Abstract
The 2014 CSAR Benchmark Exercise was the last community-wide exercise that was conducted by the group at the University of Michigan, Ann Arbor. For this event, GlaxoSmithKline (GSK) donated unpublished crystal structures and affinity data from in-house projects. Three targets were used: tRNA (m1G37) methyltransferase (TrmD), Spleen Tyrosine Kinase (SYK), and Factor Xa (FXa). A particularly strong feature of the GSK data is its large size, which lends greater statistical significance to comparisons between different methods. In Phase 1 of the CSAR 2014 Exercise, participants were given several protein-ligand complexes and asked to identify the one near-native pose from among 200 decoys provided by CSAR. Though decoys were requested by the community, we found that they complicated our analysis. We could not discern whether poor predictions were failures of the chosen method or an incompatibility between the participant’s method and the setup protocol we used. This problem is inherent to decoys and we strongly advise against their use. In Phase 2, participants had to dock and rank/score a set of small molecules given only the SMILES strings of the ligands and a protein structure with a different ligand bound. Overall, docking was a success for most participants, much better in Phase 2 than in Phase 1. However, scoring was a greater challenge. No particular approach to docking and scoring had an edge, and successful methods included empirical, knowledge-based, machine-learning, shape-fitting, and even those with solvation and entropy terms. Several groups were successful in ranking TrmD and/or SYK, but ranking FXa ligands was intractable for all participants. Methods that were able to dock well across all submitted systems include MDock1, Glide-XP2, PLANTS3, Wilma4, Gold5, SMINA6, Glide-XP2/PELE7, FlexX8, and MedusaDock9. In fact, the submission based on Glide-XP2/PELE7 cross-docked all ligands to many crystal structures, and it was particularly impressive to see success across an ensemble of protein structures for multiple targets. For scoring/ranking, submissions that showed statistically significant achievement include MDock1 using ITScore1,10 with a flexible-ligand term11, SMINA6 using Autodock-Vina12,13, FlexX8 using HYDE14, and Glide-XP2 using XP DockScore2 with and without ROCS15 shape similarity16. Of course, these results are for only three protein targets, and many more systems need to be investigated to truly identify which approaches are more successful than others. Furthermore, our exercise is not a competition.
Graphical abstract
INTRODUCTION
Recognizing the importance of docking calculations for structure-based drug design, many different academic groups, software vendors, and individual scientists have written software to dock small molecules into protein binding sites. These developers have different approaches and philosophies, and they have explored various algorithms with different force fields and scoring functions. In some cases, docking is straightforward. With a small, rigid molecule and a tight binding site, there might be only one binding mode that accommodates all the hydrogen-bonding partners. Perhaps the binding mode of a new compound is obvious based on prior crystal structures of similar compounds bound to the same protein. However, other cases can be extremely challenging. For example, binding a compound might involve major conformational changes in the protein, specific water molecules in the binding site, or interactions that require quantum mechanics for accurate simulation. It would be ideal if software developers could devise universally applicable docking methods, but in reality, different algorithms and force fields perform better or worse in specific situations. In fact, software developers may be surprised by the wide range of different molecular-docking problems that arise in drug-design practice. Some home-grown software is developed for a specific type of docking problem, often when commercial software fails. These home-grown methods might later be generalized and extended, but they still perform particularly well for the original type of problem and poorly for other docking problems. Also, docking software tends to work better for the original authors of a method than for subsequent users. The original authors may have tuned their method, knowingly or unknowingly, for the specific protein-ligand complexes used in their publications. Also, it is very likely that the original authors may simply have a better understanding of how to use their own software. These complications make it difficult for users to compare the software and methods available.
Blind challenges have been used in various disciplines to compare prediction methodologies17–25. The Community Structure-Activity Resource (CSAR) at the University of Michigan, Ann Arbor has held open challenges three times before, and two have been based on blinded data.26,27 With complex methodologies, there is always the danger of over-fitting to available data or to particular problems. Blind challenges provide a good way of checking for this. Iterative cycles of challenge, analysis, and further software development can gradually improve methodologies throughout a whole research community19.
The challenges should exercise and test the software with problems that simulate actual usage. Within the pharmaceutical industry, docking software is generally used in 1) lead discovery and 2) lead optimization. In lead discovery, some collection of compounds is typically screened against a biological assay, with the most effective compounds identified as “hits.” If the 3D structure of the target protein is known, then docking calculations can be used to predict the binding geometries or “poses” of the hit compounds within the binding site of the target protein. Docking can also be used to screen compounds in silico, prior to the acquisition of compounds, for experimental screening in a biological assay. The docked structures can reveal interactions with the protein, which should help explain the Structure-Activity Relationships (SARs), and may suggest chemical modifications to improve potency or selectivity. In lead optimization, chemists make modifications to the “lead” compounds, seeking to boost potency, selectivity, solubility, and other properties. Experimental and computational chemists use docked structures and crystal structures to design new compounds. As lead optimization progresses, the team may get crystal structures for multiple analogs within the congeneric series of interest. These crystal structures can greatly facilitate subsequent docking calculations by helping to define the likely binding geometry, characterizing the movements induced in the protein by the compounds, and by revealing key water molecules. Prediction of the binding geometry may be difficult in lead discovery, but relatively easy for lead optimization within a congeneric series. Of course, prediction of potency is generally challenging in both lead discovery and lead optimization.
CSAR and Its Benchmark Exercises
CSAR was funded by the National Institute of General Medical Sciences for a five-year period to collect and curate datasets for use in improving docking and scoring. These datasets were primarily obtained from the pharmaceutical industry such as Abbvie28, Vertex28, and GlaxoSmithKline (GSK). A few datasets were obtained from academia, such as one from the Baker group that formed our 2013 exercise27. Part of the remit of the CSAR Center was to run regular exercises to actively engage the docking and scoring community in assessing the current state of the art and the impact of potential improvements. The datasets that were collected by CSAR were used to run four worldwide exercises in 201017,18, 201226, 201327, and 2014). For each of the exercises, the Journal of Chemical Information and Modeling has published a series of papers from the organizers and participants to report their outcomes. All crystal structures used in the exercises were deposited in the Protein Data Bank (PDB) for others to use in developing their methods.29 The 2014 CSAR Benchmark Exercise, conducted with GSK data, is our last exercise. There is one last CSAR dataset of Hsp90 structures and affinities that was donated by colleagues at Abbott (now AbbVie) and augmented by in-house experiments at Michigan. This data has been passed on to the Drug Design Data Resource (D3R, www.drugdesigndata.org), a new effort for docking and scoring data that is housed at UCSD.
GlaxoSmithKline and the CSAR 2014 Exercise
To provide suitable data for blind challenges, GSK reviewed available unpublished crystal structures and their electron densities from in-house lead discovery and lead optimization projects. Three targets emerged from the review: tRNA (m1G37) methyltransferase (TrmD), Spleen Tyrosine Kinase (SYK), and Factor-Xa (FXa). TrmD is a target for antibiotic discovery30. SYK is a target for autoimmune diseases. FXa is an established target for blood clotting.
TrmD is a relatively new target where GSK has carried out lead discovery work using Encoded Library Technology (ELT) and fragment-based design31. GSK determined crystal structures for more than 30 fragments and several ELT hits bound to TrmD. The fragment-based compounds are relatively low molecular weight, with correspondingly low potency. Most of the compounds have some similarity to the S-adenosyl-L-methionine substrate, but are otherwise relatively diverse. This TrmD dataset provides one of the very first fragment-based challenges for docking software.
SYK is a kinase, and it exemplifies the lead optimization problem for kinases. SYK has been the target in drug discovery efforts at a number of pharmaceutical and biotech companies. Several SYK inhibitors have emerged from these efforts32. In particular, fostamatinib has shown some effectiveness in clinical trials for arthritis33,34 and lymphoma35. For the CSAR blind challenge, GSK identified 8 unpublished, in-house, crystal structures of SYK with bound ligands, together with affinity data on 267 unique compounds from closely-related congeneric series that had not previously been disclosed. A few ligands had affinity data for different salt forms which resulted in 276 reported affinities. The SYK dataset includes a large number of closely-related compounds that simulate the challenges encountered in lead optimization in the kinase target class.
FXa has been the target in drug discovery efforts at many pharmaceutical companies, and at least three FXa inhibitors have reached the market (Rivaroxaban36, Apixaban37, and Edoxaban38). Structure-based design has been used extensively in these FXa projects, and FXa has been used as a testbed for new methodologies in structure-based design39. Although GSK and other pharmaceutical companies have published extensively around FXa40–51, there are still many crystal structures and much assay data that has never been disclosed. For the CSAR blind challenge, there were three complexes for which binding affinities and structures were available. There were 163 affinity values for 106 unique ligands in closely-related congeneric series that had not been disclosed previously. This dataset uses new compounds to exercise docking software on an old, familiar target. This dataset should provide a good test of docking software working in lead optimization mode.
The CSAR 2014 Exercise opened on March 31st, 2014 and the last submissions were taken on July 25th, 2014. In Phase 1 of the CSAR 2014 Exercise, participants were given several protein-ligand complexes, each with 200 docked poses for the ligands. Participants were asked to score the poses and identify the one near-native pose over the 199 docking decoys. In Phase 2, the groups had to dock and rank/score a set of small molecules given only a set of SMILES strings for the ligands and crystal structure of the target with a congeneric ligand bound. A particularly strong feature of the data used in the CSAR 2014 Exercise is the large number of ligand affinities provided by GSK from their SYK and FXa projects. Large numbers are needed to identify statistically significant differences across the various computational approaches52. Few datasets have this critical feature. Furthermore, some of the affinity data from GSK is redundant for SYK and FXa ligands; this comes from multiple assay measurements or different salt forms for some molecules. All ligand sets were given to participants in their “raw” format, so they contained the redundant ligands. This very unique feature allows us to evaluate the computational reproducibility in the participants’ submissions and compare it to the experimental variability.
METHODS
CSAR – Phase 1
Some of the donated crystal structures were not used in Phase 1 because they were held in reserve for the second phase of the exercise. In Phase 1, we used structures of 14 ligands bound to TrmD (gtc000445-gtc00448, gtc000451-gtc000453, gtc000456-gtc000460, gtc000464, and gtc000465), five ligands bound to SYK (gtc000224, gtc000225, gtc000233, gtc000249, and gtc000250), and three ligands bound to FXa (gtc000101, gtc000398, and gtc000401). All sets of electron density from GSK were examined by CSAR colleagues, and all of these crystal structures satisfied the criteria for high-quality (HiQ) crystal structures as defined in the release of the 2012 CSAR data set.28
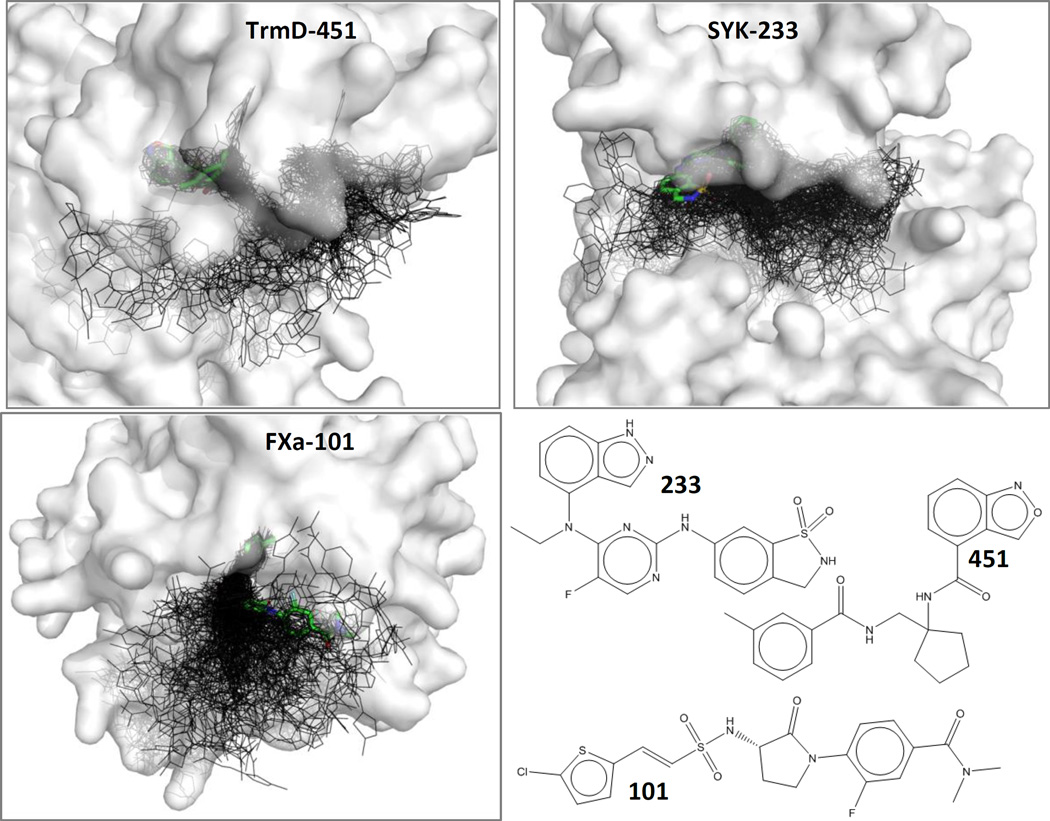
Those structures were used to generate a near-native pose and 199 decoys for each system. Each crystal structure was set up for docking and scoring using MOE 2011.10 (force field: MMFF94x with AM1-BCC charges for the ligands)53. To set up the protein, hydrogens were added in MOE 2011.10. The ligands and near-by (≤6Å) asparagines, glutamines, and histidines were inspected to select the appropriate tautomer and/or charge state. A few residues far from the binding sites had missing sidechains that were added with the “Mutate” option within the sequence editor of MOE. Any breaks in the chains were capped with ACE or NME residues. All added caps, sidechains, and hydrogens were minimized using MOE’s default minimization parameters and the mmff94x forcefield. Each ligand was removed from each structure, and 500 docked poses were generated with DOCK (version 6.5)54. A subset of 200 poses was chosen for each protein-ligand pair (a self-docking scheme). One of the poses was <1 Å RMSD of the actual crystal structure pose, and the other 199 decoys were chosen systematically, guided by diversity analysis in MOE. All of the decoy poses were >2 Å RMSD of the crystal pose. Participants were asked to score/rank the 200 poses for each system to test their method’s ability to identify a near-native pose from a set of decoys. Figure 1 shows a representative distribution of decoys for each system.
