Abstract
Poly(ethylene glycol) hydrogels with disulfide linkages are functionalized through applied force. Compression or tension induces bond rupture at the relatively weak disulfide linkages, which will subsequently react through Michael-type addition with an acceptor molecule within the gel. We demonstrate the utility of this approach by patterning cell adhesion proteins through compression of a lithographically structured stamp, where cells predominately adhere to the compressed regions.
Synthetic hydrogels represent an important biomaterial for a host of applications including contact lenses, sensor interfaces, drug delivery vehicles, and scaffolds for regenerative medicine and tissue engineering.1 Natural hydrogels in organisms are constantly remodelled, through redox reactions and enzymatic means,2 to impart dynamically changing mechanical, diffusional and biochemical properties. Designing synthetic hydrogel materials that recapitulate natural systems and are amenable to temporal modification of properties and chemistry is an area of significant interest.3–5
Various approaches have been developed to change the chemistry and properties within a hydrogel over time,6 and include pH and temperature responsive materials,7 enzyme-degradable hydrogels,8 photodegradable and photoactivatable crosslinks,9 the addition of secondary crosslinking and polymerization motifs,10 and inclusion of reactive units within the hydrogel for subsequent conjugation.11 In particular, the simplicity of “click-type” reactions,12 where incorporation of a bioactive moiety can be accomplished in the presence of cells, has proved effective using a number of modular chemistries including: azide-alkyne cycloaddition,13 diels-alder cycloaddition,14 Michael-addition of thiols,15 and photo-mediated thiol-ene type reactions.16
In this communication we present a novel hydrogel functionalization approach, where mechanical force mediates bond rupture in a disulfide-linked hydrogel to catalyse a Michael-type addition reaction in the presence of a functional biomolecule. The use of disulfide bonds was motivated by natural systems, where the properties of the relatively weak disulfide bond lends itself to reduction and exchange reactions for building protein structures and remodelling the extracellular matrix.17–19 In the presence of maleimide or acryloyl tagged molecules, we show that compression and tension will mediate bond rupture and Michael addition, and we demonstrate the use of force to immobilize a cell ad hesion ligand for micropatterning cells.
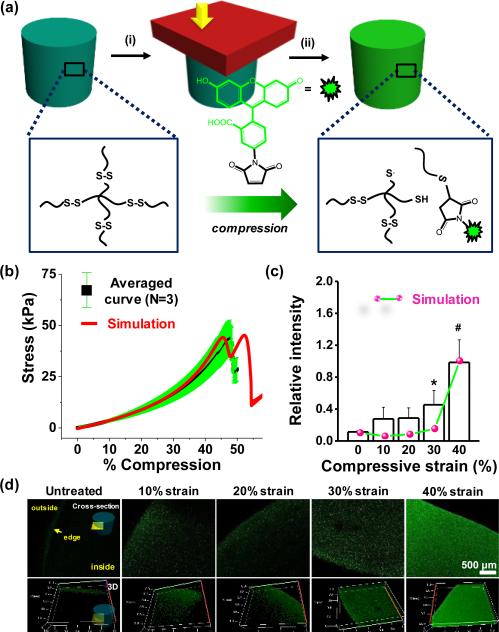
Previous work by Anseth and colleagues demonstrated how disulfide-linked hydrogels can be remoulded through photo-mediated fragmentation of the network.20 Inspired by this work, we postulated that mechanical force could rupture disulfide bonds, and the generated radicals will likely abstract hydrogens from water to form thiols.21 The reaction in the presence of an appropriate acceptor may occur through either a thiol radical or free thiol after abstraction(Figure 1a). To test this hypothesis we optimized the gelation time of 4 armed poly(ethylene glycol) (PEG)-thiol (MW 5,000) based on measurements of gel density and swelling (Table S1 & S2 and Figure S1) to ensure complete gelation. Based on these results, 10% disulfide hydrogels were fabricated in a cylindrical mould for 3 hours with initiators. The stress-strain behaviour of disulfide gels was obtained with dynamic mechanical analysis (DMA) at room temperature (Figure 1b). The disulfide gels failed around ~47% compression (compression rate 5 μm /sec), thus we exposed gels to a range of compressive strains from 0 to 40% for 30 seconds. After applying strain to the material, the gels were incubated with fluorescein-5-maleimide for 1 hour to allow reaction with ruptured disulfide bonds. The gels were rinsed and stored in distilled water to ensure removal of unreacted fluorophore. Confocal laser scanning microscopy was employed tonon monitor fluorescence introduced through reaction. Fluorescence increased with increasing compressive strains with highest intensity at 40% strain (Figure 1c and 1d). Without compression, the edge of the gels displayed some fluorescence, presumably due tonon-specific adsorption at gel-glass interface inhomogeneities, or through physical adsorption during partial drying while processing samples . To explore whether the forces from compressive strain are on the order required to rupture disulfide bonds, we employed a computational model for disulfide bond-based hydrogels. The gel model with mechanochemically dissociated disulfide bonds was fit to the monotonic stress-strain response of the gels. The model was able to capture the typical hyperelastic gel response and the moderately strain rate dependent fracture strain (Figure 1b and S2). Based on the polymer chain force at fracture (Figure S2), the disulfide energy barrier parameters required to fit the monotonic behaviour appear to be reasonable for disulfide bonds in an aqueous environment (~0.1 nN).22–24 The model predicts that a noticeable force driven disulfide dissociation (for a 30 second hold time, corresponding to experimental conditions) will occur for 30% and 40% strain, with significantly more dissociation at 40% strain - also corresponding to the experimental results. However, this is somewhat inconsistent with the low strain experimental results which do show a surprising jump between 0% and 10% followed by no change from 10% to 20%. This inconsistency at low strain is likely due in part to polydispersity in length between cross-links inherent to nearly all gels.
