Abstract
Correct establishment of the left/right (L/R) body asymmetry in the mouse embryo requires asymmetric activation of the evolutionarily conserved Nodal signaling cascade in the left lateral plate mesoderm (L-LPM). Furthermore, the presence of Nodal in the node is essential for its own expression in the L-LPM. Here, we have characterized the function of cerl-2, a novel Nodal antagonist, which displays a unique asymmetric expression on the right side of the mouse node. cerl-2 knockout mice display multiple laterality defects including randomization of the L/R axis. These defects can be partially rescued by removing one nodal allele. Our results demonstrate that Cerl-2 plays a key role in restricting the Nodal signaling pathway toward the left side of the mouse embryo by preventing its activity in the right side.
Keywords: L/R asymmetry, nodal signaling, Cerberus, nodal flow, Cerl-2
Development of the internal organs proceeds across the left/right (L/R) axis and becomes apparent during organogenesis as a result of asymmetric activation of the conserved Nodal signaling cascade in the left lateral plate mesoderm (L-LPM) (for review, see Beddington and Robertson 1999; Capdevila et al. 2000; Hamada et al. 2002). Nodal signaling is a crucial player in the correct establishment of the vertebrate L/R body axis (Capdevila et al. 2000; Wright 2001; Hamada et al. 2002). Nodal expression in the perinodal region of the embryonic day 7.0 (E7.0) mouse embryo has been shown to be required for its own activation in L-LPM and thus generate the asymmetric expression of Nodal's downstream genes (Brennan et al. 2002; Saijoh et al. 2003). Leftward flow in the mouse node (nodal flow) generated by specialized cilia (Nonaka et al. 1998) and intracellular calcium signaling (McGrath et al. 2003) has been recently implicated in the initial steps of lateralization. However, the exact mechanism behind the asymmetric Nodal activity in the node remains largely unexplained (for review, see Hamada et al. 2002).
We have identified a novel Cerberus/Dan family member, mouse cerberus-like2 (cerl-2), that is asymmetrically expressed on the right side of the node. cerl-2 encodes a secreted protein with the capability to bind directly to Nodal and to inhibit its signaling pathway in Xenopus assays. Inactivation of mouse cerl-2 resulted in a wide range of laterality defects including randomization of Nodal expression domain in the LPM. These findings are consistent with an important role of cerl-2 in early events of L/R axis specification. In addition, the observed abnormalities can be partially rescued by the removal of one Nodal allele. Our results demonstrate that Nodal antagonism in the node, mediated by Cerl-2, is essential for proper specification of the mouse L/R axis.
Results and Discussion
Using a sequence-similarity-based search, we identified an incomplete EST sequence (GenBank accession no. AA289243), later also designated Dante (Pearce et al. 1999) and mouse Coco (Bell et al. 2003) as the cDNA most related to mouse cerberus-like (Belo et al. 1997) in the mammalian database. After cloning of the full-length cDNA, we observed that this gene, here designated cerberus-like2 (cerl-2), is located on mouse Chromosome 8, and is genomically organized into two exons, separated by an intron of 5.92 kb. It encodes a 20-kDa protein with a predicted signal peptide sequence and a cysteine-rich domain (CRD) containing nine cysteines characteristic of the Cerberus/DAN family (Supplementary Fig. S1). The CRD domain is essential for the biological function of these proteins (Belo et al. 1997; Hsu et al. 1998), so it is important to note that only in Cerl-2 is this complete domain present, in contrast to what has been previously described for the incomplete sequence of Dante (Pearce et al. 1999).
Cerl-2 has close similarities to mouse cerberus-like (I = 35%, P = 47%), cCaronte (I = 34%, P = 51%), Xcoco (I = 38%, P = 53%), and to a hypothetical human protein (I = 57%, P = 65%; Supplementary Fig. S1). The latter is probably the human homolog of mouse Cerl-2, and we named it human Cer2. It also shares some similarities with the recently described zebrafish Charon (I = 33%, P = 55%).
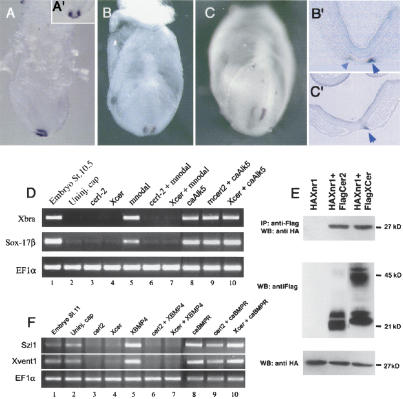
As shown by whole-mount in situ hybridization (WISH), cerl-2 transcripts can be first detected in a horseshoe-shaped expression pattern in the perinodal region of the early head-fold stage of the mouse embryo (E7.0; Fig. 1A,A′), resembling Nodal expression at this stage (Collignon et al. 1996; Lowe et al. 1996). However, by late head-fold stage (E7.5), expression of cerl-2 begins to decrease in intensity on the left side (Fig. 1B,B′), and by early somitogenesis (E8.0), it can be strongly detected in the right side of the node (Fig. 1C,C′), assuming a complementary expression pattern to that of Nodal (Collignon et al. 1996; Lowe et al. 1996). After a thorough in situ hybridization analysis, cerl-2 transcripts could not be found in later stages of mouse development. Thus, cerl-2 is expressed at the proper time and place to be involved in an early L/R symmetry-breaking event in the mouse gastrula.