Figure 1.
Examples are given for TrmD, SYK, and FXa, showing the near-native poses (thick sticks with green carbons) among each set of 199 decoys (black lines). Protein surfaces are shown in white and are partially transparent. Ligands are labeled with a short-hand notation above; the complexes are TrmD-gtc000451, SYK-gtc000233, and FXa-gtc000101. These three ligands have the most favorable binding affinity, out of the ligands that have an available crystal structure.
For each protein-ligand pair in the 22 structures, the set-up protein was provided in a “.mol2” file, and the 200 ligand poses were provided in a “multi .mol2” file. It should be noted that participants were cautioned that any setup introduces some unavoidable bias toward the chosen force field. Participants were encouraged to modify the decoys to remove bias if necessary. It is likely that some participants minimized the hydrogens for each protein (no heavy atoms were moved from the crystal coordinates in our setup). Some may have also chosen to minimize each ligand pose to its closest local minimum under their force field.
CSAR – Phase 2
Though crystal structures were not available for all of the ligands in Phase 2, affinity data was known. There were 31 unique ligands for TrmD and 267 unique ligands for SYK that were provided to the participants for docking and ranking/scoring. A small number of redundancies (different salt forms) lead to a total of 276 affinity measurements for SYK. There were 106 unique ligands for FXa. Of the 106 ligands, 55 had binding affinities available from two different assays, and one ligand had affinity available in three different assays for a total of 163 independent binding affinity values. When multiple affinity measurements were available for a ligand, our analysis used the median value when comparing the submitted scores/ranks to affinities.
In order to provide a thorough assessment of docking and scoring functions, the ligands need to possess a wide range of affinities, preferably spanning at least 3 log units. For TrmD, the minimum pIC50 is 3.5 and the maximum is 8.3 log units. For SYK, the range spans from 5.2 to 8.9 log units, and for FXa, the span is 4.9 to 9.2 log units. The distribution of pIC50 values can be seen Figure S1 of the Supplemental Information. TrmD ligands had lower affinities (p < 0.0001 two-tailed Wilcoxon rank-sum), as would be expected for a fragment-based set of ligands. The median pIC50 for TrmD was 5.7, as opposed to 7.5 and 7.3 for SYK and FXa, respectively. Figure S1 also shows the distribution of molecular weight, number of rotatable bonds, SlogP, and the number of oxygens and nitrogens as an estimate of hydrogen-bonding capabilities. These physiochemical properties of the ligands were calculated using MOE2013.0855. All experimental data and methods are provided in the Supplemental Information as well as the CSARdock.org website. The crystal structures and structure factors from GSK have been deposited in the PDB, and the ID codes for each complex are given in the Supplemental Information.
Participants were asked to dock and score each ligand with the method(s) of their choice, given the SMILES strings for the ligands and crystal structure coordinates for the target with congeneric ligands bound in the active site. Ideally, the participant would test multiple methods to systematically identify improvements for their methods. Participants provided scores/ranks and docked poses for each ligand. Babel56 was used to convert all submitted coordinate files into a consistent format. Crystal structures were not available for every ligand, but when they were, the submitted poses for ligands were compared to the crystallographic pose using symmetry-corrected RMSD. This value was calculated by an SVL script implemented using MOE. The script was generously provided by the Chemical Computing Group (CCG). To compare the submitted scores/ranks to the measured binding affinities, the R-squared, Pearson (r), Spearman (ρ), and Kendall (τ) were calculated using JMP Pro 1057. The 95th percent confidence intervals were calculated for the Pearson (r) and Spearman (ρ) correlations using the Fisher transform58, while the 95th percent confidence interval for the Kendall (τ) coefficient59 is approximated by τ + 95% = τ + σ*1.96 and τ - 95% = τ - σ*1.96, where
It should be noted that some methods used a negative score for the most favorable pose, and others used positive scores. It was necessary to translate all the participants’ scores/ranks into a “common frame of reference.” The maximum and minimum scores were translated to a scale of 0.0 to 1.0, with 0.0 as the most favorable score. This translation was done in conference with participants to ensure the conversions were correct and properly represented the participants’ interpretation of their scores.
Programs used by participants in each phase of the CSAR 2014 Exercise
It should be noted that some of the participants used different methods in Phase 1 and Phase 2. Because of this, it is confusing to discuss the results based on each participant; instead, it is clearer to discuss the results based on the method used in each phase independently. The convention for labeling groups is simply based on the order in which the participants submitted their predictions, and the labels for the submissions are different for the two phases. In Phase 1, submissions from participants are noted by numbers, p1-p30 (there is no group p3 because of a submission error). In Phase 2, participating groups are named by letters, A-Y. If a group submitted more than one set of results to compare multiple approaches, they were labeled with numbered extensions (e.g., p12-1 and p12-2 or B-1 and B-2). Some groups chose not to disclose their general approach or specific methods.
The submissions for Phase 1 consisted of ranks or scores for the sets of 200 docked poses provided for each ligand complex. Submissions were evaluated based on identifying the near-native pose with the top score and within the top-3 scores. The methods used for submissions to Phase 1 and their results are given in Table 1. A few of the methods are described here because they are too detailed to give sufficient descriptions in the table. Group p20 used a hybrid, empirical scoring function with a Lennard-Jones potential computed from AMBER, a desolvation term based on atomic polarities derived from the molecule’s AlogP, a hydrogen-bond energy term adapted from SLICK, and penalty terms to account for atomic clashes and constraints.60 Table 1 notes when groups used the Vina scoring function within the Autodock12 software or used Vina in the SMINA6,13 program. Group p26 used a hybrid scoring function with terms from both Autodock61 and Autodock-Vina12 with coefficients fit using data from PDBBind62. Group p9 utilized SZMAP63 (p9-1 and p9-8), MMPBSA from Szybki64 with varying parameters (p9-2 through p9-7), and Chemgauss465 from FRED66 with or without rigid ligand optimization (p9–10 and p9-9, respectively). Group p2 used a regression of boosted decision tree models for scoring, based on atomic contacts.67–69
Table 1.
Results for Phase 1 are given for all 52 methods submitted by the 29 participating research groups. The columns note the number of structures for which each method was able to identify the near-native pose as the top-scoring pose and in the top-3 poses. Bold numbers in the last two columns highlight the methods that were able to score at least 20 of the structures correctly. The list of Phase 1 participants is ordered by the total number of structures where the near-native pose is their top score, but the IDs in the first columns simply reflects the order that submissions were received.
| Participants IDa |
TrmD (14 Structures) |
SYK (5 Structures) |
FXa (3 Structures) |
Composite (All 22) |
Pose-Scoring Method The order of this list is not intended to declare one method better than another. More structures are needed for a statistically significant assessment. |
||||
|---|---|---|---|---|---|---|---|---|---|
| Top Score |
Top 3 | Top Score |
Top 3 | Top Score |
Top 3 | Top Score |
Top 3 | ||
| p5 | 14 | 14 | 5 | 5 | 3 | 3 | 22 | 22 | Autodock-Vina12 implemented in SMINA6,13 |
| p6 | 14 | 14 | 5 | 5 | 3 | 3 | 22 | 22 | GoldScore5 (TrmD, Syk), RMSD-based function (FXa)78 |
| p7 | 14 | 14 | 5 | 5 | 3 | 3 | 22 | 22 | PELE7 + GB solvation energy83 + ligand-strain term + conformational entropy term84 |
| p14 | 14 | 14 | 5 | 5 | 3 | 3 | 22 | 22 | GBVI/WSA (MOE)91 / ChemPLP3 |
| p17 | 14 | 14 | 5 | 5 | 3 | 3 | 22 | 22 | SIE4,77 |
| p19 | 14 | 14 | 5 | 5 | 3 | 3 | 22 | 22 | Rosetta Talaris201392,93 |
| p20 | 14 | 14 | 5 | 5 | 3 | 3 | 22 | 22 | Customized empirical scoring60 |
| p8 | 13 | 14 | 5 | 5 | 3 | 3 | 21 | 22 | MedusaScore75,79 |
| p12-2 | 13 | 14 | 5 | 5 | 3 | 3 | 21 | 22 | Unknownb |
| p22 | 13 | 14 | 5 | 5 | 3 | 3 | 21 | 22 | Glide-XP2 |
| p26 | 14 | 14 | 5 | 5 | 2 | 3 | 21 | 22 | Hybrid Autodock61/Autodock-Vina12 |
| p27-4 | 14 | 14 | 5 | 5 | 2 | 3 | 21 | 22 | Autodock-Vina12,81 |
| p9-9 | 13 | 13 | 5 | 5 | 3 | 3 | 21 | 21 | Chemgauss465/FRED66 |
| p9-10 | 13 | 13 | 5 | 5 | 3 | 3 | 21 | 21 | Chemgauss4/FRED65 with rigid optimization of ligand |
| p4 | 12 | 13 | 5 | 5 | 3 | 3 | 20 | 21 | Machine Learning scoring model MARS94 |
| p27-2 | 13 | 14 | 4 | 4 | 3 | 3 | 20 | 21 | ITScore1,10 (refit to refined PDBbind 2012)81 |
| p13 | 12 | 12 | 5 | 5 | 3 | 3 | 20 | 20 | Unknownb |
| p30-2 | 14 | — | 3 | — | 3 | — | 20 | — | HYDE14 + visual inspection (lab provided only top poses) |
| p27-3 | 11 | 14 | 5 | 5 | 3 | 3 | 19 | 22 | ITScore1,10 (modified)81 |
| p9-8 | 10 | 14 | 5 | 5 | 3 | 3 | 18 | 22 | SZMAP63 (formal charge/element/bonding for atom typing) |
| p28 | 10 | 14 | 5 | 5 | 3 | 3 | 18 | 22 | SPA95 |
| p21 | 8 | 14 | 5 | 5 | 3 | 3 | 16 | 22 | ITScore1,10 (plus flexible ligand term)11 |
| p9-1 | 9 | 13 | 4 | 5 | 3 | 3 | 16 | 21 | SZMAP63 (AM1BCC charge for atom typing) |
| p29 | 8 | 12 | 5 | 5 | 3 | 3 | 16 | 20 | Consensus80 (GalaxyDock96, X-Score74, DrugScore73) |
| p15-1 | 11 | 12 | 2 | 3 | 3 | 3 | 16 | 18 | ROCS15 shape similarity (best score)16 |
| p25-5 | 9 | 12 | 4 | 4 | 2 | 3 | 15 | 19 | Pharmacophore-based scoring with LigandScout97 |
| p25-1 | 6 | 12 | 5 | 5 | 3 | 3 | 14 | 20 | DSX-DrugScore73 |
| p30-1 | 12 | 13 | 2 | 3 | 0 | 1 | 14 | 17 | HYDE14 |
| p27-1 | 7 | 10 | 4 | 4 | 2 | 3 | 13 | 17 | ITScore1,10,81 |
| p9-3 | 10 | 13 | 2 | 4 | 0 | 1 | 12 | 18 | MMPBSA/Szybki64 (intRlx options) |
| p9-4 | 10 | 13 | 2 | 3 | 0 | 1 | 12 | 17 | MMPBSA/Szybki64 (intRst options) |
| p9-7 | 10 | 11 | 2 | 5 | 0 | 1 | 12 | 17 | MMPBSA/Szybki64 (totRst options) |
| p9-6 | 10 | 11 | 2 | 4 | 0 | 1 | 12 | 16 | MMPBSA/Szybki64 (totRlx options) |
| p23-1 | 5 | 7 | 4 | 5 | 3 | 3 | 12 | 15 | FLM98,99 |
| p9-5 | 7 | 11 | 3 | 4 | 1 | 1 | 11 | 16 | MMPBSA/Szybki64 (totInp options) |
| p9-2 | 7 | 12 | 2 | 4 | 1 | 1 | 10 | 17 | MMPBSA/Szybki64 (intInp options) |
| p25-3 | 6 | 7 | 0 | 0 | 2 | 2 | 8 | 9 | ChemPLP3 |
| p18-2 | 0 | 0 | 5 | 5 | 3 | 3 | 8 | 8 | Rank from Glide-SP100,101 + Rank from Random Forest model102 |
| p18-3 | 0 | 0 | 4 | 5 | 2 | 3 | 6 | 8 | Score from Glide-SP100,101 + Score Random Forest model102 |
| p18-4 | 0 | 0 | 4 | 4 | 2 | 3 | 6 | 7 | Glide-SP100,101 |
| p16 | 5 | 7 | 0 | 3 | 0 | 3 | 5 | 13 | Autodock-Vina12 + occupational probability score |
| p25-4 | 2 | 3 | 2 | 4 | 1 | 2 | 5 | 9 | ChemPLP3 (only non-bonded) |
| p18-1 | 0 | 0 | 3 | 4 | 2 | 2 | 5 | 6 | QSAR (Random Forest)102 |
| p15-2 | 3 | 7 | 0 | 0 | 1 | 2 | 4 | 9 | ROCS15 shape similarity (average score)16 |
| p24 | 0 | 1 | 2 | 3 | 2 | 2 | 4 | 6 | Unknownb |
| p2 | 0 | 1 | 0 | 1 | 3 | 3 | 3 | 5 | Boost Decision Tree scoring model67–69 |
| p1 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 3 | Scoring functions from Agostino et. al103 |
| p23-2 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | SOL98,99 |
| p10 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 3 | Unknownb |
| p11 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | Unknown b |
| p12-1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Unknownb |
| p25-2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Elekit104 |
Dashed numbers denote alternate methods from the same group. Many participants submitted more than one set of predictions to compare different approaches and parameters.
