Abstract
We present a microfluidic aptamer-based biosensor for detection of low-molecular-weight biomarkers in patient samples. Using a microfluidic device that integrates aptamer-based specific analyte extraction, isocratic elution, and detection by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, we demonstrate rapid, sensitive and label-free detection of arginine vasopressin (AVP) in human plasma ultrafiltrate. AVP molecules in complex matrices are specifically captured by an aptamer that is immobilized on microbeads via affinity binding in a microchamber. After the removal of unbound, contaminating molecules through washing, aptamer-AVP complexes are thermally disrupted via on-chip temperature control. Released AVP molecules are eluted with purified water and transferred to a separate microchamber, and deposited onto a single spot on a MALDI plate via repeated, piezoelectrically actuated ejection, which enriches AVP molecules over the spot area. This integrated on-chip sample processing enables the quantitative detection of low-abundance AVP by MALDI-TOF mass spectrometry in a rapid and label-free manner. Our experimental results show the detection of AVP in human plasma ultrafiltrate as low as physiologically relevant picomolar concentrations via aptamer-based selective preconcentration, demonstrating the potential of our approach as a rapid (~ 1hr), sensitive clinical AVP assay.
Graphical Abstract

Introduction
Biosensors are widely used for the detection and analysis of biomolecules that are disease relevant biomarkers such as genes, proteins, and peptides. Typically, they consist of a molecular recognition element and a transducer converting the binding event into a measurable physical signal.1 A particularly important class of biosensors comprises affinity biosensors, which rely on highly selective affinity receptors, such as aptamers,2 to recognize target biomolecules. Aptamers are single-stranded oligonucleotides that bind to a biological target with high specificity and affinity, and have received much attention since they can be synthetically developed for a wide range of targets (e.g., small molecules,3 peptides,4 and proteins5) with minimal batch-to-batch variability. Biosensors that use aptamers as molecular recognition elements, called aptasensors, have been used extensively to detect analytes such as proteins, viruses, bacteria, and cells in conditioned media,6–8 and also in human bodily fluids.9–11
Despite these advances, aptasensors for analytes of low-molecular-weight (e.g., ions, small molecules, oligopeptides) have been challenging due to their small size, low electrical charge, and limited availability of functional groups and structural motifs for interaction with aptamers. Various strategies such as adaptive binding-driven signaling,12 affinity chromatography,13 and nanomaterial-based sensing14 have been employed in aptasensors for detection of low-molecular-weight analytes in conditioned media15 or human bodily fluids such as whole blood,16 plasma,17–20 serum,21–27 saliva,28 and urine.29–33 For example, based on target binding-induced aptamer folding, a microfluidic electrochemical aptasensor, which integrates aptamer-functionalized electrodes within a microchamber, was constructed for rapid detection of cocaine from serum samples.34 Aptamer-based extraction of adenosine and its analogues from tissue extract samples was demonstrated via affinity chromatography in a fused silica capillary packed with aptamer-functionalized microbeads.35 Carbon nanotube36 and graphene37 were used for detection of low-molecular-weight species in conditioned media. While these aptasensors have shown promise for clinical applications, they are still limited by a low sensitivity, complicated biochemical preparation steps, and molecular labeling requirements.
Arginine vasopressin (AVP) is an anti-diuretic hormone, and is known to be a clinically important low-molecular-weight biomarker (molecular weight: 1084.5 Da) for severe hemorrhage, septic shock and congestive heart failure. For instance, the plasma level of AVP normally ranges from 0 to 10 pM. In response to a decrease in blood volume and low arterial pressure by hemorrhage, the plasma AVP level markedly increases up to hundred picomolar levels, inducing vasoconstriction. As this state is prolonged, the AVP concentration gradually declines below 30 pM within 1 hr. Further decreases in AVP levels lead to the failure of vasoconstriction, which may result in a low blood pressure that is insufficient for proper blood perfusion into organs, i.e., late-phase hemorrhagic shock. At this stage, the administration of exogenous AVP is essential to raise the arterial pressure. While rapid and sensitive measurement of plasma AVP levels is vital for timely clinical intervention to prevent a late-phase hemorrhagic shock,38 the clinical utility of conventional AVP assays such as enzyme-linked immunosorbent assay (ELISA) and radioimmunoassay (RIA) are severely impeded as they require labor-intensive work and time-consuming procedures (exceeding 24 hrs). Several aptasensors have been previously developed for AVP detection,39–43 although their clinical applicability is limited by labeling requirement and insufficient sensitivity.
