Abstract
The Schizosaccharomyces pombe genome encodes only one of each of the three major classes of proteins implicated in RNA silencing: Dicer (Dcr1), RNA-dependent RNA polymerase (RdRP; Rdp1), and Argonaute (Ago1). These three proteins are required for silencing at centromeres and for the initiation of transcriptionally silent heterochromatin at the mating-type locus. Here, we show that the introduction of a double-stranded RNA (dsRNA) hairpin corresponding to a green fluorescent protein (GFP) transgene triggers classical RNA interference (RNAi) in S. pombe. That is, GFP silencing triggered by dsRNA reflects a change in the steady-state concentration of GFP mRNA, but not in the rate of GFP transcription. RNAi in S. pombe requires dcr1, rdp1, and ago1, but does not require chp1, tas3, or swi6, genes required for transcriptional silencing. Thus, the RNAi machinery in S. pombe can direct both transcriptional and posttranscriptional silencing using a single Dicer, RdRP, and Argonaute protein. Our findings suggest that these three proteins fulfill a common biochemical function in distinct siRNA-directed silencing pathways.
Keywords: RNAi, PTGS, TGS, siRNA, Argonaute, Dicer
In many eukaryotic cells, exogenous long double-stranded RNA (dsRNA) triggers the specific degradation of cellular mRNAs with corresponding sequences, a phenomenon termed RNA interference (RNAi) (Fire et al. 1998). The dsRNA is cleaved by the multidomain ribonuclease III enzyme, Dicer, into a population of 21-27-nt dsRNAs termed small interfering RNAs (siRNAs) (Bernstein et al. 2001). siRNAs are the specificity determinants of the RNAi pathway (Hamilton and Baulcombe 1999; Hammond et al. 2000; Zamore et al. 2000). siRNAs are assembled into a protein-RNA complex, the RNA-induced silencing complex (RISC), which directs cleavage of the target RNA (Hammond et al. 2000; Zamore et al. 2000; Nykänen et al. 2001). Among the various protein factors required for RNAi, members of the Argonaute family of proteins are universally associated with siRNA-directed gene silencing (Cogoni and Macino 1997; Tabara et al. 1999, 2002; Cerutti et al. 2000; Fagard et al. 2000; Catalanotto et al. 2002; Kennerdell et al. 2002; Mochizuki et al. 2002; Pal-Bhadra et al. 2002; Tijsterman et al. 2002; Williams and Rubin 2002; Shi et al. 2004). Plants and animals contain multiple Argonaute paralogs; the Drosophila genome encodes at least five distinct Argonaute proteins; humans have eight; and worms, 27! The remarkable diversity of Argonaute proteins suggests that each Argonaute paralog plays a specific role in RNA silencing. The Ago2 protein is a core-component of biochemically purified RISC from both Drosophila (Hammond et al. 2001) and mammals (Hutvágner and Zamore 2002; Martinez et al. 2002). In fact, among the human Argonaute proteins, only Ago2 can mediate microRNA (miRNA)- or siRNA-directed endonucleolytic cleavage of target RNA (Liu et al. 2004; Meister et al. 2004). The recent three-dimensional structure of an archeal Argonaute protein, together with experiments evaluating the importance of predicted catalytic residues in human Ago2, suggests that human Ago2 itself is the small RNA-guided endoribonuclease that cleaves target RNA (Liu et al. 2004; Song et al. 2004). Other human Argonaute proteins may be specialized to direct translational repression of mRNA or transcriptional silencing of DNA sequences by the small RNA-directed production of silent heterochromatin. Such functional specialization might also extend to the animal miRNA pathway, in which small RNAs typically direct translational repression, but not destruction, of their target mRNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). In Caenorhabditis elegans, the Argonaute protein Rde-1 is required for RNAi but not for miRNA function, whereas the Argonaute proteins Alg-1 and Alg-2 are required for miRNA function but not RNAi (Tabara et al. 1999; Grishok et al. 2001). In Drosophila, Ago2 function is restricted to the siRNA-directed posttranscriptional gene silencing (PTGS) pathway, whereas miRNA function is associated with Ago1 (Okamura et al. 2004). In Arabidopsis thaliana, AGO1 is required for miRNA function and PTGS, whereas AGO4 acts in a silencing pathway that targets chromatin, rather than mRNA (Fagard et al. 2000; Morel et al. 2002; Zilberman et al. 2003; Vaucheret et al. 2004).
In contrast to the multiplicity of Argonaute proteins in higher organisms, the sequenced fungal genomes appear to encode one (Neurospora crassa and Schizosaccharomyces pombe) or no Argonaute proteins (Saccharomyces cerevisiae). The Neurospora Argonaute protein QDE-2 is required for quelling, an RNAi-like phenomenon (Cogoni and Macino 1997) and is a component of an siRNA-containing complex (Catalanotto et al. 2002). In contrast, Ago1 is required for the silencing of transcription at centromeres in the fission yeast S. pombe (Volpe et al. 2002) and initiation of silent heterochromatin at the mating type locus (Hall et al. 2002). S. pombe Ago1 is a component of the RNA-induced initiation of transcriptional silencing (RITS) complex (Verdel et al. 2004). In addition to Ago1, the RITS contains siRNAs derived from centromeric sequences, as well as the chromodomain protein Chp1 and the protein Tas3, whose function is unknown.