Some participants were unable to provide details about their methods.
In Phase 2, the groups had to dock and rank/score a set of small molecules to the three proteins, given only the SMILES string of the ligand and at least one protein structure with a congeneric ligand bound. The docking and scoring/ranking methods used by the participants are given in Table 2. A few points are noted here for clarity. Group R used a combination of SHAFTS11,70 to align ligand conformations and MDock1 to place the ligands in the protein structure. Group M clustered the available ligand structures from the PDB and our crystal structures from Phase 1 to develop pharmacophore models for each structure. Ligands were then matched to the pharmacophore model using Pharmer.71,72 Group I used a consensus score incorporating DSX-DrugScore73, X-score74, MedusaScore75, and ChemPLP53. Table 2 notes when people used the Vina scoring function from Autodock-Vina12 or from SMINA.6 Group T used a hybrid scoring function with terms from both Autodock61 and Autodock-Vina12 with coefficients fit using data from PDBBind62 (same as p26 in Phase 1). Group X used a variety of scoring methods to rank the docked compounds. In method X-1, the Vina12 scoring function was used and manual inspection of the score was done based on known crystallographic information. Methods X-2 through X-5 were all scored with the Vina scoring function using different choices for parameters. Methods X-6 and X-7 used a ligand-based method with either a K-nearest neighbor cluster to compare to known binding ligands (X-6) or a support vector machine to compare to known binders (X-7).13
Table 2.
Docking and scoring results for Phase 2 were evaluated by Spearman ρ for relative ranking across all the ligands for a target and by the median, symmetry-corrected RMSD for the top-pose ligands (based on all ligands with crystal structures available). Numbers in bold denote good performance (median RMSD ≤ 2.0Å or ρ ≥ 0.5). For docking, methods with all median RMSD ≤ 2Å are listed in bold with blue background, and italics with green backgrounds mark those with all median RMSD ≤ 3Å. For ranking, methods with at least one ρ ≥ 0.5 are listed in italics with green background, and those in bold with blue background have a second system with ρ ≥ 0.4. The statistical significance is discussed at length in the text.
| IDa | TrmD | SYK | FXa | Evaluationb | Docking Method |
Scoring/Ranking Method (More than one scoring method can be applied to the same docked poses) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Median RMSD |
ρ | Median RMSD |
ρ | Median RMSD |
ρ | Dock | Rank | |||
| R | 1.56 | 0.59 | 2.00 | 0.52 | 1.62 | −0.15 | 9 | 4 |
SHAFTS70 + MDock1 |
ITScore1,10(plus flexible ligand term)11 |
| B-1 | 0.95 | 0.47 | 0.55 | 0.60 | 1.43 | −0.13 | 9 | 3 | Glide-XP2 | XP DockScore2,16 |
| B-2 | 0.95 | 0.34 | 0.55 | 0.78 | 1.43 | 0.00 | repeat | Bl2 | Repeat of B-1 | XP DockScore2+ ROCS15shape similarity16 |
| S-2 | 0.90 | 0.64 | 0.50 | 0.28 | 1.62 | 0.15 | 9 | 2 | Glide-XP2 | MMGBSA2,100,101 |
| I | 1.08 | 0.64 | 0.81 | 0.25 | 1.81 | −0.13 | 9 | 2 | PLANTS3 |
Consensus (MedusaScore75X-score74DSX- DrugScore73+ ChemPLP3) |
| G-2 | 1.21 | 0.51 | 1.25 | 0.09 | 1.32 | 0.15 | 9 | 2 | Wilma4 | SIE-FISH77,105 |
| C | 1.34 | 0.47 | 1.59 | 0.30 | 1.73 | −0.14 | 9 | 1 | Gold5 | GoldScore5,78 |
| S-1 | 0.90 | 0.20 | 0.50 | 0.38 | 1.62 | −0.05 | repeat | 0 | Repeat of S-2 | XP DockScore2 |
| X-5 | 1.32 | 0.29 | 0.95 | 0.27 | 1.74 | 0.34 | 9 | 0 |
SMINA6– Any Xtalc |
Autodock-Vina12,13 |
| J cross- dock |
1.22 | 0.36 | 1.73 | 0.08 | 1.77 | 0.12 | 9 | 0 |
Glide XP2/PELE7 |
PELE7 + GB solvation energy83 + ligand-strain term + conformational entropy term84 |
| G-1 | 1.21 | 0.06 | 0.61 | 0.10 | 1.31 | 0.13 | 9 | 0 | Wilma4 | SIE4,77 |
| V | 1.07 | 0.23 | 1.67 | 0.54 | — | — | 8.5 | 2 | FlexX8 | HYDE14 |
| N | — | — | 1.75 | −0.03 | 1.57 | 0.00 | 8.5 | 0 | MedusaDock9 | Consensus model of Random Forest + SVM106 |
| E | 2.09 | 0.59 | 0.91 | 0.04 | 1.85 | 0.25 | 8 | 2 | Unknownd | Unknownd |
| A-1 | 1.51 | −0.31 | 2.17 | −0.06 | 1.74 | −0.06 | 8 | 0 | SOL98,99 | FLM98,99 |
| A-2 | 1.84 | 0.41 | 2.13 | 0.29 | 2.07 | 0.26 | 7 | 1 | SOL98,99 | SOL98,99 |
| D | 2.22 | 0.61 | 2.23 | 0.12 | 2.15 | 0.19 | 6.5 | 2 | Unknownd | Unknownd |
| K | 2.21 | 0.18 | 1.16 | 0.31 | 2.75 | 0.11 | 6.5 | 0 | IMG Dock107 | Customized empirical60 |
| X-1 | 1.23 | 0.41 | 0.78 | 0.63 | 4.77 | 0.25e | 6 | 3 | Built in by hand g – Most Sim f |
Autodock-Vina12+ visual inspection13 |
| X-2 | 1.23 | 0.41 | 0.78 | 0.63 | 4.77 | 0.27e | repeat | 3 | Repeat of X-1 | Autodock-Vina12,13 |
| H | 1.10 | 0.51 | 1.89 | 0.40 | 6.65 | 0.13e | 6 | 3 | MedusaDock9 | MedusaScore75,79 |
| X-4 | 1.32 | 0.54 | 4.46 | 0.07e | 1.74 | 0.36 | 6 | 2 | SMINA6 – Most Sim f |
Autodock-Vina12,13 |
| X-8 | 1.23 | −0.18 | 0.78 | 0.32 | 4.77 | 0.23e | repeat | 0 | Repeat of X-1 | Autodock-Vina12 divided by size13 |
| X-3 | 1.10 | 0.18 | 1.07 | 0.04 | 4.69 | 0.28e | 6 | 0 | Built in by hand g – Any Xtal c |
Autodock-Vina12,13 |
| U-3 | 1.54 | 0.38 | 7.99 | 0.40e | 1.88 | 0.16 | 6 | 0 | MDock1 | ITScore1,10 (refit with Phase 1 complexes added to the refined PDBbind 2012 training data)81 |
| Q | 10.34 | 0.52e | 1.57 | 0.23 | 1.67 | −0.02 | 6 | 0 | Autodock-Vina12 | ConvexPL76 |
| U-4 | 1.61 | 0.58 | 6.27 | 0.40e | 2.22 | 0.15 | 5 | 2 | Autodock-Vina12 | Autodock-Vina12,81 |
| U-1 | 1.42 | 0.67 | 9.20 | 0.31e | 2.29 | −0.04 | 5 | 2 | MDock1 | ITScore1,10,81 |
| U-2 | 1.41 | 0.16 | 3.73 | 0.54e | 2.27 | 0.11 | 5 | 0 | MDock1 | ITScore1,10 (refit to refined PDBbind 2012)81 |
| F | 2.22 | 0.61 | 2.61 | 0.08 | 2.50 | 0.32 | 4.5 | 2 | Unknownd | Unknownd |
| L | 1.95 | 0.60 | 6.02 | −0.01e | 2.72 | 0.20 | 4 | 2 | Autodock-Vina12 | Autodock-Vina12 |
| P | 1.71 | 0.39 | 3.48 | 0.18e | 7.98 | 0.20e | 3 | 0 | Rosetta Ligand92,93 |
Rosetta talaris201392,93 |
| W | — | — | 2.41 | −0.47 | — | — | 3 | 0 | Gold5 | Chemscore (kinase parameters)108,109 |
| T | 2.61 | 0.25 | 4.12 | −0.13e | 2.55 | −0.08 | 2 | 0 | Autodock61 | Hybrid Autodock61/Autodock-Vina12 |
| M cross- dock |
2.73 | 0.13 | 3.83 | −0.17e | 6.03 | 0.07e | 1 | 0 | Pharmer71 | Autodock-Vina12 (in SMINA6) + fit to receptor pharmacophore model72 |
| M-2 | — | — | 3.83 | 0.21e | — | — | repeat | 0 | Repeat of M-1 | Fit to receptor pharmacophore model72 |
| Y cross- dock |
6.14 | 0.46e | 5.73 | 0.13e | 8.92 | −0.06e | 0 | 0 | SPA Dock95 | SPA95 |
| O | — | 0.37 | — | 0.23 | — | −0.19 | — | 0 | Not applicable | QSAR (unknown designd) |
| X-7 | — | 0.05 | — | 0.05 | — | 0.29 | — | 0 | Not applicable | Ligand-based (Support Vector Machine)13 |
| X-6 | — | −0.03 | — | −0.06 | — | 0.19 | — | 0 | Not applicable | Ligand-based (K-nearest neighbors)13 |
| MW | 0.39 | 0.21 | 0.12 | Null model for ranking18 | ||||||
| SlogP | 0.44 | −0.10 | 0.13 | Null model for ranking18 | ||||||
Dashed numbers denote alternate methods from the same group. Many participants submitted more than one set of predictions to compare different approaches and parameters.