We here present an integrated microfluidic aptasensor for detection of low-molecular-weight analytes in complex biological samples. The device integrates, on a single microfluidic chip, the entire process of aptamer-based specific analyte extraction from complex matrices and piezoelectric-based, repeated analyte ejections onto a MALDI plate for subsequent analyses by mass spectrometry. AVP, a low-molecular-weight hormone, is chosen as a target analyte in this study. While the rapid measurement and frequent monitoring of AVP is of great clinical importance for prevention of life-threatening hemorrhagic and septic shocks, the utility of current clinical AVP assays in these clinical situations is still limited particularly due to their long turnaround time (> 24 hrs). Our integrated microfluidic aptasensor enables rapid and sensitive detection of AVP in a label-free manner. Our early-stage experimental results demonstrate detection of AVP from human plasma ultrafiltrate within ~1hr of analysis time. We further show enhancement of the detection sensitivity for measurements of AVP in the clinically-relevant picomolar range (1–100 pM) by incorporating aptamer-based preconcentration. Thus, the aptasensor can potentially be of utility for rapid AVP measurements in clinical applications.
Experimental
Microfluidic Device Design
The integrated microdevice consists of an aptamer-based affinity purification chamber (volume: 2 μl), a PZT spotting chamber (volume: 5 μl), a PZT stack actuator and a set of resistive microheater and temperature sensor (Cr/Au) on a glass substrate (Figs. 1a, b and e). Three inlets are used to separately infuse sample, washing buffer/water and eluting water into the chip. A supplementary bead-packing inlet is used to introduce microbeads which are functionalized with an AVP-specific aptamer44 into the purification chamber. The introduced microbeads (diameter: 50–80 μm) are retained in the purification chamber (height: 150 μm) (Fig. 1c) by two dam-like weir structures formed by the connection to thin microchannels (height: 20 μm). An analyte-eluting solution is transferred to the spotting chamber through a secondary channel (Fig. 1d). The device was fabricated using standard microfabrication techniques such as photolithography and soft lithography, and its fabrication processes are described in Fig. S1.
Fig. 1.
Schematic drawings of the integrated microdevice, (a) the side and (b) top views. (c) A micrograph of the purification chamber packed with aptamer-functionalized microbeads. (d) Demonstration of the pressure-driven fluid flow change. A flow of a fluorescein solution to the waste outlet (left) is guided to the spotting chamber when the waste outlet is plugged (right). (e) A photo of the packaged device.
The principle of AVP detection is schematically described in Fig. 2. A sample solution containing AVP and other biological impurities is first introduced into the purification chamber packed with aptamer-functionalized microbeads while the temperature inside is maintained at 37 °C, the optimum temperature for aptamer-AVP binding.45 In the purification chamber, the aptamer selectively captures AVP molecules on the microbead surface (Fig. 2a). The chamber is then flushed with a buffer solution at 37 °C to remove unbound impurities and nonspecifically adsorbed AVP molecules (Fig. 2b). After the residual salt removal with brief water washing, the chamber temperature is raised up to 60 °C, which disrupts the aptamer-AVP complex for isocratic elution of released AVP with purified water (Fig. 2c). The eluted AVP solution is transferred to the spotting chamber via the secondary channel and is then spotted onto a thin layer of matrix crystal on a MALDI plate by using a PZT nanoliter-droplet ejector (Fig. 2d). After the spotted droplet is evaporated, a subsequent droplet is spotted onto the same site (Fig. 2e), and these repeated depositions induce the concentration of analyte molecules on the spot, called on-spot analyte enrichment (Fig. 2f). A fast evaporation method, by which the MALDI plate is pre-coated with a uniform and thin layer of matrix crystal prior to experiments, is employed to improve resolution, sensitivity and uniformity of mass spectral acquisition.46, 47
Fig. 2.
Principle of AVP detection using the integrated aptasensor. (a–c) Aptamer-based specific extraction of AVP from sample. (d–f) On-spot analyte enrichment.