Fig. 1.
(a) Schematic of disulfide bond rupture by compression and subsequent Michael addition. (b) Average stress-strain relationship and simulation for the disulfide gels (MW 5000) with 5 μm/s compression. (c) Fluorescence intensity of gels as a function of compressive strain.(d) Representative laser scanning confocal microscope images of disulfide hydrogels exposed to compressive strain (0-40%) in the presence of fluorescein-5-maleimide.(*P<0.05, #P<0.01, t-test)
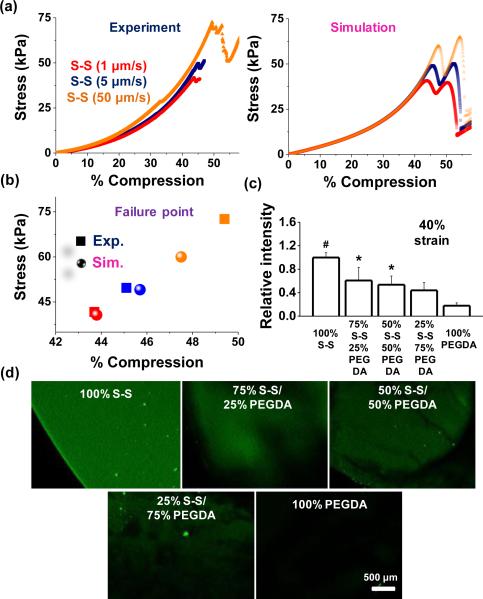
To explore whether failure characteristic is dependent on the compression rate, 4 arm-PEG-thiols were tested using 1 μm /sec, 5 μm /sec, and 50 μm /sec, and compared with the simulated results (Figure 2a).; Although both experimental and simulation results show hardly discernible effect on the compression at failure but the strain leading to the mechani cal rupture of disulfide bonds slightly increases with increasing the compression rate and the ultimate stress was increased around 150% when the compression rate was changed from 5 μm /sec to 50 μm /sec. These results demonstrate good agreement with simulation (Figure 2a and 2b) and we employed the compression rate, 5 μm /sec, for the remaining experiments. To explore the response to compression when the disulfide linkages are diluted within the gel network, we co-polymerized poly(ethylene glycol) diacrylate (PEGDA) with the PEG disulfide hydrogels. We chose 10% poly(ethylene glycol) diacrylate (PEGDA) of average molecular weight 10,000 because their elastic modulus was similar to that of 10% disulfide hydrogels (~10 kPa). We fabricated various gels with 100% PEGDA, 75% PEGDA + 25% 4arm-PEG-thiols, 50% PEGDA + 50% 4arm-PEG-thiols, 25% PEGDA + 75% 4arm-PEG-thiols, and different molecular weight of 4arm-PEG-thiols (MW5,000 & 10,000). Since the rate of Michael addition between thiols and acrylates is considerably slower than hydrogel formation (Figure S3), we do not believe Michael addition products will significantly contribute to the bulk gel network. As shown in the representative curves, the failure strain was influenced by PEGDA composition (compression rate 5 μm /sec) (Figure S4). The 100% PEGDA specimen did not fail until ~85% compression and incorporation of the 4 arm-PEG-thiols led to failure at lower strain, consistent with the disulfide-linked network contributing to the bulk mechanical properties. However, the effects of the mixing ratio and the molecular weight of 4 arm-PEG-thiols on failure characteristic were not significant. Moreover, we also investigated the effects of molecular weight to probe disulfide cross-linking density on bond rupture (Figure S5), however, we did not see a significant change in the failure strain within the range of the molecular weight (5,000 & 10,000 Da) of 4arm-PEG-thiols. PEGDA and disulfide hydrogels and their mixtures were compressed with 40% compressive strains for 30 sec and then treated with fluorescein-5-maleimide. Fluorescence within the gel decreased as a function of the PEG component: 1.6–fold (25% PEG & 75% S-S), 1.9–fold (50% PEG & 50% S-S), 2.3–fold (75% PEG & 25% S-S), and 5.5–fold (100% PEG), compared to¬ 100% disulfide gels (Figure 2c and 2d).