The overall evaluation of docking was calculated by adding points for each median RMSD across all three targets: 3 points for ≤2Å, 2 points for ≤2.4Å, 1 point for ≤3Å, or 0 for >3Å. The overall evaluation of ranking was calculated by adding points for each ρ across all three targets: 2 points for ≥0.5, 1 point for ≥0.4, or 0 for <0.4. Adjustments were made 1) for participants who submitted results for only two systems (groups V and N are not penalized), 2) for methods where docking was less than optimal for all three targets but none had RMSD >3Å (groups D, K, and F), and 3) for good rankings when the median RMSD was >3Å (no points for group U on SYK, no points for groups Q and Y for TrmD).
The crystal structure used for docking each ligand was randomly chosen from among those in the set provided to the applicants (compare to f below).
Some participants were unable to provide details about their methods.
ρ values are italicized when coupled with a median RMSD > 3Å. In these cases, ρ values are suspect.
Each ligand in the set was specifically docked to the receptor of the crystal structure with the most similar ligand bound.
The docked pose was built into the binding site using substructure alignment and minimization in SMINA(Vina).
Many groups gave basic descriptions of their methods, but an in-depth, comprehensive discussion of all 50+ individual methods is difficult because we do not have the full details for each submission. For more information about the various methods, the reader is encouraged to read the manuscripts that the participants have submitted to this special issue. Their papers properly describe the unique features of their methods and what they have learned from the CSAR 2014 exercise.11,13,16,72,76–82
RESULTS AND DISCUSSION
Here, we provide a general overview of the performance of the docking and scoring methods, based on the three systems used in the exercise. We focus on points where most programs appear to be having success or difficulties as a whole. Overall, no single class of methodologies appears to perform consistently better or worse on the protein targets provided. Furthermore, some submissions from different participants had different outcomes despite using the same methods.
Phase 1. Identify the near-native pose within a set of docking decoys
A total of 22 crystal structures from GSK were chosen for Phase 1: 14 for TrmD, 5 for SYK, and 3 for FXa. Broad participation resulted in 29 groups submitting scores/ranks using 52 different methods. Each of the methods was analyzed independently. Figure 2 and Table 1 give an overview of the results for Phase 1.
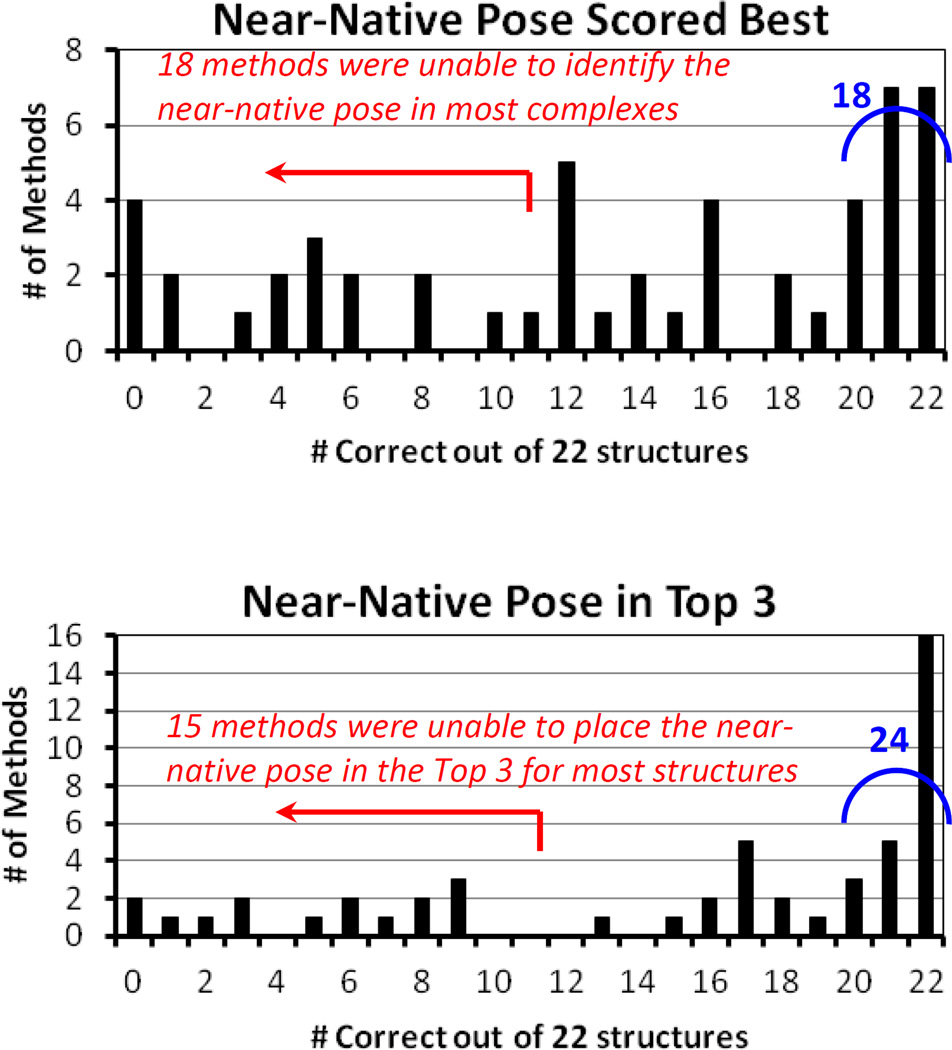
Figure 2.
Histograms of the results of Phase 1 of the 2014 CSAR Exercise. A total of 22 crystal structures were used, and 52 methods were submitted. Participants were given 199 decoys and one near-native pose for each structure. The histograms show how many methods predicted the near-native pose with their top score and within the top-3 scores across all the structures.
Assessment of top pose and top-3 poses
Analyzing the top-scoring poses from each participant show that 18 of the 52 submissions (35%) had excellent performance. Seven of the 18 methods correctly identified the near-native pose with the top score across all 22 complexes, and the other 11 methods missed only one or two complexes. If we look among each participant’s top-3 scored poses, 24 submissions (46%) had good performance. Of those, 16 identify all 22 of the near-native poses, and eight miss only one or two complexes. The list of successful methods includes basically every type of scoring approach used: empirical, knowledge-based, machine-learning, shape-fitting, and even those with advanced electrostatics methods for solvation. The slightly higher number of empirical scoring functions simply reflects the fact that more were submitted than any other type of scoring.
Unfortunately, almost all of these methodological approaches also appear in the lowest performing submissions, with the exception of knowledge-based scoring
For our top-score analysis, 18 other submissions performed poorly and failed to identify the near-native pose for half or more of the complexes. Though expanding our analysis to the top-3 poses increased the number of successful methods, there was little change in the poorly performing methods. Only 3 methods were improved enough to move out of this lowest category. Analysis of the poor methods was significantly hindered because several groups did not provide their computational details. There were several reasons: details were lost when a co-worker had moved on from the lab, one method was unpublished and still in development, others had proprietary reasons, etc. However, the correlation between poor performance and unavailable details is notable.
Docking decoys and other possible reasons for difficulties in Phase 1
Many participants were comparing two or more methods, so some techniques were expected to have poor performance. This may be one reason that 15 methods (29%) did not capture 12 or more of the near-native poses within their top-3 choices. Another issue may be the use of pre-generated docked poses. The choice to use docking decoys had overwhelming grass-roots support in the community. In fact, participants in our previous exercises requested that we separate the “docking problem” from the “scoring problem” by giving everyone the same poses in CSAR 2014. While this seems reasonable, it introduces an inherent bias. Though the poses looked appropriate and performed well for many submissions, it is always possible that poor performance simply identifies when a method is incompatible with our setup protocol. In later sections, we show that success rates were higher for docking in Phase 2 where participants set up the proteins and generated the protein-ligand poses, consistent with their methodology.
If we break down the results by protein target, scoring the near-native poses was most tractable for FXa; 27 submissions (52%) correctly predicted all three FXa structures with their top-scoring pose and 34 methods (66%) placed the near-native pose in the top 3 for all FXa structures. For SYK, 23 submissions (46%) correctly identified the near-native pose with the top score in all five structures used in Phase 1, and 27 methods (54%) identified them all in the top 3. However, the TrmD series appears to have been the most challenging in Phase 1. Only 10 methods (19%) identified the near-native pose with the top score for all 14 TrmD structures used in Phase 1, and only 18 methods (35%) placed the near-native in the top 3 for all structures.
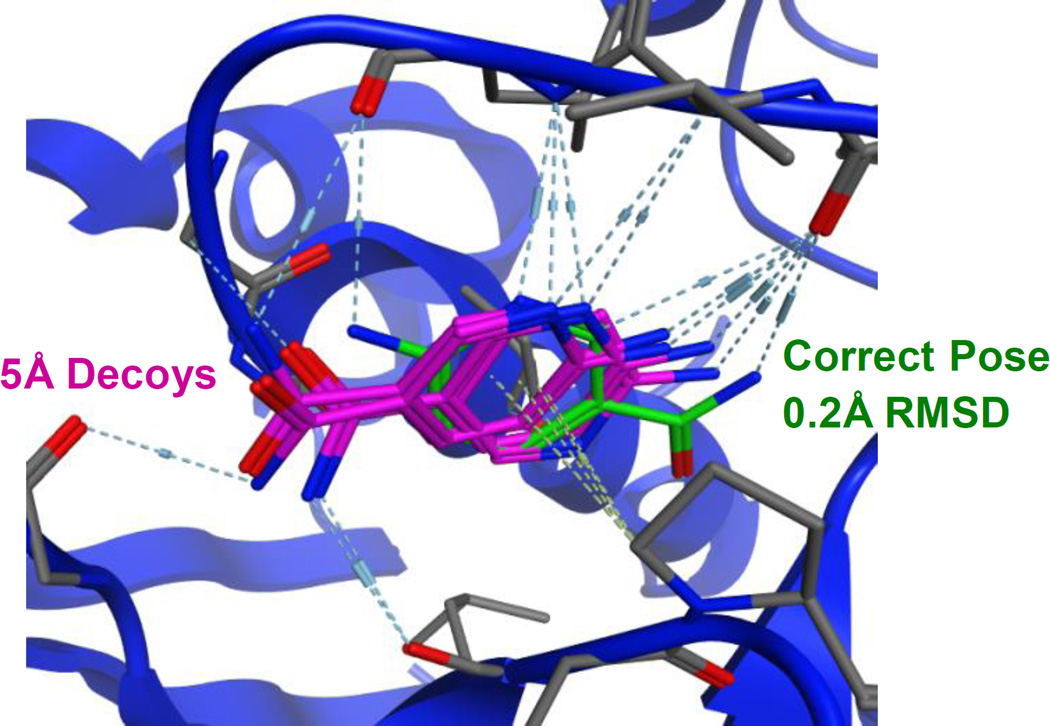
We propose two reasons why TrmD decoys may be more frequently misscored. First, it may simply reflect the fact that getting the right answer 14 times for TrmD is harder than doing it three or five times for FXa and SYK. The second factor clearly comes from the decoy poses. It appears there is a second local minimum for poses at ~5Å RMSD. This same set of poses was consistently identified across many methods. Figure 3 shows those poses for ligand gtc000445. The difficulty many methods faced was mis-ranking this local minimum as more favorable than the near-native pose, which is the true global minimum. We stress that the structures used in Phase 1 were those with the most pristine electron density in their binding sites. The B-factors are very low. The crystallographic position and orientation of the ligand is certain, at least at the low temperatures used in protein crystallography. At room temperature, it is likely that this second minimum is occasionally occupied, but it should still be less favorable.
Figure 3.
The poses that comprise a second, local minimum for gtc000445 are shown. The decoys (colored purple) are 5Å RMSD from the crystal pose, but they have significant overlap with the correct, near-native pose (colored green). The decoys are flipped over backwards with many favorable hydrogen bonds that lead to good scores.
Phase 2. Dock and score a congeneric series, starting from a PDB structure and a list of SMILES
Of the 52 methods from Phase 1, 29 were also submitted to Phase 2. This included 16 of the blue, successful methods in Figure 2’s Top-3 results, but also 9 from the red “poor performance” category. In Phase 2, some participants submitted one set of docked poses with multiple sets of relative rankings calculated with different scoring functions. For these participants, the docking data gives one RMSD measure with multiple Spearman ρ for the different rankings from each scoring function. These were counted once in our assessment of docking, but all unique ρ were included in the ranking assessment. A total of 25 groups provided results, based on 32 docking methods and 40 scoring/ranking methods.