Current evidence suggests that the RITS uses the sequence information in its siRNA component to direct the methylation of Lys 9 (K9) of histone H3 bound to centromeric DNA (Verdel et al. 2004). Histone H3 K9 methylation, in turn, triggers the formation of transcriptionally silent heterochromatin, a process dependent on the histone methyltransferase Clr4 and a second chromodomain protein, Swi6 (Elgin and Grewal 2003). The initiation of silent heterochromatin requires not only the RITS complex, but also Dcr1, the S. pombe homolog of Dicer, and a putative RNA-dependent RNA polymerase, Rdp1 (Hall et al. 2002). By analogy to silencing in higher organisms, Dcr1 is presumed to generate siRNA from dsRNA derived from centromeric DNA. Rdp1 has been postulated either to generate this dsRNA itself or to amplify the production of siRNA, but the precise function of RNA-dependent RNA polymerases in siRNA-directed silencing is not yet understood.
In S. pombe, as in other eukaryotes, silencing can be triggered by the introduction of exogenous long dsRNA transcribed from transgenic DNA. Such dsRNA can silence both endogenous (Schramke and Allshire 2003) and exogenous genes (Raponi and Arndt 2003), although it has not been determined whether this silencing is transcriptional—like the endogenous silencing of centromeric DNA—or posttranscriptional, as is observed in the RNAi pathway in animals and plants. Grewal, Martienssen, and colleagues (Volpe et al. 2002) have shown that the production of heterochromatin at the S. pombe centromere contains features characteristic of both transcriptional and posttranscriptional silencing. Forward transcription at this locus appears to be repressed by the formation of heterochromatin, yet reverse transcription through the same locus is not, suggesting that reverse transcripts are degraded posttranscriptionally. To address the question of whether PTGS exists in S. pombe in the absence of transcriptional gene silencing (TGS), we developed a simplified silencing system based on green fluorescent protein (GFP) expression. Here, we show that the introduction of a dsRNA hairpin corresponding to a GFP transgene triggers classical RNAi in S. pombe: introduction of GFP dsRNA causes a change in the steady-state concentration of GFP mRNA, but not the rate of GFP transcription. RNAi in S. pombe requires Ago1, Dcr1, and Rdp1, but does not require Chp1, Tas3, or Swi6, which are required for transcriptional silencing. Our data suggest that Argonaute, Dicer, and RdRP play common biochemical roles in functionally distinct silencing pathways.
Results
To test whether dsRNA triggers RNAi in S. pombe, we engineered a strain containing the enhanced GFP (gfp) protein coding sequence fused in-frame to the endogenous adh1 gene. PCR analysis (data not shown) verified that the strain contained the adh1:gfp fusion gene integrated at the adh1 locus on chromosome 3. The fusion mRNA encoded by this locus was the only source of adh1 mRNA in the strain. The strain also contained the kanamycin resistance gene (aph, encoding the enzyme aminoglycoside-3′-phosphotransferase) downstream from adh1:gfp, with 628 bp separating the start of aph open reading frame (ORF) from the end of the gfp ORF.
To trigger silencing of adh1:gfp, we engineered a plasmid-borne GFP hairpin (Fig. 1). The 760-bp gfp ORF was cloned as an inverted repeat, with the sense and antisense arms of the repeat separated by a 67-bp spacer containing the first intron of the rad9 gene. The intron was included, because intron-containing hairpin RNAs induce PTGS in plants with nearly 100% efficiency (Smith et al. 2000). The construct used here, when spliced, is presumed to leave a loop of 14 unpaired nucleotides (nt). Transcription of the GFP hairpin was under the control of the thiamine-repressible nmt1 promoter; in the absence of thiamine, nmt1 is among the strongest promoters in S. pombe (Moreno et al. 1991). The plasmid also contained the ura4 gene, to permit selection for retention of the plasmid in the absence of uracil. (The reporter strain contains a ura4 mutation and thus cannot synthe-size uracil.) Expression of the GFP hairpin in the ura4-strain had no effect on cell morphology or growth rate (data not shown).
Figure 1.
Experimental strategy. The coding sequence of GFP was fused in frame with the adh1 locus on chromosome 3. The kanamycin resistance gene (aph) was inserted adjacent to the adh1:gfp transgene as an insertion marker under the control of the Ashbya gossypii translation elongation factor 1α gene promoter. Red arrows indicate the position of adh1:gfp-specific primers used in the quantitative RT-PCR assays in Figure 3. The silencing trigger was expressed from an episomal plasmid encoding a hairpin transcript corresponding to GFP expressed from the nmt1 promoter. The GFP dsRNA hairpin contained within the loop sequences the 67-bp intron 1 from rad9 to facilitate hairpin expression.