Fig. 2.
(a) Stress–strain behaviour for disulfide gels with different compression speeds and (b) failure strain–stress points for experiment and simulation. (c) Fluorescence intensity of gels containing different fractions of PEGDA and 4arm-PEG-thiols in the presence of fluorescein-5-maleimide,and (d) representative laser scanning confocal microscope images of these hydrogels when exposed to 40% compression. (*P < 0.05, #P < 0.01,t-test).
We also explored the effect of tension on the mechanical rupture of disulfide bonds. Taking advantage of the strong chemisorption of thiols onto gold films, disulfide hydrogels were bound to polydimethylsiloxane (PDMS) substrates coated with a thin layer of sputtered gold. After applying tension to the PDMS substrate for 30 sec, the gels were incubated in the presence of fluorescein-5-maleimide for 1 hour. Tensile stress leads to increased fluorescence intensity, particularly at > 30% elongation in good correspondence with the simulation results, which mirrors the results from compression testing (Figure S6b and c). Based on these results, we propose that force applied through either compression or tension will mechanically reveal active moieties that are available for reaction with an appropriate acceptor.
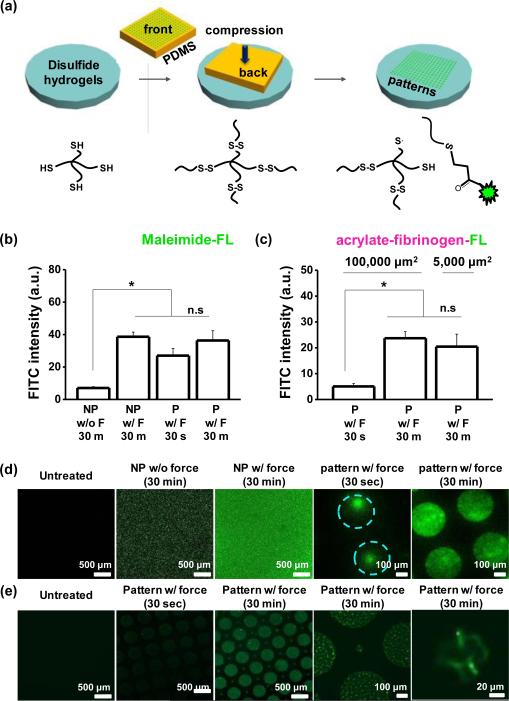
Next we sought to explore whether force could be used to pattern soft materials through compression of a structured mould. We prepared PDMS stamps using photolithography to present geometric features in relief (patterned) or flat surfaces without structure (non-patterned). Fluorescein-5-maleimide or acrylated-fluorescent fibrinogen soluti ons were pipetted on the stamps for 30 min, dried under a stream of air, and compressed onto the gel surfaces (Figure 3a). First, we compared fluorescein-5-maleimide intensity on the non-patterned gels with or without compression of approximately 10 kPa of stress. After 30 min, the fluorescence intensity from gels with applied compression was 5.5–fold higher (Figure 3b). Next, to examine the role of the compression time applied to the gels on the conjugation and patterning accuracy of fluorescein-5-maleimide or acrylated-fluorescent fibrinogen, we patterned 100,000 μm2 area circular features on the gels while applying compression for 30 sec or 30 min. The fluorescence intensity was slightly higher (1.3 –fold) for fluorescein-5-maleimide, while for acrylated-fluorescent fibrinogen, the fluorescence shows a 5.5–fold increase from 30 sec to 30 min holding time (Figure 3c). Furthermore, patterns of fluorescein-5-maleimide generated through compression for 30 min had much higher pattern fidelity compared to those fabricated through 30 sec of applied force (Figure 3d). Pat-tern fidelity did not depend on holding time when using acrylated-fluorescent fibrinogen (Figure 3e). This may be due to multiple dye molecules for each protein monomer, or differences in reaction rate or diffusion between the small molecule and the globular protein.25 Fluoresceinamine will not react with thiols, and was used as a negative control for the fluorescein-5-maleimide; after 30 minutes of compression the intensity of fluorescein-5-maleimide treated gels was much higher than those treated with fluoresceinamine (Figure S7a). Moreover after applying compressi on we see some residual fluores cence even after treatment with strong detergent (Triton X-100; Figure S7b), suggesting some chemical conjugation of acrylated-fluorescent fibrinogen may occur through ex-change with disulfides on fibrinogen.