Processing the submissions
The congeneric series of ligands were given in SMILES strings, along with the 22 crystal structure “answers” from Phase 1 (plain PDB format of the ligand-bound complexes). Participants were told to setup the protein and small molecules as needed for best performance of their method. Submissions included a file of ranks/scores for ligands and the top poses for each small molecule docked in the target. For our docking assessment, we evaluated the poses based on symmetry-corrected RMSD after a weighted overlay of the protein backbones. New crystal structures were reserved to evaluate blinded cross-docking in Phase 2 (17 for TrmD, 3 for SYK, and 2 for FXa). However, submissions also included some unblinded self- and cross-docking poses for the ligands of the 22 crystal structures given to the participants from Phase 1. The supplemental information presents docking analysis for the blinded structures, the unblinded structures, and the full set of both combined. The results are similar across the categories, but the small number of structures makes it difficult to properly assess the statistical significance in the docking results. As such, the results for the largest set of all docked poses for all crystal structures are given in Table 2 as median RMSDs. Three groups chose to cross-dock the whole ligand set against all Phase-1 crystal structures given for each target. For this ensemble-docking, each groups’ results were consistent across all the protein conformations used, and treating each conformation separately as an independent “method” imposes a heavy bias for the ensemble-dockers vs groups that submitted one set of predictions. Therefore, each set of results for each conformation in the ensemble was evaluated like all other submissions, but then, the median of median RMSDs from all conformations were used to provide one, inclusive result for the same approach from the same group based on multiple, but similar, crystal structures.
All affinity data from GSK were reserved for Phase 2, and we used Spearman ρ to evaluate the submitted rankings (median ρ for the ensemble-dockers). For FXa, participants were asked to dock and score each subset of small molecules as a separate dataset, labeling them as set1, set2, or set3. There was some overlap in the small molecules in each set, so participants processed a few molecules two or three times. We analyzed each of the sets separately and together as one large set. For brevity, only the one-large-set analysis is shown and discussed here, but the conclusions are the same for each individual subset. The data in the Supplemental Information gives both the one-large-set result and the individual results for each of the three subsets. The results of Phase 2 and the methods used by the participants are given in Table 2. Scoring and docking across all participants is also summarized in Figure 4, separated into each independent protein system. Histograms of the data in Figure 4 are given in the Supplemental Information.
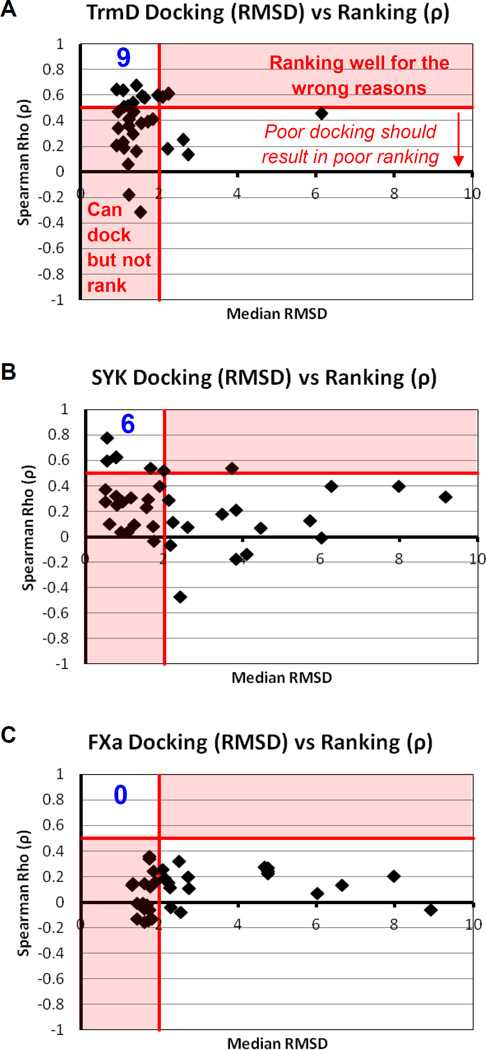
Figure 4.
Comparison of docking and ranking performance for each method submitted for Phase 2. The region in the upper left is the area where the most successful submissions are found. The value in blue is the number of methods with median RMSD ≤ 2 Å and ρ ≥ 0.5. Median ρ are calculated using all the unique ligands for each system. Median RMSD are calculated with the set of all Phase-2 ligands that have crystal structures.
Evaluation of docking in Phase 2
Docking results could only be assessed on the subset of unique ligands that had crystal structures available: 31 complexes for TrmD, 8 for SYK, and 3 for FXa. With the exception of a few outlying methods for each protein system, docking was a success for most participants, and success rates were higher for docking in Phase 2 than in Phase 1 where we provided setup protein-ligand poses. Overall, the top poses produced by the majority of all docking methods had median RMSDs ≤ 2 Å: 22 out of 30 submissions (73%) for TrmD, 17 out of 32 (53%) for SYK, and 15 out of 30 (47%) for FXa. The most difficult task was consistently docking well over all three targets. Only 11 submissions (34%) had median RMSD ≤ 2 Å for all targets examined, see blue and green highlights in Table 2. In particular, submission J (Glide-XP2/PELE7 with scoring based on PELE7 + GB solvation energy83 + ligand-strain + conformational entropy84) deserves special recognition for docking well to all three targets. This group cross-docked all the ligands against all the given crystal structures. To see robust performance across an ensemble of protein structures for multiple targets is an impressive accomplishment.
Our docking criteria are particularly stringent because we are only counting the top poses submitted for each docking method, not the top-3 poses like Phase 1. Also, the submissions are primarily cross-docking results, not self-docking poses. The docking success is even more apparent if we include the large number of structures with median RMSD between 2–3 Å, which are clearly on the right track. For submissions with median RMSD ≤ 3 Å, the success rate is 28 out of 30 methods (93%) for TrmD, 22 out of 32 methods (69%) for SYK, and 24 out of 30 methods (80%) for FXa. Relaxing the cutoff to ≤ 3 Å also increases the number that successfully dock all three targets to 16 of the 32 methods (50%).
We were surprised to find that participants had the most success with docking TrmD in Phase 2, especially given its difficulty in Phase 1. Across the participants, TrmD submissions had much smaller median RMSDs than the SYK and FXa submissions from the same groups. The conformational search space available to the TrmD ligands is restricted because the ligands are smaller and have fewer rotatable bonds (Figure S1), which may explain part of its success in Phase 2. Furthermore, crystal structures were given to participants as starting points for Phase 2, and at least one example for each congeneric series in each protein was provided. These crystal structures could be used to correct any mis-dockings of TrmD (such as the 5 Å minimum seen in Phase 1), which would also contribute to better docking outcomes for TrmD in Phase 2 than in Phase 1. On the contrary, FXa’s ligands were the largest and most flexible (p <0.0001 for comparisons to both TrmD and SYK sets, see the distributions of rotatable bonds in Figure S1). This results in a larger conformational space to sample for the FXa ligands which makes the sampling problem more difficult. This could explain why participants found that docking was the most difficult for FXa.
Evaluation of scoring/ranking in Phase 2: Scoring/rank-ordering the ligands was much more difficult than docking
We used Spearman ρ to calculate the agreement in the rank-order of the submitted scores vs the experimental affinities provided by GSK. These ρ are calculated using the entire set of unique ligands provided to participants for Phase 2: 31 for TrmD, 267 for SYK, and 106 for FXa (median affinities were used for any ligand with multiple affinity measurements). It is important to note that ρ values are suspect when the median RMSD > 3Å. In those cases, methods with ρ values ≥ 0.5 are getting the right answers for the wrong reasons, and any poor ρ values may be due to poorly docked poses, rather than a failure of the ranking method itself.
In Figure 4, the relationship between the docking and scoring results is given for each submission that included both docked poses and ranks. For TrmD, 50% of the submissions appear in the red, lower-left section in Figure 4A, showing that they are able to dock, but not rank, the ligands. There is little data in the public domain for TrmD, which should make this system more challenging for a blinded exercise. Conversely, large amounts of information exist in the public domain for FXa, yet it was the toughest system for participants to dock and score, see Figure 4C. In fact, scoring/ranking ligands for FXa was intractable for all methods.
Of the 37 scoring methods submitted for TrmD, 9 (32%) were successful with ρ ≥ 0.5 (and median RMSD ≤ 2Å). Regarding scoring for TrmD, there is a correlation between affinity and MW (ρ=0.39) for the ligands. Most scoring functions are based on two-body interactions, which are heavily influenced by the number of atoms and contacts. Two TrmD ligands appear to establish the correlation, gtc000449 and gtc000450. Each have a pIC50 of 8.3 while no other ligand has a pIC50 greater than 6.8. These two ligands are also the largest and most flexible of the TrmD ligands. For the ligands of SYK and FXa, there is no meaningful correlation between affinity and MW, which may be why the scoring/ranking for TrmD was more successful than for the other two targets.
Of the 40 scoring methods submitted for SYK, six (15%) were successful. However, only one method had ρ ≥ 0.5 for both TrmD and SYK, and that was group R’s use of MDock1 and ITScore with an added penalty term for reduced ligand flexibility in the bound state.11 Submission B-1’s use of Glide-XP2 and its XP DockScore came very close with ρ = 0.47 for TrmD and ρ = 0.60 for SYK.
What is concerning is that submission S-1 also used Glide-XP and XP DockScore, but the rankings are much poorer (ρ = 0.20 and ρ = 0.38 for TrmD and SYK, respectively). The dockings are very similar for B-1 and S-1, but they are not exactly the same. It is possible the two groups used different crystal structures for their docking. Also, both groups said they used the standard setup protocol, but clearly, some small differences likely exist. Setup is usually considered straightforward, and we trust that colleagues in our scientific community make the best choices for their studies. However, many of these choices are arbitrary, and it is easy to justify changes if difficulties are encountered during a retrospective study. In CSAR 2014, those choices could not be pre-tailored, and this likely explains the varied outcomes also seen for several different submissions based on Vina scoring. In a prospective, blinded exercise, the choices must stand on their own. Setup might be the unseen lynchpin that compromises the efforts to implement and compare methods because each method is inherently wedded to its own setup protocol. In our first exercise in 2010, one group’s changes to system setup improved their correlation to experimental ΔGbind from R2 = 0.188 to 0.375.85 When those same changes were given to all the other participants, it made no difference to their R2 values despite some important corrections to ligand tautomers and protonation states.
Statistical significance and “null models” in Phase 2
It should be noted that the number of data points in the set directly effects the size of the standard deviations (σ) and 95%-confidence intervals (95% CI), with more data leading to smaller σ and tighter confidence intervals. SYK is our largest set with the best statistical significance. The successful methods (ρ ≥ 0.5 and RMSD ≤ 2Å) for SYK were MDock1 using ITScore1,10 + lig flex11 (group R), SMINA6 using Autodock-Vina12,13 (submissions X-1 and X-2), FlexX8 using HYDE14 (group V), and Glide-XP2 using XP DockScore2 with and without ROCS15 shape similarity16 (B-2 and B-1, respectively). All of these methods have no overlap with other methods based on ρ ± σ; furthermore, they were statistically significant in their performance over the nulls based on the 95% CI. The most common null models are based on the correlation between experimental affinities and the ligands’ molecular weights (MW) or calculated SlogP.86 Valid scoring functions should add more value to the predictions and result in better correlation to the affinity data than these simple, physicochemical properties. The nulls are given in Table 2 and in the Supplemental Information. The Supplemental Information also provides parametric and non-parametric measures of ranking: R2, Pearson R, Spearman ρ, and Kendall τ with σ and 95% CI.
It is disappointing that the rankings for TrmD are within error of one another and not significantly different than our null models for ρ.18 All methods have significantly overlapping 95% CIs. Examining ρ ± σ for all methods shows that each is within error of ρ for SlogP vs affinity. The most optimistic assessment we can give is that the highest ρ of 0.67 (MDock1 with ITScore1,10 in submission U-1) has error bars (± σ) that do not overlap with those of the MW null. Overall, these nulls highlight the fundamental problem with small data sets: a valid correlation cannot be identified as statistically significant from a random correlation. With 31 ligands and affinities for TrmD, this set is larger than many used in the literature for training and testing scoring functions, but it is still limited. We have noted before that data sets must have hundreds of data points to distinguish between different scoring methods in a statistically significant way.17,18,52
This is why these large datasets for SYK and FXa are so valuable as a testing and development resource. Unfortunately for FXa, ρ for all methods overlap the nulls’ 95% CIs. This is simply because of the poor predictive performance for the scoring functions. The two methods with the highest ρ use Autodock-Vina12 implemented in SMINA6 (X-4 and X-5), and they have ±σ error bars that do not overlap with any of the nulls’ ±σ error bars. For those submissions, we are confident that they are better than a null approach even though they did not score very well.
Despite the large amount of existing data in the literature for FXa and the frequent use of the system in docking/scoring development, scoring was intractable for this system
Many methods were able to dock the ligands of FXa, but the real difficulty was scoring. The best agreement with experimental affinities for FXa was ρ = 0.36 (submission X-4). It was particularly disappointing that 11 methods produced anti-correlated results for FXa. It should be noted that the submitted raw scores/ranks included definitions of their metric. The anti-correlation is not a misinterpretation of negative numbers for ΔGbind; it is truly a negative correlation to experimental binding affinities.