Materials
AVP, desmopressin (dDAVP), vasotocin and lysine-modified arginine-vasopressin (Lys-AVP) were purchased from American Peptide Company Inc. (Sunnyvale, CA). An AVP-specific spiegelmer or mirror-image aptamer (5′-GGG GUA GGG CUU GGA UGG GUA GUA CAC GUG UGC GUG GU-3′) was custom synthesized and biotinylated by ChemGenes Corporation (Wilmington, MA). The aptamer was originally isolated in its (D) natural RNA form via a selection process targeting enantiomeric (D) vasopressin and then changed into its mirror image (L) RNA form which binds natural (L) vasopressin. An adenosine triphosphate (ATP)-specific aptamer (5′-ACC TGG GGG AGT ATT GCG GAG GAA GGT AAA AAA A-3′) was obtained from Integrated DNA Technologies Inc. (Coralville, IA). A streptavidin-coated polyacrylamide microbead was acquired from Thermo Fisher Scientific Inc. (Rockford, IL) and functionalized with aptamers via streptavidin-biotin links. Plasma ultrafiltrate samples were obtained from patients on CVVH (continuous veno-venous hemofiltration), and all patients provided informed consent to participate in the study, which was approved by the Institutional Review Board of Columbia University. All experiments were done in accordance with the guidelines of the Human Research Protection Program (HRPP) and Institutional Review Board (IRB) of Columbia University. A binding buffer solution (pH 7.4) was prepared in deionized water with Tris-HCl (20 mM), NaCl (150 mM), KCl (5 mM), CaCl2 (1 mM), MgCl2 (1mM) from Sigma-Aldrich (St. Louis, MO). Cyano-4-hydroxycinnamic acid (CHCA), a MALDI matrix, was purchased from Protea Biosciences, Inc. (Morgantown, WV) and solvated in a volume ratio of 80 % acetone, 20 % isopropyl alcohol and 0.1 % trifluoroacetic acid. A PZT stack actuator (5 × 5 × 10 mm; widths × height) inducing 6 μm displacement with 80 V applied was obtained from Steiner & Martins, Inc. (Miami, FL), and a solid state relay for switching on/off PZT actuation was purchased from Omega Engineering Inc. (Stamford, CT) respectively.
Experimental Procedure
Streptavidin-coated microbeads were incubated with a biotinylated aptamer solution in a tube for 1 hr, allowing the aptamer immobilization onto the bead surface. Then, the aptamer-functionalized microbeads were introduced into the purification chamber through a supplementary bead-packing inlet using manual pressure, followed by sealing the inlet. Devices were rinsed with a binding buffer solution (flow rate: 10 μl/min) prior to each experiment. Sample solutions at various AVP concentrations were prepared in binding buffer or human plasma ultrafiltrate. A sample solution (volume: 10 μl) was infused into the chip using a syringe pump (KD210P, KD Scientific Inc.) and incubated with aptamer-functionalized microbeads in the purification chamber for 3 min at 37 °C. In the experiment for aptamer-based preconcentration, 1 ml of sample solution was infused into the chip over a period of approximately 65 min (flow rate: 15 μl/min) while the chamber temperature was kept at 37 °C. The temperature inside the chamber was controlled using the integrated microheater and temperature sensor connected to closed-loop control with LabVIEW. After the chamber was thoroughly rinsed by flushing with a buffer solution (volume: 50 μl; flow rate: 10 μl/min) and purified water (volume: 10 μl; flow rate: 10 μl/min) sequentially, a plug of purified water (volume: 10 μl; flow rate: 2 μl/min) was slowly flowed through the chamber at 60 °C while the other two inlets and the waste outlet were plugged. The eluted AVP solution was then transferred and filled the spotting chamber followed by 50 times deposition onto the matrix-coated MALDI plate.
For the PZT actuation at a predetermined frequency, the square wave-like function was generated by switching on and off power supply (N5768A, Agilent Technologies) with a solid state relay (SSRDC100VDC, Omega Engineering) controlled by a function generator. A schematic of the experimental setup is illustrated in Fig. S2, and the PZT nanoliter-droplet ejector was characterized as described in Fig. S3.
The MALDI plate was pre-coated with a thin layer of matrix crystal by spin-coating (1000 rpm) prior to experiments. After the sample deposition onto the matrix-coated MALDI plate, the plate was inserted into a Voyager DE™ MALDI-TOF mass spectrometer (Applied Biosystems®, Life Technologies, Grand Island, NY) for mass spectrometry.
Results and discussion
Effects of PZT Spotting on Mass Spectrometric AVP Detection
We first investigated effects of PZT spotting on MALDI-TOF mass spectrometry for AVP detection. A 100 nM AVP solution prepared in purified water was deposited once on the MALDI plate using pipetting (1 μl) or PZT spotting (volume: ~ 7 nl), which remained dried MALDI spots with diameters of 1.5 mm and 300 μm, respectively (Figs. 3a and b). Manually controlled UV laser shots (the diameter of the laser irradiation region, hereafter referred to as the laser irradiation diameter, was ~150 μm) were irradiated onto five different irradiation regions on each spot for mass spectral acquisitions (the mass spectrometer allowed operation in manual mode only), and AVP peak intensities from five measurements were averaged for comparison. Experimental results showed that the pipetted spot produced three times larger spatial variation of AVP peak intensities (27 %), compared to a PZT spotting generated spot (9.2 %) (Fig. 3c), concluding that nanoliter-volume spotting can reduce the spatial variation of signals, which in turn improves the reproducibility of mass spectra. This beneficial effect arises from the fact that the MALDI spot size becomes comparable to the laser irradiation diameter, thus a larger portion of the spot can be analyzed simultaneously.48
Fig. 3.