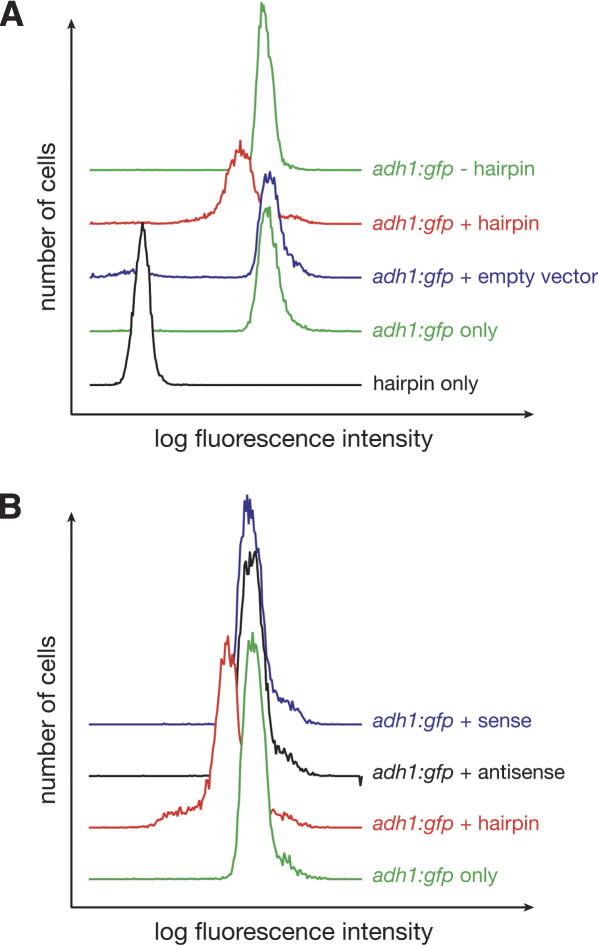
When the plasmid encoding the GFP hairpin was introduced into the adh1:gfp target strain, the GFP fluorescence intensity was reduced by more than twofold (adh1:gfp + hairpin; Figs. 2, 5 [below]). Silencing was observed only in the presence of the GFP hairpin plasmid. No silencing was observed in a adh1:gfp strain transformed with the same plasmid lacking the hairpin (Fig. 2A, empty vector), nor was silencing triggered by the same plasmid expressing only sense or antisense GFP transcript (Fig. 2B). Furthermore, when the silenced strain was grown in the presence of uracil, the plasmid expressing the GFP hairpin was lost (adh:gfp-hairpin), as expected, and GFP fluorescence was restored to that of the original adh1:gfp strain (Fig. 2A). The loss of silencing in nonselective conditions argues against epigenetic (i.e., heritable) GFP silencing. Thus, silencing in our GFP/GFP hairpin system is distinct from that observed previously at both the centromeres and the mating-type locus, where dsRNA is proposed to trigger assembly of epigenetically heritable, repressive chromatin structures (Volpe et al. 2002).
Figure 2.
The GFP hairpin triggers adh1:gfp silencing. (A) Representative FACS data demonstrating that GFP silencing occurs only in the presence of the GFP hairpin. Silencing of GFP expression, measured by fluorescence intensity, was not observed for the empty vector alone, and silencing was lost when the silenced strain was grown in the presence of uracil to cause loss of the hairpin-encoding plasmid. Ten-thousand cells were analyzed for each genotype. See also Figure 5. (B) Representative FACS data demonstrating that GFP silencing is triggered by the GFP hairpin but not by either sense or antisense GFP transcripts.
Figure 5.
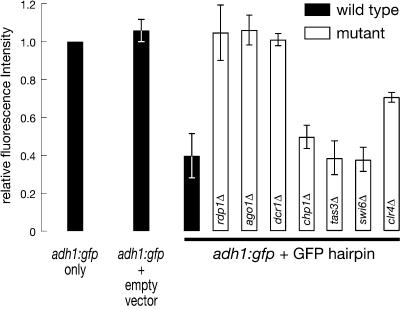
GFP silencing depends on the RNAi machinery but not the transcriptional silencing proteins Chp1, Tas3, or Swi6. The geometric mean of fluorescence intensity was determined for each strain from experiments like that in Figure 2, and normalized to the geometric mean for the wild-type adh1:gfp strain. The data are the average ± standard deviation for three trials.
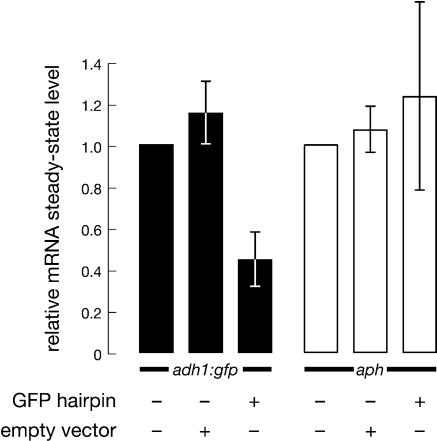
We could imagine two possible mechanisms for the silencing of adh1:gfp by the GFP hairpin. Silencing might reflect either unstable transcriptional repression of the locus or bona fide posttranscriptional gene silencing, that is, RNAi. To distinguish between these possibilities, we measured both the steady-state level (Fig. 3) and the rate of nuclear transcription (Fig. 4) of the adh1:gfp mRNA. To measure steady-state mRNA abundance, we performed quantitative RT-PCR using primers that spanned the fusion site in the adh1:gfp transgene (Fig. 1); actin (act1) mRNAs levels were measured as a control. In the presence of the GFP hairpin, the level of gfp mRNA was reduced more than twofold. The steady-state level of aph mRNA was unchanged, demonstrating the specificity of the silencing triggered by the GFP hairpin.
