanation is that the mutated ATM is dominant negative, but ATM haploinsufficiency has not been ruled out.36 Aberrations in multiple other DNA repair genes associated with PARP-inhibitor sensitivity may be present at a low frequency in metastatic, castration-resistant prostate cancer, as reported in other cancers.21,37 We observed clinical responses to olaparib in patients with metastatic, castration-resistant prostate cancer and defective PALB2 or HDAC2. HDAC inhibition has been reported to result in sensitivity to PARP inhibition,28,38 but the clinical relevance of HDAC genomic loss is not known.
Platinum-based chemotherapy is generally not used for the treatment of metastatic, castration-resistant prostate cancer, since phase 3 studies have failed to show a survival benefit in unselected patients. Nevertheless, responses to single-agent chemotherapy with a platinum analogue such as satraplatin have been reported.39 Considering the mechanisms of action of platinum and data emerging from studies of ovarian cancer in which platinum and PARP-inhibitor sensitivity were correlated with defects in homologous recombination DNA repair,40,41 we hypothesize that the DNA-repair defects we are reporting in metastatic, castration-resistant prostate cancer may be associated with platinum sensitivity.
In conclusion, we report that PARP inhibition has antitumor activity in sporadic cases of metastatic, castration-resistant prostate cancer and that these responses are associated with DNA-repair defects in tumor cells that can be identified through next-generation sequencing assays.
Supplementary Material
Acknowledgments
Funded by Cancer Research UK and others; ClinicalTrials.gov number, NCT01682772; Cancer Research UK number, CRUK/11/029.
Supported by grants from Cancer Research UK (C12540/A12829, C12540/A13230, C1491/A9895, and C1491/A15955, for trial CRUK/11/029), Stand Up To Cancer–Prostate Cancer Foundation (a Prostate Dream Team Translational Cancer Research Grant), and Prostate Cancer UK. Also supported by grants from Cancer Research UK and the National Institute for Health Research (to the Cancer Research UK Center, the Experimental Cancer Medicine Center, and the Biomedical Research Center of the Institute of Cancer Research and the Royal Marsden NHS Foundation Trust), as well as a Medical Research Council–Prostate Cancer UK fellowship (to Dr. Mateo), a grant from the Swiss Cancer League (BIL KLS-02592-02-2010, to Dr. Omlin), and a Marie Curie International Incoming Fellowship (625792, to Dr. Boysen). Dr. Perez-Lopez participated in this work in the Medicine Doctorate framework of Universitat Autonoma de Barcelona. This study was conducted with support from the Investigator-Sponsored Study Collaboration between Astra-Zeneca and the National Institute for Health Research Cancer Research Network.
Dr. de Bono reports receiving consulting fees and travel support from Astellas, Genentech, GlaxoSmithKline, Janssen, Sanofi-Aventis, and Medivation and royalties to his institution from a patent related to PARP inhibition in BRCA cancers (WO2004GB05025). Dr. Omlin reports receiving consulting fees from Bayer, Sanofi, Astellas, and Janssen; travel support from Bayer, Sanofi, and Astellas; and grant support from Janssen. Dr. Jain reports receiving fees for serving on an advisory board from Ferring Pharmaceuticals, lecture fees from Janssen, travel support from Astellas, and support from AstraZeneca to host a conference. Dr. Ralph reports receiving honoraria from Pfizer and Bristol-Myers Squibb and travel support from Novartis, Roche, GlaxoSmithKline, Viralytics, and Oncolytics Biotech. Dr. Protheroe reports receiving honoraria from Astellas and Novartis. Dr. Hussain reports receiving fees for serving on advisory boards from Astellas; consulting fees from Bayer and Pierre Fabre; travel support from Bayer, Janssen, and Boehringer Ingelheim; and grant support from Bayer, Pierre Fabre, and Boehringer Ingelheim. Dr. Jones reports receiving grant support from AstraZeneca. Dr. Bianchini reports receiving fees for serving on an advisory board from Janssen. Dr. Eeles reports receiving travel support from Janssen. Dr. Attard reports receiving consulting fees from Janssen–Cilag, Veridex, Ventana Medical Systems, Astellas, Medivation, Novartis, Millennium, and Abbott; lecture fees from Ventana Medical Systems, Astellas, Janssen, Takeda, and Sanofi-Aventis; honoraria from Astellas and Janssen; travel support from Ventana Medical Systems, Astellas, Medivation, Abbott, and Janssen; and grant support from Janssen and AstraZeneca; his employer, the Institute of Cancer Research (ICR), developed abiraterone acetate and has a commercial interest in this drug, and he is on the ICR’s list of rewards to inventors of abiraterone acetate. Dr. Lord reports holding patents related to the use of PARP inhibitors (WO2008020180 [A2] and WO2009027650 [A1]). Dr. Ashworth reports receiving fees for serving on advisory boards from Genentech and GlaxoSmithKline and consulting fees from Merck Serono, Novartis, and Celgene; he also reports receiving royalties from patents related to PARP inhibitors (US7449464, US7981889, US7692006, and US8247416), which are licensed to AstraZeneca. Dr. Feng reports receiving fees for serving on advisory boards from Medivation–Astellas, Celgene, and GenomeDx; he also reports founding and serving on the board of directors of PFS Genomics. Dr. Hall reports receiving grant support from Astra-Zeneca and Bayer and receiving drug supplies for clinical trials from AstraZeneca, Bayer, and Astellas.