Photos of dried MALDI spots deposited by (a) pipetting (diameter: 1.5 mm) and (b) PZT spotting (diameter: 300 μm). Approximately 25 times smaller spot (area) was produced when using the PZT nanoliter-droplet ejector (Scale bar = 500 μm). (c) Averages of peak intensities from 5 measurements at different irradiation regions within a spot were compared between two sample deposition manners, coefficient of variations (C.V.= standard deviation/mean value) were indicated above the error bars.
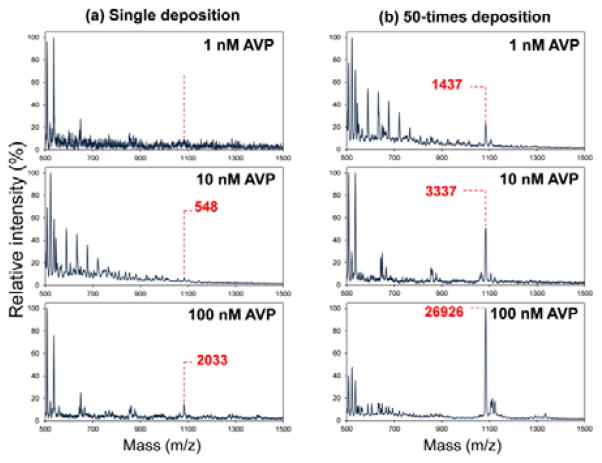
To study the effect of multiple depositions on a spot, an AVP solution (1–100 nM) prepared in purified water was deposited by single or 50 times spotting using the PZT nanoliter-droplet ejector. The optimal number of deposition was determined by the results presented in Fig. S4. Repeated depositions more than 50 times frequently dissolved a matrix layer, thus precluding MALDI measurements. Intensities of the AVP peak were significantly amplified at all concentrations with an enrichment factor of up to 13 (Fig. 4), indicating that AVP was enriched on the spot after multiple depositions. It is noteworthy that at low concentrations (1–10 nM), only peaks comparable to background noises (signal-to-noise ratio: S/N < 5) were detected via single deposition, while an appreciable AVP peak appeared by repeated depositions due to the on-spot analyte enrichment effect. These results suggest that multiple depositions can significantly lower the detection limit in MALDI-TOF mass spectrometric detection of AVP.
Fig. 4.
The effect of multiple depositions using the PZT nanoliter-droplet ejector in the integrated device: mass spectra at various AVP concentrations (1/10/100 nM from the top) when (a) an AVP solution in purified water is deposited once and (b) 50 times on the same spot.
AVP Detection
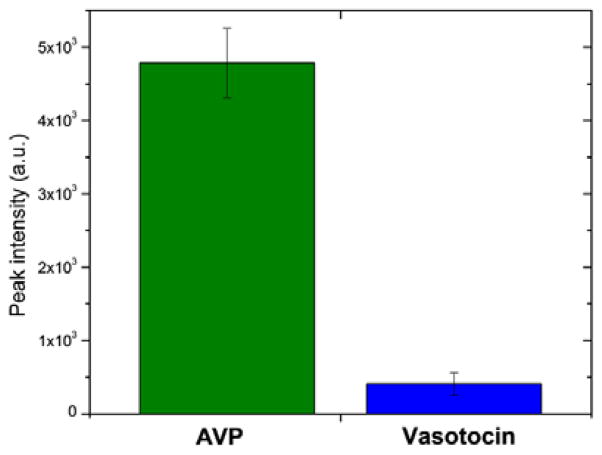
For AVP detection, the specificity of the aptasensor is derived from that of the aptamer.41, 43, 44 We demonstrate this specificity using vasotocin as a control analyte due to its high similarity to AVP in terms of the size (AVP: 1084.5 Da vs. Vasotocin: 1050 Da) and amino acid composition (AVP: Cys-Tyr-Phe-Gln-Asn-Cys-Pro-Arg-Gly-NH2 vs. Vasotocin: Cys-Tyr-Ile-Gln-Asn-Cys-Pro-Arg-Gly-NH2). The device, functionalized with the AVP-specific aptamer, was first tested for detection of AVP at 100 nM in binding buffer. A strong peak at 1084.5 m/z indicative of AVP was observed in the mass spectra (Fig. 5, green bar). On the other hand, vasotocin, an oligopeptide that differs from AVP by a single amino acid, yielded a mass spectral peak (at 1050 m/z) that was small compared with that of AVP (Fig. 5, blue bar) and could be attributed to the nonspecific physical adsorption of vasotocin molecules to microbead and microfluidic device surfaces. Thus, while aptamer-based preconcentration and on-spot enrichment resulted in a significant amount of AVP molecules for mass spectrometric detection, the binding between the aptamer and the control molecule vasotocin was insignificant. In addition, when the device was functionalized with a control aptamer (an aptamer that binds to ATP), testing of AVP detection did not yield a distinctive mass spectral peak (data not shown). These results demonstrated that the aptasensor was specific to AVP.