We have argued before that FXa is not a good test system for SBDD (in our analysis paper from the first exercise in 2010).18 Many PDB structures of FXa have sub-nM ligands bound in the active site, but the pockets are largely solvent-exposed and the complementarity appears poor (Figure 1). This may underscore a more important role for solvation effects or perhaps structural water in scoring/ranking the ligands. However, it is possible that the complexes available are limited by the crystallography. All FXa crystal structures are missing several regulatory domains that must be truncated from FXa to make the crystal form of the protein. It is possible that some of these domains provide parts of the pocket for the inhibitors, effect the electrostatics, or change the conformational behavior of the catalytic domain. All of the domains of FXa are present in the assays for the inhibitors, but some are missing in the crystal structures, so the data could be simply mismatched.
As further support for the proposed mismatched between the affinities and crystal structures for FXa, we should emphasize that the experimental error in the affinity data is minimal and not the limiting factor. The standard deviation for the “standards” used in the FXa assays average 0.1, 0.4, and 0.8 pIC50 for Sets 1–3, respectively (see Supplemental Information). The range of pIC50 for FXa inhibitors is 4.3, which is much larger than any errors from the assays. Understanding the variance in the biological data is a key component to improving docking and scoring.87–90 A model is only as good as the data used for training, and it is bad science to expect higher precision from a computational method than the measured affinities actually warrant.
Reproducibility in Phase 2
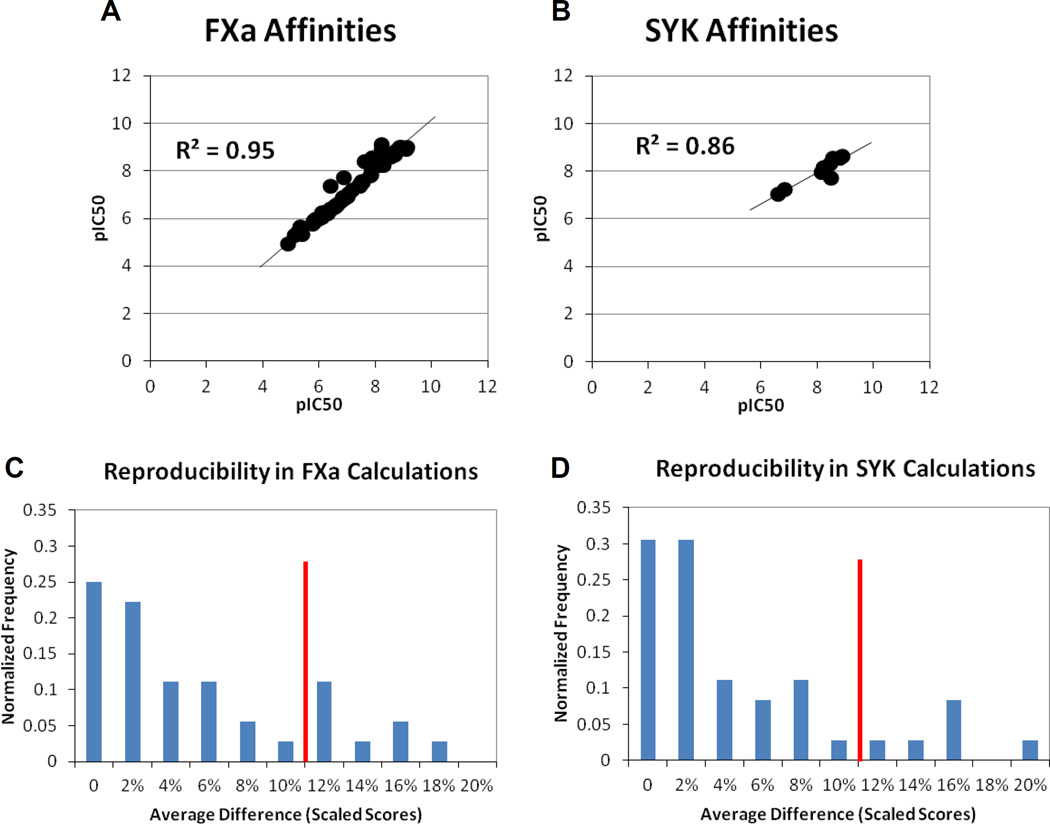
The format of data submissions allowed us to compare computational reproducibility to experimental reproducibility. In the Supplemental Information, the standard deviations of the experiments are discussed with the descriptions of the assays, but that is the reproducibility of the same compound in the same assay. For 58 inhibitors of FXa, the pIC50 has been measured in two or more independent and slightly different assays. This gives us a measure of reproducibility from assay to assay. For SYK, we only have data on one assay, but seven SYK inhibitors have multiple salt forms, which gives us slightly different conditions for those measures too. Figure 5A,B shows that the agreement in the assay data is impressive with R2 of 0.95 and 0.86 for FXa and SYK, respectively. The average difference in the pIC50 measurements is 0.2 for FXa and 0.3 for SYK.
Figure 5.
There is very tight agreement in the IC50 data from different assays for FXa and different salt forms of SYK. (A) Across all the duplicate measurements for FXa, the average unsigned difference is 0.15 pIC50 and the standard deviation is only 0.24 pIC50. The slope is 1. (B) For SYK, the average unsigned difference is 0.27 pIC50 and the standard deviation is 0.21 pIC50. The slope deviates from 1.0, but the smaller range of data makes this less relevant. (C) For calculations of repeat FXa inhibitors, 9 methods produced the exact same scores/ranks for all the inhibitors. Another 20 methods had average differences less than 2σexpt (red line). (D) For calculations of SYK repeats, 11 methods gave the same scores/ranks, and 23 other methods had average differences less than 2σexpt (red line).
This examination of the experiments warrants a similar inspection of the calculated submissions. Did the participants get the same scores for these “repeat” molecules in the data sets? One might expect all methods to produce the exact same value each time, but some methods use random sampling and can produce slightly different poses and scores each time. Of the 37 methods for FXa, 14 had well correlated values for the repeats (R2 ≥ 0.9) and 6 had R2 ≥ 0.8. Of the 40 methods for SYK, 20 had well correlated values for the repeats (R2 ≥ 0.9) and 2 had R2 ≥ 0.8.
For a more detailed comparison, we must convert the experimental standard deviations to relative values as we did for the calculations. Based on the range of affinities, the standard deviation of the FXa experiments is 5.5%, and SYK is 5.6%. For each method, the differences in the values for each repeat compound were calculated, and the unsigned difference for all repeats were averaged, see Figure 5C,D. For the FXa calculations, 9 methods produced the same scores/ranks for all repeats (0% difference). Another 19 methods had average differences under 11% (eg, within twice the standard deviation of the experiments). For the SYK calculations, 11 methods produced the same scores/ranks, and 23 methods had average differences under 11%. Note that it is unreasonable to expect less variation in the calculations than exist in the experiments, so average differences that are within 2σ are acceptable and over 3σ are not. Only a few of the “unknown” methods from Table 2 had average differences larger than 3σ.
CONCLUSIONS
Successful methods used basically every type of scoring approach known: empirical, knowledge-based, machine-learning, shape-fitting, and even those with advanced electrostatics methods for solvation. In Phase 1, 35% of the submitted methods scored the near-native poses with the top score, and 46% placed the near-native pose in the top-3. Scoring the docking decoys was easiest for FXa and hardest for TrmD. In Phase 2, the pattern was reversed. When participants setup their own proteins and submitted docked poses, they had the most success with TrmD and the least with FXa. Methods that were able to dock well include MDock1, Glide-XP2, PLANTS3, Wilma4, Gold5, SMINA6, Glide-XP2/PELE7, FlexX8, and MedusaDock9. In particular, the ensemble-docking results from Group J (Glide-XP2/PELE7 with scoring based on PELE7 + GB solvation energy83 + ligand-strain + conformational entropy84) deserve special recognition for performing well against all three targets. To see robust performance for cross-docking to an ensemble of protein structures for multiple targets is an impressive accomplishment.
The most difficult task was relative ranking in Phase 2. When submitted ranks/scores were compared to experimental data, few methods were able to rank with Spearman ρ ≥ 0.5. Despite FXa’s large dataset and frequent use in method development, scoring was intractable for this system. The most statistically significant results were possible with SYK, and a few stand-out submissions deserve recognition: MDock1 using ITScore1,10 + lig flex11, SMINA6 using Autodock-Vina12,13, FlexX8 using HYDE14, and Glide-XP2 using XP DockScore2 with and without ROCS15 shape similarity16.
The reader can reproduce this benchmark exercise based on the data and PDB list given in the supplemental information.
Acknowledgments
We would like to thank all participants in the benchmark exercise! We would also like to thank Dr. Robert Young at GSK for assistance in reviewing the FXa chemical structures, chemical strategy, and SAR. The CSAR Center is funded by the National Institute of General Medical Sciences (U01 GM086873). We also thank the Chemical Computing Group and OpenEye Scientific Software for generously donating the use of their software. AA would like to thank Michigan Institute for Clinical and Health Research (funded by National Center for Advancing Translational Sciences (NCATS, UL1 TR000433) for the training grant.