Figure 1.
Biological activity of the asymmetrically expressed cerl-2. (A-C) cerl-2 expression pattern during early mouse development. (A,A′) Lateral and anterior views, respectively, of cerl-2 expression in the node at E7.0. At E7.5 (B), cerl-2 starts to be asymmetrically up-regulated on the right side of the node, and at E8.0 (C) the asymmetry becomes more evident. B′ and C′ show frontal sections of the embryos in B and C, respectively, and provide a detailed view of the perinodal region where cerl-2 is expressed (arrowheads). (D-F) Inhibitory effects of Cerl-2 on Nodal and BMP signaling. (D) cerl-2 inhibits mNodal but not caALK5 mRNA, as assayed by the induction of their target genes Xbra and Sox-17β. (E) Coimmunoprecipitation experiments showing direct binding of Cerl-2 and Xcer to Xnr1. (F) cerl-2 inhibits XBMP4 but not caBr mRNA, as assayed by the induction of their target genes Szl1 and Xvent1.
Cerl-2 belongs to a family of secreted antagonists that are inhibitors of TGF-β proteins like Nodal and BMPs, and also of Wnts, whose specificity can be unveiled by heterologous assays in the frog embryo (Piccolo et al. 1999; Belo et al. 2000). To assay Cerl-2 activity against endogenous signals, we injected cerl-2 mRNA (750 pg) in the marginal zone of Xenopus embryos at the four-cell stage and then performed WISH for a mesodermal marker (Xbra) at stage 11. We found that embryos injected with cerl-2 mRNA fail to gastrulate properly and form mesoderm (data not shown), indicating a possible interference with endogenous nodal signals. To test this hypothesis, cerl-2 and mNodal mRNAs were coinjected in the animal pole of four-cell-stage embryos, animal caps were explanted at blastula stage, harvested at stage 10.5, and analyzed by RT-PCR for Xbra and Sox17β (a pan-endodermal marker), two prototypic nodal target genes (Belo et al. 2000). To test whether inhibition of mNodal took place upstream or downstream of the Nodal receptor, we performed epistatic experiments with a constitutively active form of the activin receptor (caAlk5). Microinjection of animal caps with either mNodal (50 pg) or caAlk5 (800 pg) (Fig. 1D, lanes 5,8) induced expression of both Xbra and Sox17β. Coinjection of mNodal with cerl-2 mRNA (1 ng) completely blocked mNodal signal (Fig. 1D, lane 6). However, these inductions could not be prevented when cerl-2 was coinjected along with caAlk5, showing that it acts upstream of this Nodal receptor (Fig. 1D, lane 9). A similar experiment was performed to test if cerl-2 could also inhibit BMP4 signaling (Fig. 1F). For this experiment, animal caps were harvested at stage 11, and the downstream targets Szl1 and Xvent1 were analyzed by RT-PCR. Microinjection of XBMP4 (300 pg) or caBr (a constitutively active BMP4 receptor used for the epistatic experiment) alone induced the expression of both Szl1 and Xvent1 (Fig. 1F, lanes 5,8). Coinjection of cerl-2 (1 ng) with caBr (480 pg) did not prevent Szl1 and Xvent1 expression (Fig. 1F, lane 9), whereas coinjection of Cerl-2 with XBMP4 inhibited the expression of both markers (Fig. 1F, lane 6). In both experiments Xcer (800 pg) was injected as a control as it is known to inhibit both Nodal and BMP4 signaling. Taken together, these results show that full-length cerl-2 inhibits nodal and BMP4 but not caAlk5 or caBr signaling (Fig. 1D,F), suggesting that Cerl-2 might antagonize Nodal and BMP4 extracellularly.
To determine whether Cerl-2 is able to physically interact with TGFβ proteins, we coinjected synthetic mRNAs encoding a Flag-tagged version of Cerl-2 together with an HA-tagged version of Xnr1 into animal poles. Extracts were immunoprecipitated with anti-Flag antibody and the coprecipitating proteins were analyzed by anti-HA Western blotting. As shown in Figure 1E, HAXnr1 protein was found in a complex with FlagCerl2. All the in vitro experiments—(1) the inhibition of Nodal and its downstream targets by Cerl-2; (2) its activity being upstream of the Nodal receptor; and (3) the biochemical assay showing a physical interaction between the two proteins—suggest that Cerl-2 is a novel Nodal antagonist.