Fig. 5.
Specificity of the aptasensor: the device, functionalized with the AVP-specific aptamer, was tested respectively on AVP and vasotocin (control), both at 100 nM. Experiments for each condition were run in triplicates.
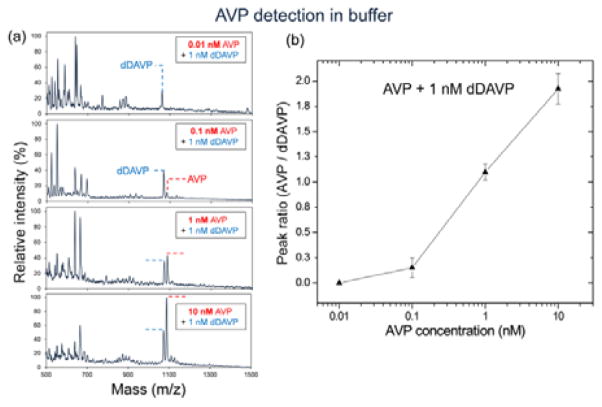
We first tested our aptasensor for AVP detection in binding buffer. To reduce the experimental variability which can arises from chip processing (binding/elution/spotting) and MALDI measurements (distribution/ionization), a constant amount of an internal standard was added into AVP samples. Desmopressin (dDAVP), a synthetic replacement for AVP with a molecular weight of 1069 Da, was chosen as an internal standard through the investigation of similarity in aptamer binding and MALDI behavior among various polypeptides (Fig. S5). An AVP sample at varying concentrations (0.01–10 nM) with 1 nM dDAVP was prepared in binding buffer, and then processed through the integrated device followed by MALDI-TOF mass spectrometry. Resulting mass spectra displayed two peaks at 1069 m/z and 1084.5 m/z, which respectively represent dDAVP and AVP, while no AVP peak appeared at 10 pM (Fig. 6a). This can be attributed to that the binding affinity of the aptamer towards AVP is insufficiently low for its detection below 100 pM as the dissociation constant (Kd) reported elsewhere is 1.7 nM at 37 °C.44 Peak intensity ratios of AVP to dDAVP were calculated for relative quantification, and the ratio was found to proportionally increase with the AVP concentration in the range between 100 pM and 10 nM (Fig. 6b). This implies that higher AVP concentrations resulted in more AVP captured by aptamer on the microbead surface in the aptasensor, thus inducing the larger amount of AVP on a MALDI spot.
Fig. 6.
AVP detection in buffer: (a) Mass spectra obtained from experiments with AVP samples at various concentrations (0.01–10 nM) containing dDAVP at a fixed concentration (1 nM). (b) MALDI peak ratios (AVP/dDAVP) vs. AVP concentrations, showing the concentration-dependent responses in the range of 100 pM to 10 nM. Experiments at each concentration were run in triplicates.
To further characterize the potential utility of our device in clinical settings, we also tested the AVP detection from human plasma ultrafiltrate. (The ultrafiltration process is in general believed to preserve the amount of AVP in plasma while removing other larger molecules.49) AVP at various concentrations (1–100nM) and dDAVP at a fixed concentration (10 nM) were spiked into a human plasma ultrafiltrate solution, which was then processed by the integrated aptasensor for MALDI-TOF mass spectrometric detection. Resulting mass spectra showed that absolute intensities of the AVP peak and the AVP concentration did not correlate (Fig. 7a), indicating that biological impurities interfered with both the aptamer-AVP binding and the ionization of AVP molecules during the MALDI process, consequently increasing the experimental variability. However, the effects of such variations could be effectively suppressed by relative quantification with dDAVP, in which case the mass spectral peak ratio (AVP/dDAVP) monotonically increased with the AVP concentration in the nanomolar range (Fig. 7b). These results suggest that the integrated aptasensor specifically extracted AVP from human plasma ultrafiltrate via aptamer-based affinity binding, and further enriched the extracted analyte on a MALDI spot to aid effective mass spectrometric detection of AVP.
Fig. 7.
AVP detection in human plasma ultrafiltrate (UF): (a) Mass spectra obtained from experiments with plasma ultrafiltrate samples spiked with AVP at various concentrations (1–100 nM) and dDAVP at a fixed concentration (10 nM). (b) MALDI peak ratio (AVP/dDAVP) vs. AVP concentration. Experiments at each concentration were run in triplicates.