Figure 3.
GFP silencing reflects a reduction in the steady-state level of adh1:gfp mRNA. The steady-state level of mRNA was measured for gfp and aph, relative to the expression level of the act1 mRNA. For each strain three independent cultures were assayed; each assay was analyzed by quantitative RT-PCR in triplicate. The data are presented as the average of the three independent trials (using the average value for the triplicate quantitative RT-PCR reactions) ± the standard deviation of the three independent trials.
Figure 4.
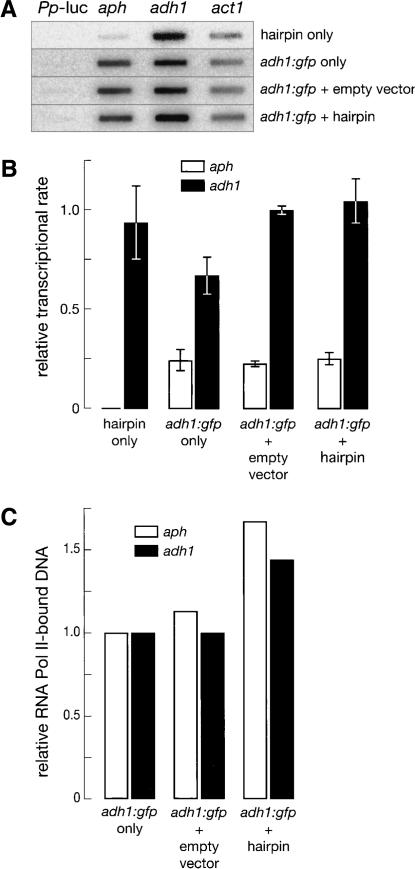
GFP silencing triggered by the GFP hairpin is posttranscriptional. (A) Representative nuclear run-on experiment. (B) The average value ± standard deviation for three independent run-on experiments is presented. For each strain, the rates of transcription for both aph and adh1 were measured and normalized to the rate of transcription of act1. Firefly luciferase (Pp luc) served as a negative control. No decrease in transcription of adh1:gfp was observed when GFP was silenced by introduction of the GFP hairpin-expressing plasmid. The rate of aph transcription was essentially constant in all aph-containing genotypes. (C) RNA Pol II chromatin immunoprecipitation (ChIP). The density of RNA Pol II at the adh1:gfp locus was not decreased by the introduction of the GFP hairpin, reinforcing the view that silencing triggered by this hairpin is posttranscriptional. Data are the average of two independent trials. The data for adh1:gfp and those for aph were separately normalized to their respective values in the absence of the empty or hairpin vector.
In contrast to its effect on steady-state mRNA levels, the GFP hairpin caused no observable change in the transcription of the adh1:gfp mRNA. Transcription was assessed by the nuclear run-on method. Briefly, yeast cells were permeabilized with detergent to permit the introduction of α-32P-UTP to label transcripts from elongating RNA polymerase II (RNA Pol II) (Volpe et al. 2002). This technique measures the density of RNA Pol II on a gene at the time of lysis, a reflection of the transcriptional rate of the gene in vivo. To distinguish transcription of the adh1:gfp mRNA from that of the GFP hairpin, we measured the rate of transcription for the adh1:gfp fusion mRNA using a probe for adh1. The rate of transcription of the act1 locus provided a normalization control. The GFP hairpin did not detectably decrease the rate of transcription of adh1:gfp (Fig. 4B). Control experiments demonstrate that the nuclear run-on assay can detect changes in the rate of transcription at least as small as 25% (Supplementary Fig. 1). Thus, the nuclear run-on data suggest that the reduction of steady-state level of gfp mRNA triggered by the hairpin was posttranscriptional.
To test our hypothesis by an independent method, we performed quantitative chromatin immunoprecipitation (ChIP) using an antibody to RNA Pol II. Like the nuclear run-on method, this method measures the relative density of RNA Pol II on a DNA sequence, a reflection of its transcriptional rate. Again, we could detect no decrease in the association of RNA Pol II with adh1:gfp in the presence of the GFP hairpin, relative to its association in the absence of the hairpin or in the presence of the plasmid lacking the hairpin sequences (empty vector) (Fig. 4C). We conclude that silencing of the adh1:gfp locus by the GFP hairpin occurs posttranscriptionally.