We thank the staff at the Prostate Targeted Therapy Group and the Tumor Profiling Unit, Institute of Cancer Research; the members of the independent steering committee and independent data monitoring committee; and all the patients who agreed to participate in the study.
APPENDIX
The authors’ full names and academic degrees are as follows: Joaquin Mateo, M.D., Suzanne Carreira, Ph.D., Shahneen Sandhu, M.D., Susana Miranda, B.Sc., Helen Mossop, M.Math.Stat., Raquel Perez-Lopez, M.D., Daniel Nava Rodrigues, M.D., Dan Robinson, Ph.D., Aurelius Omlin, M.D., Nina Tunariu, M.D.Res., Gunther Boysen, Ph.D., Nuria Porta, Ph.D., Penny Flohr, B.Sc., Alexa Gillman, B.Sc., Ines Figueiredo, B.Sc., Claire Paulding, B.Sc., George Seed, M.Sc., Suneil Jain, M.D., Christy Ralph, M.D., Andrew Protheroe, M.D., Ph.D., Syed Hussain, M.D., Robert Jones, M.D., Ph.D., Tony Elliott, M.D., Ph.D., Ursula McGovern, M.D., Ph.D., Diletta Bianchini, M.D., Jane Goodall, B.Sc., Zafeiris Zafeiriou, M.D., Chris T. Williamson, Ph.D., Roberta Ferraldeschi, M.D., Ph.D., Ruth Riisnaes, F.I.B.M.S., Bernardette Ebbs, B.T.E.C., Gemma Fowler, B.Sc., Desamparados Roda, M.D., Wei Yuan, Ph.D., Yi-Mi Wu, Ph.D., Xuhong Cao, M.S., Rachel Brough, Ph.D., Helen Pemberton, Ph.D., Roger A’Hern, Ph.D., Amanda Swain, Ph.D., Lakshmi P. Kunju, M.D., Rosalind Eeles, M.D., Ph.D., Gerhardt Attard, M.D., Ph.D., Christopher J. Lord, Ph.D., Alan Ashworth, Ph.D., Mark A. Rubin, M.D., Karen E. Knudsen, Ph.D., Felix Y. Feng, M.D., Ph.D., Arul M. Chinnaiyan, M.D., Ph.D., Emma Hall, Ph.D., and Johann S. de Bono, M.B., Ch.B., Ph.D.
The authors’ affiliations are as follows: the Institute of Cancer Research (J.M., S.C., S.S., S.M., H.M., R.P.-L., D.N.R., A.O., N.T., G.B., N.P., P.F., A.G., I.F., C.P., G.S., D.B., J.G., Z.Z., C.T.W., R.F., R.R., B.E., G.F., D. Roda, W.Y., R.B., H.P., R.A., A.S., R.E., G.A., C.J.L., A.A., E.H., J.S.B.), the Royal Marsden NHS Foundation Trust (J.M., S.S., R.P.-L., A.O., N.T., D.B., Z.Z., R.F., D. Roda, R.E., G.A., J.S.B.), and University College London Hospital (U.M.), London, Queen’s University, Belfast (S.J.), University of Leeds, Leeds (C.R.), Churchill Hospital, Oxford (A.P.), University of Liverpool, Liverpool (S.H.), Beatson West of Scotland Cancer Centre, Glasgow (R.J.), and Christie Hospital, Manchester (T.E.) — all in the United Kingdom; the University of Michigan, Ann Arbor (D. Robinson, Y.-M.W., X.C., L.P.K., F.Y.F., A.M.C.); Weill Cornell Medical College, New York (M.A.R.); and Thomas Jefferson University, Philadelphia (K.E.K.).
Footnotes
Presented in part at the Annual Congress of the European Society for Medical Oncology, Madrid, September 26–30, 2014; the 21st Annual Scientific Retreat of the Prostate Cancer Foundation, Carlsbad, CA, October 23–25, 2014; and the Annual Meeting of the American Association for Cancer Research, Philadelphia, April 18–22, 2015.