AVP detection at clinically relevant concentrations
To demonstrate AVP detection in the clinically relevant range (0–300 pM38, 50, 51), we combined aptamer-based preconcentration of AVP with on-spot analyte enrichment, for which a dilute AVP solution was continuously infused into the device followed by washing, elution and repeated depositions on the MALDI plate. We first tested this approach for AVP detection in binding buffer, in which AVP concentrations were varied while the dDAVP concentration was kept constant (10 pM). Results showed that the peak ratio of AVP to dDAVP increased with the AVP concentration, confirming the ability of the aptasensor to measure AVP levels in the picomolar range (Fig. 8a). In particular, the aptasensor was capable of reliably detection AVP as low as 1 pM in concentration. This indicates that the continuous sample infusion caused aptamer-based selective preconcentration of AVP on microbead surfaces followed by further on-spot analyte enrichment, thereby enabling detection of AVP at such low picomolar concentrations.
Fig. 8.
Picomolar AVP detection via aptamer-based preconcentration coupled to on-spot analyte enrichment. (a) In buffer: MALDI peak ratio (AVP/dDAVP) vs. AVP concentration in the range between 1 and 100 pM with 10 pM dDAVP. Experiments at each concentration were run in triplicates. (b) In human plasma ultrafiltrate: Mass spectra obtained from a plasma ultrafiltrate sample spiked with 10 pM (top) or 100 pM (bottom) AVP.
Using this approach, we also conducted preliminary tests for AVP detection in human plasma ultrafiltrate at clinically relevant concentrations. In the resulting mass spectra, a distinct AVP peak was observed at each of the concentrations tested (10 and 100 pM) in the presence of significant background peaks (Fig. 8b). On the other hand, mass spectral peaks of AVP were not distinguishable from background peaks when aptamer-based preconcentration was not used (data not shown), indicating that the repeated selective extraction of AVP effectively increased the sensitivity of the aptasensor even in the presence of biological impurities in the human plasma ultrafiltrate sample. Impurities and ions that nonspecifically adsorbed to beads likely contributed significantly to the background peaks, whose reduction would enable the device to measure AVP at further lower concentrations in a more reproducible manner. For example, modification of bead surfaces with an anti-adsorption coating (e.g., polyethylene glycol52), while not implemented in the present proof-of-concept work, should significantly reduce background peaks in the mass spectra. In addition, this issue could be further mitigated by optimization of the device design (e.g., chamber volume and shape), washing methods (e.g., choice of the buffer as well as washing time and flow rate) during the device operation, and microbead manipulation schemes.53
Conclusion
We have developed an integrated microfluidic aptasensor for label-free mass spectrometric detection of low-molecular-weight analytes in human plasma ultrafiltrate. The device integrates aptamer-based specific extraction and isocratic elution of the analyte from human plasma ultrafiltrate samples, and PZT spotting for following MALDI-TOF mass spectrometric detection. AVP, a clinically important oligopeptide biomarker, was effectively extracted from human plasma ultrafiltrate via solid phase aptamer-based affinity binding, thus allowing detection of AVP by MALDI-TOF mass spectrometry. The PZT spotting technique formed a dried MALDI spot as small as laser irradiation region, thus minimizing the spatial variation of mass spectra within a spot. The purified AVP solution was then repeatedly deposited on a spot, thus inducing on-spot analyte enrichment for improved sensitivity.