Fig. 3.
(a) Schematic illustrating the process used to pattern different molecules via compression on disulfide hydrogels. Fluorescence intensity for patterned (P) and non-patterned (NP) gels with (b) fluorescein-5-maleimide and (c) acrylate fibrinogen fluorescein with or without force and different holding time. Representative laser scanning confocal microscopen images of disulfide hydrogels when patterned with (d) fluorescein-5-maleimide and (e) acrylate fibrinogen fluorescein. (*P<0.05, t-test)
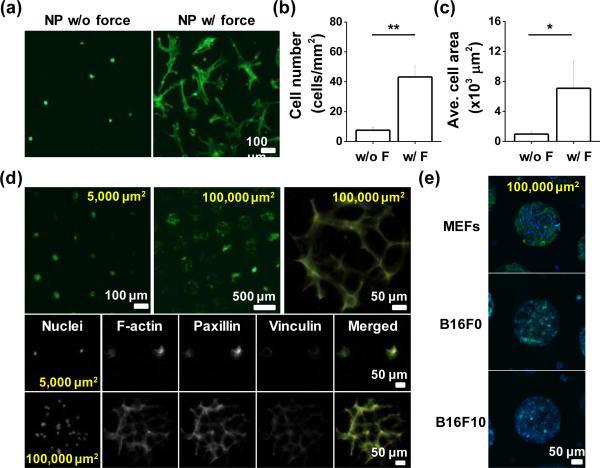
One potential application of using force for patterning hydrogels is to decorate hydrogels with ligands for cell culture applications. To characterize the utility of conjugating proteins through compression to mediate cell adhesion, we compared the number of attached cells and their degree of spreading on the gels with or without the applied force in the presence of acrylated fibronectin. Mesenchymal stem cells (MSCs) are an attractive cell type for integration with hydrogel materials for regenerative therapies.26 Cells were seeded on disulfide hydrogels patterned with acrylated fibronectin via micro- contact printing with compressive force for 30 min. After two days culture, there were 5.7–fold greater numbers of cells on compressed gels (Figure 4a & b), and the average spread cell area was 7.3–fold higher than those cultured on gels without compression (Figure 4c). We used immunofluorescence microscopy to investigate the focal adhesions and stress fibres in MSCs adherent to the mechano-functionalized gels. Exploring cell adhesion to different sized patterns (5,000 μm2 for single cells & 100,000 μm2 for multiple cells) we see robust focal adhesion and stress fibre formation in the cells adherent to the protein conjugated patterns (Figure 4d). Since cells from different tissues will adhere differently to the extracellular matrix, we also explored adhesion of other cell types: mouse embryonic fibroblasts (MEFs) and murine melanoma cells (B16F0 & B16F10). After two days of culture, MEFs and B16 cells showed strong adhesion to the protein conjugated micropatterns, demonstrating the applicability of this technique across diverse cell types.
Fig. 4.
(a) Representative images of MSCs on the unpatterned gel-protein substrate with or without compression. Plot of (b) cell number and (c) average cell area differences with or without compression. (d) Representative immunofluorescence images of MSCs captured on protein patterned islands (100,000 or 5,000 μm2) and im-munostained with Paxillin and Vinculin. (e) Immunofluorescence images of different types of patterned cells (MEFs, B16F0, and B16F10) cultured on protein conjugated disulfide hydrogels. (*P<0.05, **P<0.05, t-test)
Conclusions
We demonstrate a novel hydrogel functionalization method using mechanical force to mediate a Michael-type addition in the presence of a functional biomolecule. Combining experimental results with computational modelling, we demonstrate that >30% strain will lead to bond rupture, with the possibility to leverage this mechanochemical activation for subsequent reactions with functional molecules. This strategy opens up the possibility of a novel route to “click-type” chemistry, where thiol-ene reactions may be promoted with force. Furthermore, we believe this approach may prove amenable to other dynamic hydrogel systems where force may act as a trigger for bond cleavage or molecular reorganization in soft materials. This approach will add to the tool-box of methods to perturb the mechanics and the chemistry of soft materials.