Footnotes
SUPPLEMENTAL INFORMATION
The distributions of the ligands’ affinity and key physical properties, starting data given to participants in Phase 1, numbers for the near-native poses in the decoy sets used in Phase 1, starting data provided for Phase 2, evaluation of submitted scores/ranks from Phase 2, histograms of median RMSD and ρ from the submissions to Phase 2, experimental affinity data, PDB codes for each protein-ligand complex, and the description of the experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Huang S-Y, Zou X. Ensemble Docking of Multiple Protein Structures: Considering Protein Structural Variations in Molecular Docking. Proteins Struct. Funct. Bioinforma. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 2.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 3.Korb O, Stützle T, Exner TE. Empirical Scoring Functions for Advanced Protein−Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009;49:84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- 4.Sulea T, Hogues H, Purisima EO. Exhaustive Search and Solvated Interaction Energy (SIE) for Virtual Screening and Affinity Prediction. J. Comput. Aided Mol. Des. 2011;26:617–633. doi: 10.1007/s10822-011-9529-7. [DOI] [PubMed] [Google Scholar]
- 5.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 6.Koes DR, Baumgartner MP, Camacho CJ. Lessons Learned in Empirical Scoring with Smina from the CSAR 2011 Benchmarking Exercise. J. Chem. Inf. Model. 2013;53:1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrelli KW, Vitalis A, Alcantara R, Guallar V. PELE: Protein Energy Landscape Exploration. A Novel Monte Carlo Based Technique. J. Chem. Theory Comput. 2005;1:1304–1311. doi: 10.1021/ct0501811. [DOI] [PubMed] [Google Scholar]
- 8.Rarey M, Kramer B, Lengauer T, Klebe G. A Fast Flexible Docking Method Using an Incremental Construction Algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 9.Ding F, Yin S, Dokholyan NV. Rapid Flexible Docking Using a Stochastic Rotamer Library of Ligands. J. Chem. Inf. Model. 2010;50:1623–1632. doi: 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S-Y, Zou X. An Iterative Knowledge-Based Scoring Function to Predict Protein-ligand Interactions: II. Validation of the Scoring Function. J. Comput. Chem. 2006;27:1876–1882. doi: 10.1002/jcc.20505. [DOI] [PubMed] [Google Scholar]
- 11.Huang S-Y, Li M, Wang J, Pan Y. HybridDock: A Hybrid Protein–Ligand Docking Protocol Integrating Protein- and Ligand-Based Approaches. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00275. [DOI] [PubMed] [Google Scholar]
- 12.Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumgartner MP, Camacho CJ. Choosing the Optimal Rigid Receptor for Docking and Scoring in the CSAR 2013/2014 Experiment. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider N, Lange G, Hindle S, Klein R, Rarey M. A Consistent Description of HYdrogen Bond and DEhydration Energies in Protein-ligand Complexes: Methods behind the HYDE Scoring Function. J. Comput. Aided Mol. Des. 2012;27:15–29. doi: 10.1007/s10822-012-9626-2. [DOI] [PubMed] [Google Scholar]
- 15.ROCS v. 3.2.1. Santa Fe, NM, USA: OpenEye Scientific Software, Inc; 2015. [accessed February 23 2015]. [Google Scholar]
- 16.Kumar A, Zhang KYJ. Application of Shape Similarity in Pose Selection and Virtual Screening in CSARdock2014 Exercise. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00279. [DOI] [PubMed] [Google Scholar]
- 17.Dunbar JB, Smith RD, Yang C-Y, Ung PM-U, Lexa KW, Khazanov NA, Stuckey JA, Wang S, Carlson HA. CSAR Benchmark Exercise of 2010: Selection of the Protein-Ligand Complexes. J. Chem. Inf. Model. 2011;51:2036–2046. doi: 10.1021/ci200082t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith RD, Dunbar JB, Ung PM-U, Esposito EX, Yang C-Y, Wang S, Carlson HA. CSAR Benchmark Exercise of 2010: Combined Evaluation Across All Submitted Scoring Functions. J. Chem. Inf. Model. 2011;51:2115–2131. doi: 10.1021/ci200269q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical Assessment of Methods of Protein Structure Prediction (CASP) — Round X. Proteins Struct. Funct. Bioinforma. 2014;82:1–6. doi: 10.1002/prot.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lensink MF, Wodak SJ. Docking and Scoring Protein Interactions: CAPRI 2009. Proteins Struct. Funct. Bioinforma. 2010;78:3073–3084. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 21.Lensink MF, Wodak SJ. Docking, Scoring, and Affinity Prediction in CAPRI. Proteins Struct. Funct. Bioinforma. 2013;81:2082–2095. doi: 10.1002/prot.24428. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS. Predicting Small-Molecule Solvation Free Energies: An Informal Blind Test for Computational Chemistry. J. Med. Chem. 2008;51:769–779. doi: 10.1021/jm070549+. [DOI] [PubMed] [Google Scholar]
- 23.Skillman AG, Geballe MT, Nicholls A. SAMPL2 Challenge: Prediction of Solvation Energies and Tautomer Ratios. J. Comput. Aided Mol. Des. 2010;24:257–258. doi: 10.1007/s10822-010-9358-0. [DOI] [PubMed] [Google Scholar]
- 24.Skillman AG. SAMPL3: Blinded Prediction of Host-guest Binding Affinities, Hydration Free Energies, and Trypsin Inhibitors. J. Comput. Aided Mol. Des. 2012;26:473–474. doi: 10.1007/s10822-012-9580-z. [DOI] [PubMed] [Google Scholar]
- 25.Guthrie JP. SAMPL4, a Blind Challenge for Computational Solvation Free Energies: The Compounds Considered. J. Comput. Aided Mol. Des. 2014;28:151–168. doi: 10.1007/s10822-014-9738-y. [DOI] [PubMed] [Google Scholar]
- 26.Damm-Ganamet KL, Smith RD, Dunbar JB, Stuckey JA, Carlson HA. CSAR Benchmark Exercise 2011-2012: Evaluation of Results from Docking and Relative Ranking of Blinded Congeneric Series. J. Chem. Inf. Model. 2013;53:1853–1870. doi: 10.1021/ci400025f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith RD, Damm-Ganamet KL, Dunbar JB, Ahmed A, Chinnaswamy K, Delproposto JE, Kubish GM, Tinberg CE, Khare SD, Dou J, Doyle L, Stuckey JA, Baker D, Carlson HA. CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunbar JB, Smith RD, Damm-Ganamet KL, Ahmed A, Esposito EX, Delproposto J, Chinnaswamy K, Kang Y-N, Kubish G, Gestwicki JE, Stuckey JA, Carlson HA. CSAR Data Set Release 2012: Ligands, Affinities, Complexes, and Docking Decoys. J. Chem. Inf. Model. 2013;53:1842–1852. doi: 10.1021/ci4000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White TA, Kell DB. Comparative Genomic Assessment of Novel Broad-Spectrum Targets for Antibacterial Drugs. Comp. Funct. Genomics. 2004;5:304–327. doi: 10.1002/cfg.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AT. Identification of the First Potent Inhibitors of tRNA (m1G37) Methlytransferase (TrmD) via an Integrated Screening Approach; Abstracts, 64th Southeast Regional Meeting of the American Chemical Society; November 14–17, 2012. [Google Scholar]
- 32.McAdoo SP, Tam FWK. Spleen Tyrosine Kinase: A Novel Target in Autoimmunity. In: Kapur S, editor. Immunosuppression - Role in Health and Diseases. InTech: Rijeka, Croatia; 2012. [Google Scholar]
- 33.Scott IC, Scott DL. Spleen Tyrosine Kinase Inhibitors for Rheumatoid Arthritis: Where Are We Now? Drugs. 2014;74:415–422. doi: 10.1007/s40265-014-0193-9. [DOI] [PubMed] [Google Scholar]
- 34.Weinblatt ME, Genovese MC, Ho M, Hollis S, Rosiak-Jedrychowicz K, Kavanaugh A, Millson DS, Leon G, Wang M, Heijde D. Effects of Fostamatinib, an Oral Spleen Tyrosine Kinase Inhibitor, in Rheumatoid Arthritis Patients With an Inadequate Response to Methotrexate: Results From a Phase III, Multicenter, Randomized, Double-Blind, Placebo-Controlled, Parallel-Group Study. Arthritis Rheumatol. 2014;66:3255–3264. doi: 10.1002/art.38851. [DOI] [PubMed] [Google Scholar]
- 35.Friedberg JW, Sharman J, Sweetenham J, Johnston PB, Vose JM, LaCasce A, Schaefer-Cutillo J, Vos SD, Sinha R, Leonard JP, Cripe LD, Gregory SA, Sterba MP, Lowe AM, Levy R, Shipp MA. Inhibition of Syk with Fostamatinib Disodium Has Significant Clinical Activity in Non-Hodgkin Lymphoma and Chronic Lymphocytic Leukemia. Blood. 2010;115:2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.EINSTEIN Investigators. Bauersachs R, Berkowitz SD, Brenner B, Buller HR, Decousus H, Gallus AS, Lensing AW, Misselwitz F, Prins MH, Raskob GE, Segers A, Verhamme P, Wells P, Agnelli G, Bounameaux H, Cohen A, Davidson BL, Piovella F, Schellong S. Oral Rivaroxaban for Symptomatic Venous Thromboembolism. N. Engl. J. Med. 2010;363:2499–2510. doi: 10.1056/NEJMoa1007903. [DOI] [PubMed] [Google Scholar]
- 37.Agnelli G, Buller HR, Cohen A, Curto M, Gallus AS, Johnson M, Masiukiewicz U, Pak R, Thompson J, Raskob GE, Weitz JI. Oral Apixaban for the Treatment of Acute Venous Thromboembolism. N. Engl. J. Med. 2013;369:799–808. doi: 10.1056/NEJMoa1302507. [DOI] [PubMed] [Google Scholar]
- 38.Turpie AGG. New Oral Anticoagulants in Atrial Fibrillation. Eur. Heart J. 2008;29:155–165. doi: 10.1093/eurheartj/ehm575. [DOI] [PubMed] [Google Scholar]
- 39.Abel R, Young T, Farid R, Berne BJ, Friesner RA. Role of the Active-Site Solvent in the Thermodynamics of Factor Xa Ligand Binding. J. Am. Chem. Soc. 2008;130:2817–2831. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson NS, Brown D, Campbell M, Chan C, Chaudry L, Convery MA, Fenwick R, Hamblin JN, Haslam C, Kelly HA, King NP, Kurtis CL, Leach AR, Manchee GR, Mason AM, Mitchell C, Patel C, Patel VK, Senger S, Shah GP, Weston HE, Whitworth C, Young RJ. Design and Synthesis of Orally Active Pyrrolidin-2-One-Based Factor Xa Inhibitors. Bioorg. Med. Chem. Lett. 2006;16:3784–3788. doi: 10.1016/j.bmcl.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 41.Senger S, Convery MA, Chan C, Watson NS. Arylsulfonamides: A Study of the Relationship between Activity and Conformational Preferences for a Series of Factor Xa Inhibitors. Bioorg. Med. Chem. Lett. 2006;16:5731–5735. doi: 10.1016/j.bmcl.2006.08.092. [DOI] [PubMed] [Google Scholar]
- 42.Young RJ, Campbell M, Borthwick AD, Brown D, Burns-Kurtis CL, Chan C, Convery MA, Crowe MC, Dayal S, Diallo H, Kelly HA, Paul King N, Kleanthous S, Mason AM, Mordaunt JE, Patel C, Pateman AJ, Senger S, Shah GP, Smith PW, Watson NS, Weston HE, Zhou P. Structure- and Property-Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Acyclic Alanyl Amides as P4 Motifs. Bioorg. Med. Chem. Lett. 2006;16:5953–5957. doi: 10.1016/j.bmcl.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Chan C, Borthwick AD, Brown D, Burns-Kurtis CL, Campbell M, Chaudry L, Chung C, Convery MA, Hamblin JN, Johnstone L, Kelly HA, Kleanthous S, Patikis A, Patel C, Pateman AJ, Senger S, Shah GP, Toomey JR, Watson NS, Weston HE, Whitworth C, Young RJ, Zhou P. Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}sulfonamides. J. Med. Chem. 2007;50:1546–1557. doi: 10.1021/jm060870c. [DOI] [PubMed] [Google Scholar]
- 44.Young RJ, Brown D, Burns-Kurtis CL, Chan C, Convery MA, Hubbard JA, Kelly HA, Pateman AJ, Patikis A, Senger S, Shah GP, Toomey JR, Watson NS, Zhou P. Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa. Bioorg. Med. Chem. Lett. 2007;17:2927–2930. doi: 10.1016/j.bmcl.2007.03.080. [DOI] [PubMed] [Google Scholar]
- 45.Senger S, Chan C, Convery MA, Hubbard JA, Shah GP, Watson NS, Young RJ. Sulfonamide-Related Conformational Effects and Their Importance in Structure-Based Design. Bioorg. Med. Chem. Lett. 2007;17:2931–2934. doi: 10.1016/j.bmcl.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 46.Young RJ, Borthwick AD, Brown D, Burns-Kurtis CL, Campbell M, Chan C, Charbaut M, Chung C, Convery MA, Kelly HA, Paul King N, Kleanthous S, Mason AM, Pateman AJ, Patikis AN, Pinto IL, Pollard DR, Senger S, Shah GP, Toomey JR, Watson NS, Weston HE. Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Biaryl P4 Motifs. Bioorg. Med. Chem. Lett. 2008;18:23–27. doi: 10.1016/j.bmcl.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 47.Young RJ, Borthwick AD, Brown D, Burns-Kurtis CL, Campbell M, Chan C, Charbaut M, Convery MA, Diallo H, Hortense E, Irving WR, Kelly HA, King NP, Kleanthous S, Mason AM, Pateman AJ, Patikis AN, Pinto IL, Pollard DR, Senger S, Shah GP, Toomey JR, Watson NS, Weston HE, Zhou P. Structure and Property Based Design of Factor Xa Inhibitors: Biaryl Pyrrolidin-2-Ones Incorporating Basic Heterocyclic Motifs. Bioorg. Med. Chem. Lett. 2008;18:28–33. doi: 10.1016/j.bmcl.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 48.Abboud MA, Needle SJ, Burns-Kurtis CL, Valocik RE, Koster PF, Amour AJ, Chan C, Brown D, Chaudry L, Zhou P, Patikis A, Patel C, Pateman AJ, Young RJ, Watson NS, Toomey JR. Antithrombotic Potential of GW813893: A Novel, Orally Active, Active-Site Directed Factor Xa Inhibitor. J. Cardiovasc. Pharmacol. 2008;52:66–71. doi: 10.1097/FJC.0b013e31817e9b9e. [DOI] [PubMed] [Google Scholar]