Using the integrated device, we demonstrated label-free mass spectrometric detection of AVP in binding buffer and human plasma ultrafiltrate in the range of 0.1–10 nM and 1–100 nM, respectively. Combining aptamer-based selective preconcentration of AVP on microbeads with on-spot analyte enrichment by repeated depositions on a MALDI plate, we were able to reliably detect AVP at 1 pM in buffer and 10 pM in human plasma ultrafiltrate as the lowest measurable concentrations using the aptasensor. These early-stage results provide a basis in future work to improve the aptasensor performance for measurement of AVP over the entire clinical range of 1–300 pM in human plasma ultrafiltrate. Directions of the future work may include optimization of the device design and operation, such as using anti-adsorption modification of microbead surfaces as well as improved choice of device dimensions, chamber shape and buffer washing methods during the device operation. With these improvements, the aptasensor can ultimately be useful as a potential tool for rapid detection of AVP for the timely detection and treatment of severe hemorrhage, septic shock and congestive heart failure.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial supports from the National Science Foundation (Award No. CBET-0854030), and the National Institutes of Health (Award Nos. 8R21GM104204-03 and RGM104960).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Luong JHT, Male KB, Glennon JD. Biotechnology Advances. 2008;26:492–500. doi: 10.1016/j.biotechadv.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Bunka DHJ, Stockley PG. Nat Rev Micro. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 3.Mannironi C, Di Nardo A, Fruscoloni P, Tocchini-Valentini GP. Biochemistry. 1997;36:9726–9734. doi: 10.1021/bi9700633. [DOI] [PubMed] [Google Scholar]
- 4.Nieuwlandt D, Wecker M, Gold L. Biochemistry. 1995;34:5651–5659. doi: 10.1021/bi00016a041. [DOI] [PubMed] [Google Scholar]
- 5.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Cancer Research. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 6.Bosco F, Bache M, Yang J, Chen C, Hwu ET, Lin Q, Boisen A. Sensors and Actuators A: Physical. 2013;195:154–159. doi: 10.1016/j.sna.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui ZQ, Ren Q, Wei HP, Chen Z, Deng JY, Zhang ZP, Zhang XE. Nanoscale. 2011;3:2454–2457. doi: 10.1039/c1nr10218d. [DOI] [PubMed] [Google Scholar]
- 8.Zelada-Guillén GA, Riu J, Düzgün A, Rius FX. Angewandte Chemie International Edition. 2009;48:7334–7337. doi: 10.1002/anie.200902090. [DOI] [PubMed] [Google Scholar]
- 9.Song S, Wang L, Li J, Fan C, Zhao J. TrAC Trends in Analytical Chemistry. 2008;27:108–117. [Google Scholar]
- 10.Nguyen T, Hilton J, Lin Q. Microfluid Nanofluid. 2009;6:347–362. [Google Scholar]
- 11.Zhou W, Jimmy Huang P-J, Ding J, Liu J. Analyst. 2014;139:2627–2640. doi: 10.1039/c4an00132j. [DOI] [PubMed] [Google Scholar]
- 12.Nutiu R, Li Y. Journal of the American Chemical Society. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Q, Wu M, Chris Le X, Li X-F. TrAC Trends in Analytical Chemistry. 2012;41:46–57. [Google Scholar]
- 14.Chiu TC, Huang CC. Sensors. 2009;9:10356–10388. doi: 10.3390/s91210356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Donolato M, Pinto A, Bosco FG, Hwu ET, Chen CH, Alstrøm TS, Lee GH, Schäfer T, Vavassori P. Biosensors and Bioelectronics. 2016;75:396–403. doi: 10.1016/j.bios.2015.08.062. [DOI] [PubMed] [Google Scholar]
- 16.White RJ, Plaxco KW. 2009 [Google Scholar]
- 17.Du Y, Li B, Guo S, Zhou Z, Zhou M, Wang E, Dong S. Analyst. 2011;136:493–497. doi: 10.1039/c0an00557f. [DOI] [PubMed] [Google Scholar]
- 18.Madru B, Chapuis-Hugon F, Peyrin E, Pichon V. Analytical Chemistry. 2009;81:7081–7086. doi: 10.1021/ac9006667. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Gao D, Zhang P, Gong P, Chen C, Gao G, Cai L. Chemical Communications. 2014;50:811–813. doi: 10.1039/c3cc47649a. [DOI] [PubMed] [Google Scholar]
- 20.Kashefi-Kheyrabadi L, Mehrgardi MA. Biosensors and Bioelectronics. 2012;37:94–98. doi: 10.1016/j.bios.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JJ, Cao JT, Shi GF, Huang KJ, Liu YM, Chen YH. Analytical Methods. 2014;6:6796–6801. [Google Scholar]
- 22.Liu J, Wang C, Jiang Y, Hu Y, Li J, Yang S, Li Y, Yang R, Tan W, Huang CZ. Analytical Chemistry. 2013;85:1424–1430. doi: 10.1021/ac3023982. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Zhu Z, Bamrungsap S, Zhu G, You M, He X, Wang K, Tan W. Analytical Chemistry. 2010;82:10158–10163. doi: 10.1021/ac102277p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Xia J, Li X. Analytical Chemistry. 2008;80:8382–8388. doi: 10.1021/ac800857p. [DOI] [PubMed] [Google Scholar]