Silencing at the centromere locus can be accompanied by the spread of silencing across at least 3 kb of adjacent noncentromeric sequence; the spread of silencing correlates with the coating of the adjacent DNA with the Swi6 protein (Partridge et al. 2000). In contrast, silencing of adh1:gfp by the GFP hairpin was not accompanied by silencing of the adjacent aph locus (Fig. 4), whose promoter is less than 600 bp downstream from the end of the gfp ORF (Fig. 1). Thus, in every respect, silencing of adh1:gfp by the GFP hairpin appears to be entirely posttranscriptional. We conclude that in S. pombe, as in animals, plants, protozoa, and basal eukaryotes, dsRNA triggers RNAi.
Dicer orthologs are required for posttranscriptional gene silencing in Arabidopsis thaliana, Drosophila, C. elegans, Dictyostelium discoideum, and Neurospora (Bernstein et al. 2001; Knight and Bass 2001; Martens et al. 2002; Catalanotto et al. 2004). Similarly, genes encoding putative RdRP enzymes are required for PTGS in plants, worms, Neurospora, and Dictyostelium (Cogoni and Macino 1999; Dalmay et al. 2000; Smardon et al. 2000; Martens et al. 2002). The S. pombe dcr1 and rdp1 genes are required for the initiation of silent heterochromatin at centromeres and the mating-type locus (Hall et al. 2002; Volpe et al. 2002). In fact, the Rdp1 protein is physically associated with centromeric repeats, suggesting that it plays a direct role in generating the dsRNA silencing trigger (Volpe et al. 2002).
Small RNAs corresponding to GFP sequences accumulate in strains containing the GFP hairpin, but not the empty vector (Supplementary Fig. 2). No small RNAs were detected in either the dcr1Δ strain (Supplementary Fig. 2) or the rdp1Δ strain (data not shown). Small RNA production required only the GFP hairpin, not the adh1:gfp target RNA, suggesting that the RdRP plays a direct role in the production of the small RNAs from the hairpin trigger itself or their subsequent stability, rather than in the generation of secondary siRNAs templated from the target RNA. Small RNAs could be detected in the ago1Δ strain, consistent with a role for Ago1 in the function, but not the production, of small RNAs (data not shown).
Next, we measured GFP fluorescence in the dcr1Δ, rdp1Δ, and ago1Δ strains containing both the adh1:gfp locus and the GFP hairpin. GFP was not silenced in the absence of Dcr1 or Rdp1 (Fig. 5), consistent with the requirement for these enzymes in small RNA production. Thus, both Dicer and RdRP are required for posttranscriptional silencing of GFP in S. pombe, just as they are in C. elegans, the classical model for RNAi. Unlike plants and Drosophila, S. pombe encodes a single dicer gene. Our data, together with previous studies of silent heterochromatin in S. pombe, demonstrate that a single Dicer protein can support both TGS and PTGS. The functional specializations of Dicer proteins in plants (Xie et al. 2004) and flies (Lee et al. 2004) is unlikely to reflect an inherent biochemical limitation of the ancestral Dicer protein. Unlike C. elegans, which contains four RdRP genes, S. pombe encodes a single RdRP homolog, demonstrating that a single RdRP protein can mediate both transcriptional and posttranscriptional gene silencing. The simplest explanation is that the S. pombe RdRP protein supplies a common biochemical function to both pathways. In C. elegans, both somatic and germ-line RNAi triggered by dsRNA requires an RdRP (Smardon et al. 2000). The finding that in S. pombe RNAi triggered by dsRNA requires a functional RdRP suggests that the mechanism of S. pombe silencing is more closely related to RNAi in worms than PTGS in plants, where the RdRP homolog SDE1 is required only for silencing triggered by sense RNA-producing transgenes, not dsRNA (Dalmay et al. 2000). We note that it remains possible that one of the RdRP genes distinct from SDE1 could be required for dsRNA-triggered silencing in plants.
In contrast to the Dicer and RdRP genes, Argonaute proteins form a highly ramified family in all higher eukaryotes whose genomes have been fully sequenced. For example, the C. elegans genome encodes 27 Argonaute proteins. Current evidence suggests that Argonaute proteins are functionally specialized in higher organisms. For example, the C. elegans Argonaute protein Rde-1 is required for RNAi, but not for miRNA function. Conversely, the Argonaute proteins Alg-1/Alg-2 are required for miRNA function in worms. In contrast, miRNAs associate with at least four distinct Argonaute proteins in humans, only one of which, Ago2, can direct target mRNA cleavage (Liu et al. 2004; Meister et al. 2004). The Drosophila genome encodes at least five Argonaute proteins. In Drosophila, miRNAs associate with the Argonaute protein Ago1, whereas siRNAs associate with Ago2; both Ago1 and Ago2 can direct small RNA-guided target RNA cleavage (Okamura et al. 2004). Both of the Drosophila Argonaute protein-encoding genes, piwi and aubergine (aub) are required for H3 K9 methylation and correct localization of two heterochromatic proteins, HP1 and HP2, all processes associated with TGS (Pal-Bhadra et al. 2004). Yet, mutations in aub abrogate the RNAi-based silencing of the Stellate locus (Aravin et al. 2001), RNAi triggered by exogenous dsRNA (Kennerdell et al. 2002), and RISC assembly (Tomari et al. 2004). Furthermore, mutations in piwi partially block PTGS of Adh transgenes (Pal-Bhadra et al. 2002). Because Argonaute proteins have been implicated in both the production of the siRNA silencing trigger (Tabara et al. 2002) and the execution of target RNA (Hammond et al. 2001; Hutvágner and Zamore 2002; Martinez et al. 2002), a partial resolution to this apparent paradox would be if Argonaute proteins were required at multiple steps in both the TGS and RNAi pathways. In this view, discrete Argonaute proteins would be biochemically specialized for functions common to TGS and PTGS, and others dedicated to functions unique to each process. Alternatively, the diversity of Argonaute proteins might simply reflect their specialized patterns of spatial or temporal expression or their intracellular localization.