- 49.Kleanthous S, Borthwick AD, Brown D, Burns-Kurtis CL, Campbell M, Chaudry L, Chan C, Clarte M-O, Convery MA, Harling JD, Hortense E, Irving WR, Irvine S, Pateman AJ, Patikis AN, Pinto IL, Pollard DR, Roethka TJ, Senger S, Shah GP, Stelman GJ, Toomey JR, Watson NS, West RI, Whittaker C, Zhou P, Young RJ. Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs. Bioorg. Med. Chem. Lett. 2010;20:618–622. doi: 10.1016/j.bmcl.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 50.Young RJ, Adams C, Blows M, Brown D, Burns-Kurtis CL, Chan C, Chaudry L, Convery MA, Davies DE, Exall AM, Foster G, Harling JD, Hortense E, Irvine S, Irving WR, Jackson S, Kleanthous S, Pateman AJ, Patikis AN, Roethka TJ, Senger S, Stelman GJ, Toomey JR, West RI, Whittaker C, Zhou P, Watson NS. Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs. Bioorg. Med. Chem. Lett. 2011;21:1582–1587. doi: 10.1016/j.bmcl.2011.01.131. [DOI] [PubMed] [Google Scholar]
- 51.Watson NS, Adams C, Belton D, Brown D, Burns-Kurtis CL, Chaudry L, Chan C, Convery MA, Davies DE, Exall AM, Harling JD, Irvine S, Irving WR, Kleanthous S, McLay IM, Pateman AJ, Patikis AN, Roethke TJ, Senger S, Stelman GJ, Toomey JR, West RI, Whittaker C, Zhou P, Young RJ. The Discovery of Potent and Long-Acting Oral Factor Xa Inhibitors with Tetrahydroisoquinoline and Benzazepine P4 Motifs. Bioorg. Med. Chem. Lett. 2011;21:1588–1592. doi: 10.1016/j.bmcl.2011.01.129. [DOI] [PubMed] [Google Scholar]
- 52.Carlson HA. Check Your Confidence: Size Really Does Matter. J. Chem. Inf. Model. 2013;53:1837–1841. doi: 10.1021/ci4004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MOE2011.10; Chemical Computing Group Inc.; 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2011. [accessed: February 23, 2016]. [Google Scholar]
- 54.Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT, Case DA, Kuntz ID, Rizzo RC. DOCK 6: Impact of New Features and Current Docking Performance. J. Comput. Chem. 2015;36:1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MOE2013.08. 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: Chemical Computing Group Inc.; 2013. [accessed: February 23, 2016]. [Google Scholar]
- 56.Babel, Version 3.3. OpenEye Scientific Software, Inc; 2007. [acccessed: February 23, 2016]. [Google Scholar]
- 57.JMP Pro 10. Cary, N.C: SAS Institute, Inc; [accessed: February 23, 2016]. [Google Scholar]
- 58.Fisher RA. On the “Probable Error” of a Coefficient of Correlation Deduced from a Small Sample. Metron. 1921;1:3–31. [Google Scholar]
- 59.Bonett DG, Wright TA. Sample Size Requirements for Estimating Pearson, Kendall and Spearman Correlations. Psychometrika. 2000;65:23–28. [Google Scholar]
- 60.Schumann M, Armen RS. Systematic and Efficient Side Chain Optimization for Molecular Docking Using a Cheapest-Path Procedure. J. Comput. Chem. 2013;34:1258–1269. doi: 10.1002/jcc.23251. [DOI] [PubMed] [Google Scholar]
- 61.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R, Fang X, Lu Y, Yang C-Y, Wang S. The PDBbind Database: Methodologies and Updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 63.SZMAP, 1.0.0. Santa Fe, NM: OpenEye Scientific Software, Inc; 2011. [accessed: February 23, 2016]. [Google Scholar]
- 64.SZYBKI 1.8.0.2. Santa Fe, NM: OpenEye Scientific Software; [accessed: February 23, 2016]. [Google Scholar]
- 65.McGaughey GB, Sheridan RP, Bayly CI, Culberson JC, Kreatsoulas C, Lindsley S, Maiorov V, Truchon J-F, Cornell WD. Comparison of Topological, Shape, and Docking Methods in Virtual Screening. J. Chem. Inf. Model. 2007;47:1504–1519. doi: 10.1021/ci700052x. [DOI] [PubMed] [Google Scholar]
- 66.McGann M. FRED and HYBRID Docking Performance on Standardized Datasets. J. Comput. Aided Mol. Des. 2012;26:897–906. doi: 10.1007/s10822-012-9584-8. [DOI] [PubMed] [Google Scholar]
- 67.Friedman JH. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001;29:1189–1232. [Google Scholar]
- 68.Byron P, Yang H, Zhub J. Boosted Decision Trees, a Powerful Event Classifier. Stat. Probl. Part. Phys. Astrophys. Cosmol. Proc. PHYSTAT05. 2006;40:139. [Google Scholar]
- 69.Ballester PJ, Mitchell JB. A Machine Learning Approach to Predicting Protein–ligand Binding Affinity with Applications to Molecular Docking. Bioinformatics. 2010;26:1169–1175. doi: 10.1093/bioinformatics/btq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X, Jiang H, Li H. SHAFTS: A Hybrid Approach for 3D Molecular Similarity Calculation 1 Method and Assessment of Virtual Screening. J. Chem. Inf. Model. 2011;51:2372–2385. doi: 10.1021/ci200060s. [DOI] [PubMed] [Google Scholar]
- 71.Koes DR, Camacho CJ. Pharmer: Efficient and Exact Pharmacophore Search. J. Chem. Inf. Model. 2011;51:1307–1314. doi: 10.1021/ci200097m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prathipati P, Mizuguchi K. Integration of Ligand and Structure Based Approaches for CSAR-2014. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00477. [DOI] [PubMed] [Google Scholar]
- 73.Gohlke H, Hendlich M, Klebe G. Knowledge-Based Scoring Function to Predict Protein-Ligand interactions1. J. Mol. Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- 74.Wang R, Lai L, Wang S. Further Development and Validation of Empirical Scoring Functions for Structure-Based Binding Affinity Prediction. J. Comput. Aided Mol. Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 75.Yin S, Biedermannova L, Vondrasek J, Dokholyan NV. MedusaScore: An Accurate Force Field-Based Scoring Function for Virtual Drug Screening. J. Chem. Inf. Model. 2008;48:1656–1662. doi: 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grudinin S, Popov P, Neveu E, Cheremovskiy G. Predicting Binding Poses and Affinities in the CSAR 2013–2014 Docking Exercises Using the Knowledge-Based Convex-PL Potential. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00339. [DOI] [PubMed] [Google Scholar]
- 77.Hogues H, Sulea T, Purisima EO. Evaluation of the Wilma-SIE Virtual Screening Method in Community Structure–Activity Resource 2013 and 2014 Blind Challenges. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00278. [DOI] [PubMed] [Google Scholar]
- 78.Martiny VY, Martz F, Selwa E, Iorga BI. Blind Pose Prediction, Scoring, and Affinity Ranking of the CSAR 2014 Dataset. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00337. [DOI] [PubMed] [Google Scholar]
- 79.Nedumpully-Govindan P, Jemec DB, Ding F. CSAR Benchmark of Flexible MedusaDock in Affinity Prediction and Nativelike Binding Pose Selection. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00303. [DOI] [PubMed] [Google Scholar]
- 80.Shin W-H, Lee GR, Seok C. Evaluation of GalaxyDock Based on the Community Structure–Activity Resource 2013 and 2014 Benchmark Studies. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00309. [DOI] [PubMed] [Google Scholar]
- 81.Yan C, Grinter SZ, Merideth BR, Ma Z, Zou X. Iterative Knowledge-Based Scoring Functions Derived from Rigid and Flexible Decoy Structures: Evaluation with the 2013 and 2014 CSAR Benchmarks. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu X, Shin W-H, Kim H, Kihara D. Combined Approach of Patch-Surfer and PL-PatchSurfer for Protein–Ligand Binding Prediction in CSAR 2013 and 2014. J. Chem. Inf. Model. 2015 doi: 10.1021/acs.jcim.5b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Onufriev A, Bashford D, Case DA. Modification of the Generalized Born Model Suitable for Macromolecules. J. Phys. Chem. B. 2000;104:3712–3720. [Google Scholar]
- 84.Zhou H-X, Gilson MK. Theory of Free Energy and Entropy in Noncovalent Binding. Chem. Rev. 2009;109:4092–4107. doi: 10.1021/cr800551w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sulea T, Cui Q, Purisima EO. Solvated Interaction Energy (SIE) for Scoring Protein-Ligand Binding Affinities 2 Benchmark in the CSAR-2010 Scoring Exercise. J. Chem. Inf. Model. 2011;51:2066–2081. doi: 10.1021/ci2000242. [DOI] [PubMed] [Google Scholar]
- 86.Wildman SA, Crippen GM. Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci. 1999;39:868–873. [Google Scholar]
- 87.Kramer C, Kalliokoski T, Gedeck P, Vulpetti A. The Experimental Uncertainty of Heterogeneous Public Ki Data. J. Med. Chem. 2012;55:5165–5173. doi: 10.1021/jm300131x. [DOI] [PubMed] [Google Scholar]
- 88.Kramer C, Gedeck P. Three Descriptor Model Sets a High Standard for the CSAR-NRC HiQ Benchmark. J. Chem. Inf. Model. 2011;51:2139–2145. doi: 10.1021/ci200030h. [DOI] [PubMed] [Google Scholar]
- 89.Kramer C, Lewis R. QSARs, Data and Error in the Modern Age of Drug Discovery. Curr. Top. Med. Chem. 2012;12:1896–1902. doi: 10.2174/156802612804547380. [DOI] [PubMed] [Google Scholar]
- 90.Kalliokoski T, Kramer C, Vulpetti A, Gedeck P. Comparability of Mixed IC50 Data–a Statistical Analysis. PloS One. 2013;8:e61007. doi: 10.1371/journal.pone.0061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Corbeil CR, Williams CI, Labute P. Variability in Docking Success Rates due to Dataset Preparation. J. Comput. Aided Mol. Des. 2012;26:775–786. doi: 10.1007/s10822-012-9570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lemmon G, Meiler J. Rosetta Ligand Docking with Flexible XML Protocols. Methods Mol. Biol. 2012;819:143–155. doi: 10.1007/978-1-61779-465-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban Y-EA, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popović Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. Rosetta3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ashtawy HM, Mahapatra NR. Molecular Docking for Drug Discovery: Machine-Learning Approaches for Native Pose Prediction of Protein-Ligand Complexes. In: Formenti E, Tagliaferri R, Wit E, editors. Computational Intelligence Methods for Bioinformatics and Biostatistics. Lecture Notes in Computer Science; Springer International Publishing; 2013. pp. 15–32. [Google Scholar]
- 95.Yan Z, Wang J. Specificity Quantification of Biomolecular Recognition and Its Implication for Drug Discovery. Sci. Rep. 2012;2 doi: 10.1038/srep00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shin W-H, Seok C. GalaxyDock: Protein-Ligand Docking with Flexible Protein Side-Chains. J. Chem. Inf. Model. 2012;52:3225–3232. doi: 10.1021/ci300342z. [DOI] [PubMed] [Google Scholar]
- 97.Wolber G, Langer T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 98.Sulimov AV, Kutov DC, Oferkin IV, Katkova EV, Sulimov VB. Application of the Docking Program SOL for CSAR Benchmark. J. Chem. Inf. Model. 2013;53:1946–1956. doi: 10.1021/ci400094h. [DOI] [PubMed] [Google Scholar]
- 99.Sulimov VB, Katkova EV, Oferkin IV, Sulimov AV, Romanov AN, Roschin AI, Beloglazova IB, Plekhanova OS, Tkachuk VA, Sadovnichiy VA. Application of Molecular Modeling to Urokinase Inhibitors Development. BioMed Res. Int. 2014;2014:e625176. doi: 10.1155/2014/625176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 101.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 102.Fourches D, Politi R, Tropsha A. Target-Specific Native/Decoy Pose Classifier Improves the Accuracy of Ligand Ranking in the CSAR 2013 Benchmark. J. Chem. Inf. Model. 2015;55:63–71. doi: 10.1021/ci500519w. [DOI] [PubMed] [Google Scholar]
- 103.Agostino M, Gandhi NS, Mancera RL. Development and Application of Site Mapping Methods for the Design of Glycosaminoglycans. Glycobiology. 2014:cwu045. doi: 10.1093/glycob/cwu045. [DOI] [PubMed] [Google Scholar]
- 104.Voet A, Berenger F, Zhang KYJ. Electrostatic Similarities between Protein and Small Molecule Ligands Facilitate the Design of Protein-Protein Interaction Inhibitors. PLoS One. 2013;8:e75762. doi: 10.1371/journal.pone.0075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Corbeil CR, Sulea T, Purisima EO. Rapid Prediction of Solvation Free Energy. 2. The First-Shell Hydration (FiSH) Continuum Model. J. Chem. Theory Comput. 2010;6:1622–1637. doi: 10.1021/ct9006037. [DOI] [PubMed] [Google Scholar]
- 106.Fourches D, Politi R, Tropsha A. Target-Specific Native/Decoy Pose Classifier Improves the Accuracy of Ligand Ranking in the CSAR 2013 Benchmark. J. Chem. Inf. Model. 2015;55:63–71. doi: 10.1021/ci500519w. [DOI] [PubMed] [Google Scholar]
- 107.Schumann M, Armen RS. Systematic and Efficient Side Chain Optimization for Molecular Docking Using a Cheapest-Path Procedure. J. Comput. Chem. 2013;34:1258–1269. doi: 10.1002/jcc.23251. [DOI] [PubMed] [Google Scholar]
- 108.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical Scoring Functions: I. The Development of a Fast Empirical Scoring Function to Estimate the Binding Affinity of Ligands in Receptor Complexes. J. Comput. Aided Mol. Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 109.Verdonk ML, Berdini V, Hartshorn MJ, Mooij WTM, Murray CW, Taylor RD, Watson P. Virtual Screening Using Protein−Ligand Docking: Avoiding Artificial Enrichment. J. Chem. Inf. Comput. Sci. 2004;44:793–806. doi: 10.1021/ci034289q. [DOI] [PubMed] [Google Scholar]