- 25.Sharma AK, Kent AD, Heemstra JM. Analytical Chemistry. 2012;84:6104–6109. doi: 10.1021/ac300997q. [DOI] [PubMed] [Google Scholar]
- 26.Sun N, Guo Q, Shao J, Qiu B, Lin Z, Wong KY, Chen G. Analytical Methods. 2014;6:3370–3374. [Google Scholar]
- 27.Zheng D, Zou R, Lou X. Analytical Chemistry. 2012;84:3554–3560. doi: 10.1021/ac300690r. [DOI] [PubMed] [Google Scholar]
- 28.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. Journal of the American Chemical Society. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 29.Huang C-C, Chang H-T. Chemical Communications. 2008:1461–1463. doi: 10.1039/B718752A. [DOI] [PubMed] [Google Scholar]
- 30.Huang KJ, Liu YJ, Shi GW, Yang XR, Liu YM. Sensors and Actuators B: Chemical. 2014;201:579–585. [Google Scholar]
- 31.Lin Z, Chen L, Zhang G, Liu Q, Qiu B, Cai Z, Chen G. Analyst. 2012;137:819–822. doi: 10.1039/c1an15856b. [DOI] [PubMed] [Google Scholar]
- 32.Chandra P, Noh HB, Won MS, Shim YB. Biosensors and Bioelectronics. 2011;26:4442–4449. doi: 10.1016/j.bios.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 33.Chen SJ, Huang YF, Huang CC, Lee KH, Lin ZH, Chang HT. Biosensors and Bioelectronics. 2008;23:1749–1753. doi: 10.1016/j.bios.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT. Journal of the American Chemical Society. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng Q, German I, Buchanan D, Kennedy RT. Analytical Chemistry. 2001;73:5415–5421. doi: 10.1021/ac0105437. [DOI] [PubMed] [Google Scholar]
- 36.Pilehvar S, Rather JA, Dardenne F, Robbens J, Blust R, De Wael K. Biosensors and Bioelectronics. 2014;54:78–84. doi: 10.1016/j.bios.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 37.Feng L, Zhang Z, Ren J, Qu X. Biosensors and Bioelectronics. 2014;62:52–58. doi: 10.1016/j.bios.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Morales D, Madigan J, Cullinane S, Chen J, Heath M, Oz M, Oliver JA, Landry DW. Circulation. 1999;100:226–229. doi: 10.1161/01.cir.100.3.226. [DOI] [PubMed] [Google Scholar]
- 39.He P, Oncescu V, Lee S, Choi I, Erickson D. Analytica Chimica Acta. 2013;759:74–80. doi: 10.1016/j.aca.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 40.Qin H, Liu J, Chen C, Wang J, Wang E. Analytica Chimica Acta. 2012;712:127–131. doi: 10.1016/j.aca.2011.10.044. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen T, Pei R, Landry DW, Stojanovic MN, Lin Q. Sensors and Actuators B: Chemical. 2011;154:59–66. doi: 10.1016/j.snb.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huh YS, Erickson D. Biosensors and Bioelectronics. 2010;25:1240–1243. doi: 10.1016/j.bios.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Palla M, Bosco FG, Rindzevicius T, Alstrøm TS, Schmidt MS, Boisen A, Ju J, Lin Q. ACS Nano. 2013;7:5350–5359. doi: 10.1021/nn401199k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purschke WG, Eulberg D, Buchner K, Vonhoff S, Klussmann S. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5173–5178. doi: 10.1073/pnas.0509663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen T, Pei R, Landry DW, Stojanovic MN, Lin Q. Biomicrofluidics. 2011;5 doi: 10.1063/1.3620417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorm O, Roepstorff P, Mann M. Analytical Chemistry. 1994;66:3281–3287. [Google Scholar]
- 47.Nicola AJ, Gusev AI, Proctor A, Jackson EK, Hercules DM. Rapid Communications in Mass Spectrometry. 1995;9:1164–1171. doi: 10.1002/rcm.1290091216. [DOI] [PubMed] [Google Scholar]
- 48.Tu T, Gross ML. TrAC Trends in Analytical Chemistry. 2009;28:833–841. doi: 10.1016/j.trac.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.PRUSZCZYNSKI W, VIRON B, MIGNON F, ARDAILLOU R. The Journal of Clinical Endocrinology & Metabolism. 1987;64:383–386. doi: 10.1210/jcem-64-2-383. [DOI] [PubMed] [Google Scholar]
- 50.Landry DW, Levin HR, Gallant EM, Ashton RC, Seo S, D’Alessandro D, Oz MC, Oliver JA. Circulation. 1997;95:1122–1125. doi: 10.1161/01.cir.95.5.1122. [DOI] [PubMed] [Google Scholar]
- 51.Borchers A, Magdesian K, Schenck P, Kass P. Equine veterinary journal. 2014;46:306–310. doi: 10.1111/evj.12121. [DOI] [PubMed] [Google Scholar]
- 52.Desbuquois B, Aurbach GD. The Journal of Clinical Endocrinology & Metabolism. 1971;33:732–738. doi: 10.1210/jcem-33-5-732. [DOI] [PubMed] [Google Scholar]
- 53.Sochol RD, Lo L, Ruelos R, Chang V, Bahri D, Iwai K, Lo JC, Dueck M, Lee LP, Lin L. 2011 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.