The genome of S. pombe encodes a single Argonaute protein, Ago1, which is required for transcriptional silencing (Hall et al. 2002; Volpe et al. 2002). Is it also required for RNAi in S. pombe? We introduced the GFP hairpin into an ago1Δ strain bearing the adh1:gfp transgene. Posttranscriptional silencing of adh1:gfp by the GFP hairpin required ago1 (Fig. 5). Thus, a single Argonaute protein mediates both RNAi and TGS in S. pombe.
Ago1, Rdp1, and Dcr1 are all required for both TGS and PTGS in S. pombe. Are these two distinct silencing pathways mediated by a common complex? Ago1 is a component of the RITS complex, which also contains Chp1, Tas3, and siRNA (Verdel et al. 2004). To test whether the RITS complex mediates RNAi in S. pombe, we asked whether either Chp1 or Tas3 is required for silencing of the adh1:gfp transgene by the GFP hairpin. We also asked whether the Swi6 or Clr4 proteins, which are not components of the RITS, but are required for RITS-initiated production of silent heterochromatin, are required for posttranscriptional silencing of the adh1:gfp transgene by the GFP hairpin. Deletion of chp1, tas3, or swi6 had no effect on GFP silencing by the GFP hairpin (Fig. 5). Thus, the requirements for TGS and PTGS in S. pombe are genetically distinct. In contrast, deletion of clr4 had an effect on posttranscriptional silencing of the adh:gfp locus. This finding is consistent with a previous report that siRNAs do not accumulate to normal levels in the clr4 mutant strain (Schramke and Allshire 2003); how Clr4 functions in siRNA biogenesis is not understood. That RNAi in S. pombe does not require Chp1, Tas3, or Swi6 suggests that a complex distinct from RITS mediates siRNA-directed target mRNA degradation. Alternatively, once siRNAs are produced by the action of Dcr1 and Rdp1, only Ago1 itself may be required for PTGS in S. pombe.
Discussion
In this study, we have demonstrated that a dsRNA derived from a hairpin transcript can trigger posttranscriptional silencing of a corresponding mRNA in S. pombe. Schramke and Allshire (2003) demonstrated that a similar hairpin transcript, corresponding to the ura4 locus, could trigger transcriptional silencing. In both studies, silencing triggered by a hairpin transcript required the RNAi machinery—Dcr1, Rdp1, and Ago1. Transcriptional silencing, unlike posttranscriptional silencing, required components of the transcriptional silencing apparatus—Chp1, Tas3, or Swi6. Robust silencing by both pathways requires the chromodomain protein Clr4, which appears to play a role in siRNA biogenesis or stability. Why does the GFP hairpin construct presented here trigger exclusively posttranscriptional silencing, whereas the previously studied ura4 hairpin triggered transcriptional silencing? One possible explanation is that the GFP hairpin used here included an efficiently spliced intron between the two arms of the hairpin. We presume that splicing of the intron promotes the accumulation of GFP dsRNA in the cytoplasm. In contrast, the ura4 hairpin construct of Schramke and Allshire (2003) contained an unspliced spacer sequence between the hairpin arms. Thus, the ura4 hairpin may be localized largely to the nucleus. A difference in subcellular localization might explain the different results obtained by the two studies. Alternatively, silencing of ura4 by the ura4-specific hairpin might comprise a mixture of transcriptional and posttranscriptional silencing. In this case, transcriptional silencing might not occur at the adh1 locus, even if the GFP hairpin-derived siRNAs trigger histone modification, perhaps because the gene is strongly expressed or is in a region of the genome otherwise refractory to heterochromatin formation. Nonetheless, our data, together with those of Schramke and Allshire (2003), clearly show that at least two distinct silencing responses can be initiated by a common RNAi machinery, without resorting to specialized forms of Dicer, RdRP, or Argonaute proteins. The demonstration that fission yeast contain a functional RNAi pathway now provides a simplified, genetically tractable model in which to study how the nature of the silencing trigger or of the silencing target determines the silencing pathway evoked—posttranscriptional or transcriptional.