Supplementary Material
Conceptual insights.
Hydrogels in tissue are viscoelastic materials that are continuously remodelled, and undergo dynamic changes in chemistry. Recreating dynamic chemistry in the laboratory most often involves incorporation of stimuli-responsive motifs, or secondary polymerization routines. In this communication, we illustrate a new approach to functionalize disulfide-linked poly(ethylene glycol) hydrogels through applied compression or tension. Force-induced bond rupture at the disulfide linkages causes reaction with an appropriate acceptor molecule within the hydrogel. This approach is amenable to patterning virtually any molecule with the appropriate conjugation tag, and represents a new route to modifying the mechanics and chemistry of soft materials.
Acknowledgements
This work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health, grant number HL121757. M. N. S. was supported in part by the National Science Foundation under grant DMR-1307354.
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00x
References
- 1.Geckil H, Xu F, Zhang X, Moon S, Demirci Nanomedicine U. 2010;5:469–484. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burdick JA, Prestwich GD. Adv. Mater. 2011;23:H41–H56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutolf MP, Hubbell JA. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 4.Drury JL, Mooney DJ. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 5.Yu L, Ding J. Chem. Soc. Rev. 2008;37:1473–1481. doi: 10.1039/b713009k. [DOI] [PubMed] [Google Scholar]
- 6.Stuart MAC, Huck WTS, Genzer J, Müller M, Ober C, Stamm M, Sukhorukov GB, Szleifer I, Tsukruk VV, Urban M, Winnik F, Zauscher S, Luzinov I, Minko S. Nat. Mater. 2010;9:101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Peppas NA. Macromolecules. 2000;33:102–107. [Google Scholar]
- 8.Wachiralarpphaithoon C, Iwasaki Y, Akiyoshi K. Biomaterials. 2007;28:984–993. doi: 10.1016/j.biomaterials.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 9.Kloxin AM, Tibbitt MW, Anseth KS. Nat. Protoc. 2010;5:1867–87. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Tian Y, Ding Y, Tian Y, Wang F. Macromolecules. 2012;45:8412–8419. [Google Scholar]
- 11.Lee SH, Moon JJ, West JL. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhiannon A, Iha K, Wooley Karen L., Nyström Andreas M., Burke Daniel J., Kade Matthew J., Hawker CJ. Chem. Rev. 2009;109:5620–5686. doi: 10.1021/cr900138t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JA, Baskin JM, Bertozzi CR, Koberstein JT, Turro NJ. Chem. Commun. 2008;26:3064–3066. doi: 10.1039/b803043j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durmaz H, Dag A, Altintas OO, Erdogan T, Hizal G, Tunca U. Macromolecules. 2007;40:191–198. [Google Scholar]
- 15.Chan JW, Yu B, Hoyle CE, Lowe AB. Chem. Commun. 2008;40:4959–4961. doi: 10.1039/b813438c. [DOI] [PubMed] [Google Scholar]
- 16.Grim JC, Marozas IA, Anseth KS. J. Control. Release. 2015;219:95–106. doi: 10.1016/j.jconrel.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyarmati B, Némethy Á, Szilágyi A. Eur. Polym. J. 2013;49:1268–1286. [Google Scholar]
- 18.Keten S, Chou CC, van Duin ACT, Buehler MJ. J. Mech. Behav. Biomed. Mater. 2012;5:32–40. doi: 10.1016/j.jmbbm.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Yang WJ, Tao X, Zhao T, Weng L, Kang E-T, Wang L. Polym. Chem. 2015;6:7027–7035. [Google Scholar]
- 20.Fairbanks BD, Singh SP, Bowman CN, Anseth KS. Macromolecules. 2011;44:2444–2450. doi: 10.1021/ma200202w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitch KR, Goodwin AP. Chem. Mater. 2014;26:6771–6776. [Google Scholar]
- 22.Anjukandi P, Dopieralski P, Ribas–Arino J, Marx D. PLoS One. 2014;9:e108812. doi: 10.1371/journal.pone.0108812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iozzi MF, Helgaker T, Uggerud E. J. Phys. Chem. A. 2011;115:2308–2315. doi: 10.1021/jp109428g. [DOI] [PubMed] [Google Scholar]
- 24.Wiita AP, Ainavarapu SRK, Huang HH, Fernandez JM. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7222–7227. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.HALL CE, Slayter HS. J. Biophys. Biochem. Cytol. 1959;5:11–16. doi: 10.1083/jcb.5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CAPLAN AI. J. Cell. Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.