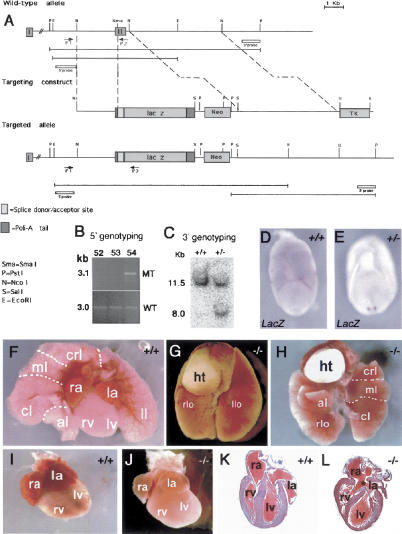
To determine the in vivo role of cerl-2, we inactivated this gene in ES cells by replacing the second exon (containing the core CRD) with a LacZ reporter cassette (Fig. 2A). WISH in E7.5 cerl-2+/- embryos using a LacZ probe revealed that its expression is also asymmetrical in the right side of the node (Fig. 2D,E). The offspring of Cerl-2+/- intercrosses were born according to the correct Mendelian ratio. We observed that 35% of the homozygous mutants (33/94) died within the first 48 h after birth. From those animals that died perinatally (n = 33), 18% (6/33) showed left pulmonary isomerism (Fig. 2G), 18% (6/33) thoracic situs inversus (Fig. 2H), and the remaining 64% (21/33) failed to show any apparent laterality defect. However, when these apparently unaffected animals were examined carefully by histological analysis, it was observed that they displayed cardiovascular malformations (Fig. 2J,L), these being their probable cause of death. These defects include incomplete atrial (Fig. 2L) and ventricular septation (data not shown). Of the 65% mutant animals that survived (61/94), 40% become normal adults (37/94), and the remaining 25% (24/94) die between weaning age and 3 mo old, most of them showing heterotaxia of the abdominal organs (Supplementary Fig. S2).
Figure 2.
Targeted inactivation of cerl-2 gene. (A) Schematic representation of the wild-type cerl-2 locus, targeting vector, and targeted allele. The positions of primers, restriction enzyme sites, and the probe used for PCR and Southern blot analysis, respectively, are shown. (B) PCR-based 5′-genotyping of wild-type and targeted ES cell DNA. (C) Genomic Southern blot of PstI-digested tail DNA, prepared from newborn offspring of a mating of heterozygous mice. (D,E) LacZ in situ hybridization in wild-type (D) and heterozygous (E) embryos. (F-H) Thoracic organs of newborn wild-type (F) and cerl-2-/- littermates displaying left lung isomerism (G) or inverted situs (H). (I,J) Hearts of newborn wild-type and cerl-2-/- littermates, respectively. (K,L) Frontal sections of the hearts depicted in I and J, respectively, showing the atrial septal defects (asterisk) in cerl-2-/-. (al) Accessory lobe; (cl) caudal lobe; (crl) cranial lobe; (ht) heart; (ml) middle lobe; (la) left atrium; (llo) left lobe; (lv) left ventricle; (ra) right atrium; (rlo) right lobe; (rv) right ventricle.