No other potential conflict of interest relevant to this article was reported.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Hellerstedt BA, Pienta KJ. The current state of hormonal therapy for prostate cancer. CA Cancer J Clin. 2002;52:154–179. doi: 10.3322/canjclin.52.3.154. [DOI] [PubMed] [Google Scholar]
- 2.Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasso CS, Wu Y-M, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 5.Beltran H, Yelensky R, Frampton GM, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 8.Fong PCP, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 11.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 12.Sandhu SK, Omlin A, Hylands L, et al. Poly (ADP-ribose) polymerase (PARP) inhibitors for the treatment of advanced germline BRCA2 mutant prostate cancer. Ann Oncol. 2013;24:1416–1418. doi: 10.1093/annonc/mdt074. [DOI] [PubMed] [Google Scholar]
- 13.Sandhu SK, Schelman WR, Wilding G, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;14:882–892. doi: 10.1016/S1470-2045(13)70240-7. [DOI] [PubMed] [Google Scholar]
- 14.Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scher HI, Halabi S, Tannock I, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–1159. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenhauer EA, Therasse P, Bogaerts J, et al. New Response Evaluation Criteria in Solid Tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Robinson D, Van Allen EM, Wu Y-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong M, Carreira S, Goodall J, et al. Validation and utilisation of high-coverage next-generation sequencing to deliver the pharmacological audit trail. Br J Cancer. 2014;111:828–836. doi: 10.1038/bjc.2014.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014;6:254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol. 2015;33:1348–1355. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 22.Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michels J, Vitale I, Saparbaev M, Castedo M, Kroemer G. Predictive biomarkers for cancer therapy with PARP inhibitors. Oncogene. 2014;33:3894–3907. doi: 10.1038/onc.2013.352. [DOI] [PubMed] [Google Scholar]
- 24.Ferraldeschi R, Nava Rodrigues D, Riisnaes R, et al. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur Urol. 2015;67:795–802. doi: 10.1016/j.eururo.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turenne GA, Paul P, Laflair L, Price BD. Activation of p53 transcriptional activity requires ATM’s kinase domain and multiple N-terminal serine residues of p53. Oncogene. 2001;20:5100–5110. doi: 10.1038/sj.onc.1204665. [DOI] [PubMed] [Google Scholar]
- 26.Beamish H, Kedar P, Kaneko H, et al. Functional link between BLM defective in Bloom’s syndrome and the ataxia-telangi-ectasia-mutated protein, ATM. J Biol Chem. 2002;277:30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- 27.Canman CE, Lim DS, Cimprich KA, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 28.Miller KM, Tjeertes JV, Coates J, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thurn KT, Thomas S, Raha P, Qureshi I, Munster PN. Histone deacetylase regulation of ATM-mediated DNA damage signaling. Mol Cancer Ther. 2013;12:2078–2087. doi: 10.1158/1535-7163.MCT-12-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adimoolam S, Sirisawad M, Chen J, Thiemann P, Ford JM, Buggy JJ. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proc Natl Acad Sci U S A. 2007;104:19482–19487. doi: 10.1073/pnas.0707828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris MJ, Molina A, Small EJ, et al. Radiographic progression-free survival as a response biomarker in metastatic castration-resistant prostate cancer: COU-AA-302 results. J Clin Oncol. 2015;33:1356–1363. doi: 10.1200/JCO.2014.55.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kote-Jarai Z, Leongamornlert D, Saunders E, et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer. 2011;105:1230–1234. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 34.Chenevix-Trench G, Spurdle AB, Gatei M, et al. Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002;94:205–215. doi: 10.1093/jnci/94.3.205. [DOI] [PubMed] [Google Scholar]
- 35.Morgan SE, Lovly C, Pandita TK, Shiloh Y, Kastan MB. Fragments of ATM which have dominant-negative or complementing activity. Mol Cell Biol. 1997;17:2020–2029. doi: 10.1128/mcb.17.4.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barlow C, Eckhaus MA, Schäffer AA, Wynshaw-Boris A. Atm haploinsufficiency results in increased sensitivity to sublethal doses of ionizing radiation in mice. Nat Genet. 1999;21:359–360. doi: 10.1038/7684. [DOI] [PubMed] [Google Scholar]
- 37.Buisson R, Dion-Côté A-M, Coulombe Y, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chao OS, Goodman OB., Jr Synergistic loss of prostate cancer cell viability by co-inhibition of HDAC and PARP. Mol Cancer Res. 2014;12:1755–1766. doi: 10.1158/1541-7786.MCR-14-0173. [DOI] [PubMed] [Google Scholar]
- 39.Sternberg CN, Petrylak DP, Sartor O, et al. Multinational, double-blind, phase III study of prednisone and either satraplatin or placebo in patients with castrate-refractory prostate cancer progressing after prior chemotherapy: the SPARC trial. J Clin Oncol. 2009;27:5431–5438. doi: 10.1200/JCO.2008.20.1228. [DOI] [PubMed] [Google Scholar]
- 40.Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 41.Ceccaldi R, O’Connor KW, Mouw KW, et al. A unique subset of epithelial ovarian cancers with platinum sensitivity and PARP inhibitor resistance. Cancer Res. 2015;75:628–634. doi: 10.1158/0008-5472.CAN-14-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.