Materials and methods
S. pombe strain construction
Fission yeast were grown and manipulated as described (Moreno et al. 1991). Unless otherwise stated, all strains (Table 1) were grown at 30°C in EMM2 media supplemented with histidine, leucine, adenine, and uracil as appropriate. Deletion and fusion strains were constructed by PCR-based cassette mutagenesis as described (Bahler et al. 1998) using the following oligonucleotides: 5′-CCTTCTCTACTCTTCCCGACGTCTACCGTCTCATGCATGAGAACAAGATTGCCGGCCGTATCGTCTTGGACCTTTCCAAGCGGATCCCCGGGTTAATTAA-3′ and 5′-AAAGATTCATGAGATGAACAGATTTGAAACCAAGCTGCAACCAAGCACACAAACTGAGATTGACAACGGCCGTACCATCGGAATTCGAGCTCGTTTAAAC-3′ for adh1:gfp; 5′-GTTTGGTATATATAAGCTTCCAACCGCCAAAGCGAATTGTCTTCAGCCAACTCGTCCTTTATGATTCAGAGTGAGTAGGGCGCGCCACTTCTAAATAAGC-3′ and 5′-TAAGGAAGTAAAAGTTGTGGGCAATCCAGTAGTCAATCGTATATCTATTTCATTACTTATTGCATGCAATCCATCAAACAGAATTCGAGCTCGTTTAAAC-3′ for ago1::kanMX; 5′-TTATTTTTACAAAAGGTTCCAAATTGATTTGTTGTCAGGGTTATAATTAAATCCAGATTTTGTATGGCAATGTAAAGCAGGCGCGCCACTTCTAAATAAG-3′ and 5′-TTCTATGTTCCATTGTCGATAATGAGTA GTACAATTACTATTAGCCTTTTACGAGCAATTAAAGCTTCGAATGTATATTCGATGAATTCGAGCTCGTTTAAAC-3′ for rdp1::kanMX; and 5′-TAGCTTAGGATTCATTATTTTTTAAGAGACAAATTTCTCGTCAATTGAATGAAACCTTCCGCCCTTTATTTTCTTTTTGAGGCGCGCCACTTCTAAATAAG-3′ and 5′-GCTTTGGAGACCCAAATTGAAAGTTTGAAAAGTTACAAGGGCCGCGGTCATAAAAAATGAAATACTGTATATTTCAAGTCGAATTCGAGCTCGTTTAAAC-3′ for dcr1::kanMX.
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| yFS106 | leu1-32 h+ura4-294 |
| yFS312 | leu1-32 h+ura4-294 pAS1 |
| yFS313 | h−leu1-32 ura4-D18 his 7-366 ade6-210 adh1:gfp |
| yFS314 | h−leu1-32 ura4-D18 his 7-366 ade6-210 adh1:gfp pAS1 |
| yFS315 | h+leu1-32 ura4-? ade6-210 adh1:gfp pRIP2 |
| yFS316 | h+leu1-32 ura4-? ade6-210 ago1::kanMX adh1:gfp pAS1 |
| yFS317 | h−leu1-32 ura4-? ade6-210 rdp1::kanMX adh1:gfp pAS1 |
| yFS318 | h−leu1-32 ura4-? dcr1::kanMX adh1:gfp pAS1 |
| yFS319 | h+leu1-32 ura4-D18 his 7-366 ade6-210 chp1::his7 adh1:gfp pAS1 |
| yFS327 | h−leu1-32 ura4-D18 ade6-210 tas3::kanMX adh1:gfp pAS1 |
| yFS328 | h+leu1-32 ura4-? his 7-366 ade6-210 arg3D swi6::arg3 adh1:gfp pAS1 |
| yFS329 | h+leu1-32 ura4-? his 7-366 ade6-210 clr4::LEU2 adh1:gfp pAS1 |
A question mark indicates it is not known if the strain contains either the ura4-294 or ura4-D18 allele of ura4.
Construction of the silencing trigger
Full-length eGFP sequence was amplified using the following primers: 5′-GGGAATTCCATATGGAGTAAAGGAGAAGAACTTTTC-3′, 5′-CACTAGCTAGCCTCTATCTTTACCAATTAGTTTCAATGTTTAGTAAGGTTTGAAAAAAGTTCCAACACACCTGATTTTATTTGTATAGTTCATCCATGCCATGTG-3′, 5′-GGCGCGGATCCGAGTAAAGGAGAAGAACTTTTC-3′, and 5′-CACTAGCTAGCTATTTGTATAGTTCATCCATGCC ATGTG-3′. PCR fragments were directly subcloned into pCR4-TOPO (Invitrogen) to produce pCR4-TOPO-B and pCR4-TOPO-N constructs. Both constructs were digested with NheI and NotI restriction enzymes; restriction fragments were gel-purified and ligated to produce pCR4-TOPO-B+N. This construct was digested with NdeI and BamHI, and the insert was subcloned into the corresponding restriction sites of pRIP2 (Maundrell 1993) to produce pAS1. A second plasmid bearing an ars1 sequence was also prepared by subcloning the EcoRI fragment of pREP2 into the corresponding site in pAS1 to produce pAS2. When transformed, both pAS1 and pAS2 produced episomal transformants that gave similar results in our experiments. Results for the pAS1 construct only are presented here.