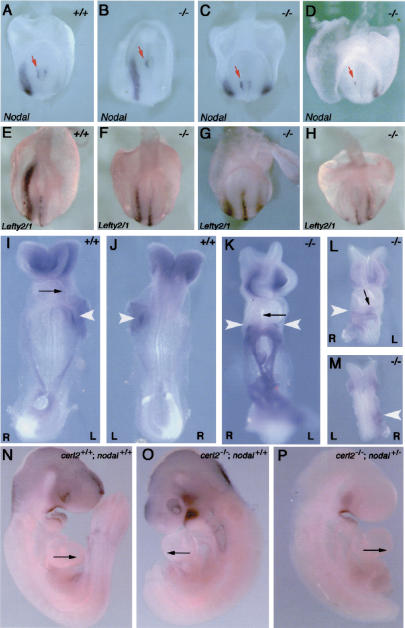
Because of its expression pattern and its Nodal inhibitory activity, together with the observed laterality defects, we decided to investigate the effect of the loss of cerl-2 in the expression of left-right determinant genes. At early somite stages, Nodal, Lefty1, Lefty2, and Pitx2 are expressed in the left side of the embryo and are part of an evolutionarily conserved signaling cascade essential for correct L/R morphogenesis (Capdevila et al. 2000; Wright 2001; Hamada et al. 2002). At E8.0, Nodal is expressed in the node and in the L-LPM of wild-type embryos (Fig. 3A). WISH in cerl-2-/- embryos (N = 60) using a Nodal probe revealed that although 40% of the embryos showed normal expression, 50% displayed bilateral expression, and 10% had an inverted pattern of expression in the R-LPM (Fig. 3B-D). Interestingly, Nodal expression in the node remains unaffected in these three situations, always being stronger in the left side (red arrows in Fig. 3A-D). This evidence further supports previously described work in which Nodal asymmetric expression in the node is reported not to be necessary or linked to its later expression domain in the L-LPM (Brennan et al. 2002; Saijoh et al. 2003). At this same developmental stage, Lefty1 is expressed along the ventral midline in the prospective floor plate of the wild-type embryo (Fig. 3E), whereas Lefty2 expression can be detected in the L-LPM (Fig. 3E; Meno et al. 1997). Both genes, Lefty1 and Lefty2, have been described to be downstream targets of Nodal and also its inhibitors (for review, see Hamada et al. 2002). By performing WISH using a riboprobe that detects both Lefty genes, we found that in cerl-2-/- embryos, Lefty2 expression in the LPM is very similar to that of Nodal, as it would be expected. It can be detected in the left LPM, bilaterally or in the R-LPM (Fig. 3F-H). We could also observe that, in cerl-2-null mutants, Lefty1 expression in the midline is not affected (Fig. 3F-H). Lefty1 was proposed to function as a midline barrier to prevent the diffusion of Nodal-induced signals emanating from the left to the right LPM (Meno et al. 1998). The proper expression of Lefty1 (Fig. 3F-H) and Shh (data not shown) in the prospective floorplate leads us to conclude that the abnormal expression of left determinant genes in cerl-2-/- embryos is not caused by midline defects. Furthermore, 10% of cerl-2-/- embryos show right-sided ectopic expression of Nodal (Fig. 3D). In addition, in embryos with bilateral Nodal expression in the LPM, this expression starts at the level of the node (Fig. 3C). Taken together, these results strongly indicate that the leaking of left-side determinants takes place at the level of the node, and not later through to a defectively patterned midline.
Figure 3.
cerl-2 null mutants display a range of L/R defects. (A-H) Posterior views of E8.0 embryos. (A) Wild-type Nodal expression in the node and left LPM. (B-D) cerl-2-/- embryos showing normal (B), bilateral (C), and right-sided (D) expression of Nodal in the LPM. (E) Wild-type Lefty2/1 expression in the left LPM and floorplate. (F-H) cerl-2-/- embryos displaying normal (F), bilateral (G), and right-sided (H) expression of Lefty2 in the LPM. (I,J) Ventral and dorsal views, respectively, of E8.5 wild-type embryos showing Pitx2 on the left LPM. (K-M) cerl-2-/- embryos presenting bilateral (K) and right-sided (L,M) expression of Pitx2 in the LPM. White arrowheads indicate the expression domain of Pitx2. (N-P) Rescue of cerl-2 phenotype by reduced Nodal activity. (N) E9.5 wild-type embryo showing rightward looping of the heart. (O) cerl-2-/- littermate with a reversed heart loop. (P) cerl-2-/-; nodal+/- compound mutant with normal looping of the heart.
Pitx2 is a downstream target of Nodal that is responsive to Nodal signaling through an asymmetric enhancer (Shiratori et al. 2001), similar to Lefty2 and Nodal (Adachi et al. 1999; Norris and Robertson 1999; Saijoh et al. 1999). At E8.5, Pitx2 expression can be detected in the left LPM (Fig. 3I,J; Ryan et al. 1998), and it has been shown to be required for asymmetric development of organ situs (Gage et al. 1999; Kitamura et al. 1999; Lin et al. 1999; Lu et al. 1999). In cerl-2-/- embryos, Pitx2 expression in the LPM (N = 11) could be detected bilaterally (38%), in the left (55%), or in the right side (9%) (Fig. 3K-M).
The L/R-determining genetic cascade leads to morphological consequences in the positioning of internal organs, and the first morphological manifestation of L/R axis determination is the orientation of embryonic heart looping (Fujinaga 1997). In wild-type mouse embryos, the linear heart tube loops rightward, whereas 54% (27/50) of cerl-2-/- mice exhibited leftward or ventral heart looping (Fig. 3K,L). This comes in agreement with the previous experiments in which we show that Cerl-2 activity is essential for the correct establishment of the left-side determinant genes, and, therefore, it is also necessary for the correct asymmetric development of the organ situs. In fact, besides the mentioned defects in heart looping, later developmentally associated phenotypes like left isomerism, situs inversus, and cardiac malformation were also observed (Fig. 2F-L).
The phenotype of cerl-2 mutants suggests that Nodal activity is increased in the node, leading to abnormal expression of Nodal and its downstream targets in the LPM. Therefore, we hypothesized that by removing one copy of the Nodal gene, in the context of the cerl-2 mutant, Nodal activity in the node would be lowered to a condition more similar to the wild type. To test this, we intercrossed cerl-2-/- with cerl-2+/-; Nodal+/- animals, recovered the embryos at E9.5, and scored them according to the heart loop direction. Indeed, we could observe that in cerl-2-/-; nodal+/- embryos (N = 20), the abnormal heart-looping phenotype was 35% (7/20) (Fig. 3P). Interestingly, from the crosses between cerl-2-/- × cerl-2+/-; Nodal+/- mutants, we also obtained 17 cerl-2-/- embryos, and of these nine (52%) showed a heart-looping phenotype, which suggests that although the number of these embryos is less than half those obtained from cerl-2-/- crosses, the percentage of defective heart loops remains fairly invariable. This observation (although not statistically significant owing to the relatively small number of embryos analyzed) suggests a qualitative tendency to partially rescue this phenotype. This tendency, as a complement of the other lines of evidence reported here, is in accordance with the proposed mechanisms for Cerl-2 activity. Taken together, our results suggest that the phenotypes in L/R axis determination observed in cerl-2-/- embryos may result from an excess of Nodal signaling because of the lack of the Cerl-2 anti-Nodal activity in the mouse node.
In vitro biochemical assays showed that the Cerl-2 molecule also has the ability of inhibiting BMP-4. The lack of this potential inhibition in the mutants is, however, unlikely to be correlated with the in vivo observed phenotypes. Other studies in which BMP-4 antagonism was absent from the node, as in the targeted inactivation of Chordin (Bachiller et al. 2003) and Noggin (McMahon et al. 1998), did not result in L/R asymmetry-related phenotypes.
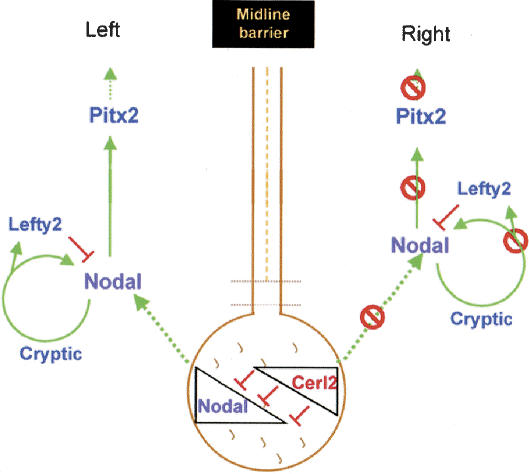
A gene named Charon that might be an ortholog of cerl-2 has recently been described in zebrafish. In zebrafish, expression of Southpaw and Charon is not asymmetric. Nevertheless, knockdown of Charon using a morpholino oligonucleotide produces a similar phenotype to that of cerl-2 mutants (Hashimoto et al. 2004). Although in zebrafish the breaking of L/R symmetry is still not very well understood, this unveils a possible conserved evolutionary mechanism of Nodal antagonism in the node, mediated by Cerberus/DAN family members, essential for the correct development of the L/R axis. In the mouse, Nodal activity in the node is required for Nodal expression in the L-LPM (Brennan et al. 2002; Saijoh et al. 2003) and subsequent activation of Lefty2 and Pitx2. The role we propose for cerl-2 is to restrict Nodal activity to the left side of the node. The consequent effect of such inhibition is to prevent additional activation of Nodal, Lefty2, and Pitx2 in the R-LPM (Fig. 4). In the absence of Cerl-2 antagonistic activity on the node, Nodal may be also activated in the R-LPM, leading to bilateral or ectopic expression of this genetic cascade in the R-LPM. In addition, we noticed that Nodal expression on the node is still asymmetric on the left side of cerl-2 mutants, indicating that cerl-2 doesn't affect the asymmetric expression of Nodal in the node. Interestingly, cerl-2 expression in the node also seems to be independent from Nodal. In fact in Nodalneo/neo mutants that lack (or have low levels of) Nodal expression in the node, cerl-2 (Dante) expression domains remain unchanged (Saijoh et al. 2003).
Figure 4.
Proposed model for the role of cerl-2 in generating asymmetric gene expression. cerl-2 restricts nodal activity to the left side of the node, preventing additional activation of Nodal, Lefty2, and Pitx2 in the R-LPM.
The maintenance of asymmetric expression of Nodal in the node of cerl-2 mutants, and later its randomized expression in the LPM, suggests that it may be uncoupled from asymmetric gene expression within the node, as previously described (Brennan et al. 2002; Saijoh et al. 2003). Although the relevance of Nodal asymmetric expression in the node as the cause of its later expression domain in the LPM remains unexplained and controversial (Saijoh et al. 2003), our data highlight the importance of tight regulation of Nodal activity in the node by the extracellular antagonism mediated by Cerl-2 as an integral part of the L/R program.
The first known event for inducing asymmetry in Nodal expression is the concerted rotational movement of cilia of the node pit cells (the nodal flow); indeed, in iv/iv mice that lack such oriented fluid flow, Nodal has a randomized asymmetric expression in the node (Supp et al. 1999).
Paradoxically, despite the different expression pattern of Nodal in the node, of the iv/iv (Supp et al. 1999) and cerl-2 mutants, they end up displaying a very similar type of randomization, characterized by left, right, or bilateral activation of the “leftness” program in the LPM. This indicates that Cerl-2 plays an important role in the early events of symmetry breaking that take place in the node. Our data suggest a possible explanation in which the L/R asymmetry is controlled by a double-assurance mechanism consisting of two parallel systems, the first relying on the leftward nodal-cilia flow, and the second, on the antagonism between Cerl-2 and Nodal described by the proposed model (Fig. 4).
Materials and methods
Cloning of the full-length CDNA
A cDNA clone (GenBank accession no. AA289245) containing the second exon and the 3′-UTR of cerl-2 was obtained by database search for proteins similar to mCer-l. A hypothetical human protein (FLJ38607) was found by searching the NCBI database for proteins homologous to cerl-2 and its corresponding cDNA BLASTed against the mouse genome. A primer designed to align in the predicted 5′-UTR (5′-CGGAATTCCGCCAGAAAACAACTCTCAAGCTGCTCTCC-3′) and a reverse primer complementary to the second exon of cerl-2 (5′-CCACACCACAGCGTCACCGATGTCCAGC-3′) were used in an RT-PCR with E7.5 mouse total RNA. The resulting 5′-cDNA portion was subcloned into a pGEMTeasy vector, and the full-length cDNA was assembled into pCS2+ plasmid and sequenced. The GenBank accession number for mouse cerberus-like2 is AY387409.
Targeted disruption of the cerl-2 gene
A mouse 129/Ola genomic library was screened for cerl-2 using a partial cDNA clone (GenBank accession no. AA289245), and two positive clones were obtained.
In our targeting vector, a 0.6-kb SmaI/NcoI DNA fragment containing the second exon was replaced with a neomycin and a lacz cassette. The linearized vector was electroporated into 129/Ola embryonic stem cells. One heterozygous embryonic stem cell clone was used to generate chimeric mice by blastocyst injection, and mutant animals were bred in both 129/Ola and 129/Ola × C57BL/6J mixed backgrounds. Genotyping was done by Southern blotting and PCR assays. For Southern blot analysis, genomic DNA was digested with PstI and hybridized with a 3′-probe. The 3′-probe was a 1.0-kb (PstI/XbaI) genomic DNA fragment downstream from the 3′ recombination arm. Primers for PCR analysis were P1 (5′-GGAACCACCTTTGTAGTCAAGACTGG-3′), P2 (5′-GGTGACTTCTTTTTTGCTTTAGCAGG-3′), and P3 (5′-CACACAGCTGTTGCAGAAGAC-3′).
Generation and genotyping of single and compound mutants
cerl-2+/- heterozygous mice were intercrossed with nodal+/- heterozygous mice (both of C57/B6 background), originating cerl-2-/- or double heterozygous animals. The latter were crossed to cerl-2+/- or cerl-2-/- mice to obtain cerl-2-/-; nodal+/- mutants. DNA preparation for genotyping was performed as described previously (Lowe et al. 1996) and analyzed by PCR using the following three oligonucleotides for cerl-2: AA289c-P3 (5′-CACACAGCTGTTGCAGAAGAC-3′), GenCer2-fwd (5′-GGAAGATTTTATGCAAGCAAGAGTGTGG-3′), and Lower2 (5′-GGTGACTTCTTTTTTGCTTTAGCAGG-3′), which resulted in bands of 300 bp and 500 bp for the wild-type and mutant alleles, respectively. nodal genotyping was performed as described previously (Bachiller et al. 2000).
Whole-mount in situ hybridization and histology
Whole-mount in situ hybridization and antisense probe preparation was carried out as described previously (Belo et al. 1997).
Detailed descriptions of the RNA probes and constructs used are available from the authors on request.
mRNA synthesis, microinjection and RT-PCR analysis
Capped sense mRNAs were synthesized using Ambion mMessage mMachine kit. In vitro fertilization, microinjection of Xenopus laevis embryos, and RT-PCR analysis were performed as described previously (Bouwmeester et al. 1996; Belo et al. 2000). The primer sets used are described in http://www.hhmi.ucla.edu/derobertis/protocol_page/oligos.PDF. For all RT-PCR reactions, ef1 was used as the loading control. Detailed descriptions of the expression constructs used are available from the authors on request.
Coimmunoprecipitation analysis
A Flag-tagged version of cerl2 was constructed by standard PCR methods and subcloned in pCS2+, using the abovementioned forward primer and the following reverse primer: 5′-CCGCTCGAGTCACTTATCGTCGTCATCCTTGTAATCTCCTCCTCCCAGCTTCGGGCGGCACTGACACTTCTGG-3′. One nanogram of HAXnr1 mRNA and 5 ng of Flagcerl-2 mRNA were injected into the animal poles of Xenopus embryos, and coimmunoprecipitation was performed as previously described (Yeo and Whitman 2001). Anti-Flag mouse monoclonal antibody (Sigma) and anti-HA rabbit polyclonal antibody (Covance) were used for immunoprecipitation and Western blot analysis. Proteins were visualized using ECLWestern blotting detection reagents (Amersham Pharmacia Biotech).
Acknowledgments
We thank M.R. Kuehn for Nodal mutant mice; J.C. Izpisúa-Belmonte and G. Persico for probes; S. Piccolo for protein work; H. Steinbeisser for plasmids and Xenopus work; and P. Vieira for help with ES cell culture. We thank S. Piccolo, A. Tavares, A.T. Tavares, M. Filipe, J. Leon, and W. Wood for critically reading of this manuscript. S.M., A.C.B., A.C.S., and S.F. are recipients of F.C.T. fellowships. This work was supported by research grants from F.C.T. and IGC/Fundação Calouste Gulbenkian to J.A.B.; J.A.B. is a Principal Investigator at the IGC/Fundação Calouste Gulbenkian.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.306504.
References
- Adachi H., Saijoh, Y., Mochida, K., Ohishi, S., Hashiguchi, H., Hirao, A., and Hamada, H. 1999. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a Lefty-2 enhancer. Genes & Dev. 13: 1589-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller D., Klinggensmith, J., Kemp, C., Belo, J.A., Anderson, R.M., May, S.R., MacMahon, J.A., McMahon, A.P., Harland, R.M., Rossant, J., et al. 2000. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature 403: 658-661. [DOI] [PubMed] [Google Scholar]
- Bachiller D., Klingensmith, J., Shaneyder, N., Tran, U., Anderson, R., Rossant, J., and De Robertis, E.M. 2003. The role of chordin/Bmp signals in mammalian pharyngeal development and DiGeorge syndrome. Development 130: 3567-3578. [DOI] [PubMed] [Google Scholar]
- Beddington R.S. and Robertson, E.J. 1999. Axis development and early asymmetry in mammals. Cell 96: 195-209. [DOI] [PubMed] [Google Scholar]
- Bell E., Munoz-Sanjuan, I., Altmann, C.R., Vonica, A., and Brivanlou, A.H. 2003. Cell fate specification and competence by Coco, maternal BMP, TGFβ and Wnt inhibitor. Development 130: 1381-1389. [DOI] [PubMed] [Google Scholar]
- Belo J.A., Bouwmeester, T., Leyns, L., Kertesz, N., Gallo, M., and De Robertis, E.M. 1997. Cerberus-like is a secreted factor with neuralizing activity expressed in the anterior primitive streak endoderm of the mouse gastrula. Mech. Dev. 68: 45-57. [DOI] [PubMed] [Google Scholar]
- Belo J.A., Bachiller, D., Agius, E., Kemp, C., Borges, A.C., Marques, S., Piccolo, S., and De Robetis, E.M. 2000. Cerberus-like is a secreted BMP and nodal antagonist not essential for mouse development. Genesis 26: 265-270. [PubMed] [Google Scholar]
- Bouwmeester T., Kim, S., Sasai, Y., Lu, B., and De Robertis, E.M. 1996. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature 382: 595-601. [DOI] [PubMed] [Google Scholar]
- Brennan J., Norris, D.P., and Robertson, E.J. 2002. Nodal activity in the node governs left-right asymmetry. Genes & Dev. 16: 2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J., Vogan, K.J., Tabin, C.J., and Izpisua-Belmonte, J.C. 2000. Mechanisms of left-right determination in vertebrates. Cell 101: 9-21. [DOI] [PubMed] [Google Scholar]
- Collignon J., Varlet, I., and Robertson, E.J. 1996. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381: 155-158. [DOI] [PubMed] [Google Scholar]
- Fujinaga M. 1997. Development of sidedness of asymmetric body structures in vertebrates. Int. J. Dev. Biol. 41: 153-186. [PubMed] [Google Scholar]
- Gage P.J., Suh, H., and Camper, S.A. 1999. Dosage requirement of Pitx2 for development of multiple organs. Development 126: 4643-4651. [DOI] [PubMed] [Google Scholar]
- Hamada H., Meno, C., Watanabe, D., and Saijoh, Y. 2002. Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 31: 103-113. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rebagliati, M., Ahmad, N., Muraoka, O., Kurokawa, T., Hibi, M., and Suzuki, T. 2004. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development 131: 1741-1753. [DOI] [PubMed] [Google Scholar]
- Hsu D.R., Economides, A.N., Wang, X., Eimon, P.M., and Harland, R.M. 1998. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell 1: 673-683. [DOI] [PubMed] [Google Scholar]
- Kitamura K., Miura, H., Miyagawa-Tomita, S., Yanazawa, M., Katoh-Fukui, Y., Suzuki, R., Ohuchi, H., Suehiro, A., Motegi, Y., Nakahara, Y., et al. 1999. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 126: 5749-5758. [DOI] [PubMed] [Google Scholar]
- Lin C.R., Kioussi, C., O'Connell, S., Briata, P., Szeto, D., Liu, F., Izpisua-Belmonte, J.C., and Rosenfeld, M.G. 1999. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401: 279-282. [DOI] [PubMed] [Google Scholar]
- Lowe L.A., Supp, D.M., Sampath, K., Yokoyama, T., Wright, C.V., Potter, S.S., Overbeek, P., and Kuehn, M.R. 1996. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature 381: 158-161. [DOI] [PubMed] [Google Scholar]
- Lu M.F., Pressman, C., Dyer, R., Johnson, R.L., and Martin, J.F. 1999. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401: 276-278. [DOI] [PubMed] [Google Scholar]
- McGrath J., Somlo, S., Makova, S., Tian, X., and Brueckner, M. 2003. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114: 61-73. [DOI] [PubMed] [Google Scholar]
- McMahon J.A., Takada, S., Zimmerman, L.B., Fan, C.M., Harland, R.M., and McMahon, A.P. 1998. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes & Dev. 12: 1438-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meno C., Ito, Y., Saijoh, Y., Matsuda, Y., Tashiro, K., Kuhara, S., and Hamada, H. 1997. Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: Their distinct expression domains, chromosomal linkage and direct neuralizing activity in Xenopus embryos. Genes Cells 2: 513-524. [DOI] [PubMed] [Google Scholar]
- Meno C., Shimono, A., Saijoh, Y., Yashiro, K., Mochida, K., Ohishi, S., Noji, S., Kondoh, H., and Hamada, H. 1998. Lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 94: 287-297. [DOI] [PubMed] [Google Scholar]
- Nonaka S., Tanaka, Y., Okada, Y., Takeda, S., Harada, A., Kanai, Y., Kido, M., and Hirokawa, N. 1998. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95: 829-837. [DOI] [PubMed] [Google Scholar]
- Norris D.P. and Robertson, E.J. 1999. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes & Dev. 13: 1575-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce J.J., Penny, G., and Rossant, J. 1999. A mouse cerberus/Dan-related gene family. Dev Biol. 209: 98-110. [DOI] [PubMed] [Google Scholar]
- Piccolo S., Agius, E., Leyns, L., Bhattacharyya, S., Grunz, H., Bouwmeester, T., and De Robertis, E.M. 1999. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397: 707-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan A.K., Blumberg, B., Rodriguez-Esteban, C., Yonei-Tamura, S., Tamura, K., Tsukui, T., de la Pena, J., Sabbagh, W., Greenwald, J., Choe, S., et al. 1998. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature 394: 545-551. [DOI] [PubMed] [Google Scholar]
- Saijoh Y., Adachi, H., Mochida, K., Ohishi, S., Hirao, A., and Hamada, H. 1999. Distinct transcriptional regulatory mechanisms underlie left-right asymmetric expression of lefty-1 and lefty-2. Genes & Dev. 13: 259-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijoh Y., Oki, S., Ohishi, S., and Hamada, H. 2003. Left-right patterning of the mouse lateral plate requires nodal produced in the node. Dev. Biol. 256: 160-172. [DOI] [PubMed] [Google Scholar]
- Shiratori H., Sakuma, R., Watanabe, M., Hashiguchi, H., Mochida, K., Sakai, Y., Nishino, J., Saijoh, Y., Whitman, M., and Hamada, H. 2001. Two-step regulation of left-right asymmetric expression of pitx2: Initiation by nodal signaling and maintenance by Nkx2. Mol. Cell 7: 137-149. [DOI] [PubMed] [Google Scholar]
- Supp D.M., Brueckner, M., Kuehn, M.R., Witte, D.P., Lowe, L.A., MacGrath, J., Corrales, J., and Potter, S.S. 1999. Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries. Development 126: 5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C.V. 2001. Mechanisms of left-right asymmetry: What's right and what's left? Dev. Cell 1: 179-186. [DOI] [PubMed] [Google Scholar]
- Yeo C. and Whitman, M. 2001. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell 7: 949-957. [DOI] [PubMed] [Google Scholar]