RNA analysis
Cells were harvested from liquid cultures at mid-exponential phase (O.D.600 = 0.1-0.4). Total RNA was extracted from cells using the hot phenol method (Lyne et al. 2003). Purified RNA was treated with DNase (RQ1, Promega) and analyzed by Quantitative RT—PCR in a DNA Engine OPTICON2 (MJ Research) using the QuantiTect SYBR Green PCR Kit (QIAGEN) according to the manufacturer's instruction Analysis was performed using Opticon Monitor (MJ Research), Excel (Microsoft), and IgorPro 5.0 (Wavemetrics) software. PCR primers were 5′-GATTGCCGGCCGTATCGTCTT-3′ and 5′-GCCCATTAACATCACCATCTA-3′ for adh1:gfp; 5′-CTGAAACATGGCAAAGGTAGC-3′ and 5′-GGGATCGCAGTGGTGAGTAAC-3′ for aph; 5′-ACTTTGCTACGTCGCTTTGGAC-3′ and 5′-CGTTTCCGATAGTGATAACTTG-3′ for act1. Relative steady-state mRNA levels were determined from the threshold cycle for amplification using the 2-ΔΔCT method (Livak and Schmittgen 2001). Control experiments measuring the change in ΔCT with template dilution demonstrated that the efficiency of amplification of the target gene (adh1:gfp or aph) and the control (act1) was approximately equal.
Nuclear run-on analysis
RNA transcriptional elongation was analyzed by nuclear run-on essentially as described (Volpe et al. 2002). Labeled RNA from permeabilized cells was purified with hot phenol as described above and hybridized to nylon membranes containing strand-specific RNA probes for firefly luciferase Photinus pyralis (Pp-luc), aph, adh1, and act1. Full-length RNA probes were transcribed with T7 RNA polymerase from PCR templates prepared with the following oligonucleotides: 5′-gcgtaatacgactcactatag GAAAAACTCATCGAGCATCAAATG-3′ and 5′-GGGTAAGGAAAAGACTCACG-3′ for aph; 5′-gcgtaatacgactcactatagggCTTGGAAAGGTCCAAGACGATAC-3′ and 5′-GACTATTCCTGACAAGCAGTTG-3′ for adh1; 5′-gcgtaatacgactcactatagGAAGCACTTACGGTAAACGATAC-3′ and 5′-GGAAGAAGAAATCGCAGCGTTG-3′ for act1, where the T7 promoter sequence is in lowercase. Pp luc RNA was transcribed from a PCR-generated DNA template amplified from the pGL-2 Control vector (Promega) using 5′-gcgtaatacgactcactatagGAGAGGAATTCATTATC-3′ and 5′-GAAGAGATACGCCCTGGTTCCTG-3′ primers.
FACS analysis
Log-phase cells were harvested, washed in PBS (10 mM Na2HPO4, 1.7 mM KH2PO4 at pH 7.4, 137 mM NaCl, and 2.7 mM KCl) and resuspended in 500 μL PBS. Fluorescence was analyzed using a FACScan Flow Cytometer (Becton-Dickinson). Constant settings were maintained for all experiments. Data were acquired from 10,000 cells in all experiments and analyzed with Cell Quest (Becton-Dickinson) and FACSPress 1.3 (Ray Hicks) software.
Chromatin immunoprecipitation (ChIP)
The density of RNA polymerase II on the adh1:gfp and aph genes was measured essentially as described (Takahashi et al. 2000). Briefly, 50-mL cultures were grown until O.D.600 = 0.8. Protein was cross-linked to DNA by adding formaldehyde to a final concentration of 1% and incubating the cultures for 10-15 min at room temperature with gentle shaking. Cross-linking was quenched by adding glycine to a final concentration of 0.125 M. Cells were harvested by centrifugation, washed three times with ice-cold PBS, frozen in liquid nitrogen, and stored at -80°C until use. Cell pellets were resuspended in 200 μL of lysis buffer (50 mM HEPES-KOH at pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% (w/v) sodium deoxycholate, 1% [w/v] Triton X-100, 0.1% [w/v] SDS) containing for each 50 mL one tablet of Complete, EDTA-free Protease Inhibitor Cocktail (Roche) and 500 μL of Protease Inhibitor Cocktail For Fungal and Yeast Cells (Sigma), then vortexed with an equal volume of silica beads for 20 min at 4°C. The volume of the cell suspension was then adjusted by adding lysis buffer to a final volume of 650 μL. After sonicating to shear the chromosomal DNA to 250-500 bp, cell suspensions were centrifuged at 16,000 × g for 10 min at 4°C, and the supernatants were divided into three equal portions. The volume of each portion was adjusted to 500 μL, anti-RNA polymerase II (Pol II) antibody (Covance), anti-HA antibody (Covance), or no antibody was added, and the samples were incubatedon a rotating wheelfor 4 h at 4°C. Fifty microliters salmon sperm DNA/protein A agarose beads (Upstate) was added and the incubation continued for 1.5-2 h at 4°C. Beads were pelleted by centrifugation at 500 × g for 2 min, supernatants discarded, then the beads were washed once for 5 min with 1 mL of each of the following buffers: buffer 1 (lysis buffer with 1% sodium deoxycholate), buffer 2 (lysis buffer with 1% sodium deoxycholate and 1M NaCl), buffer 3 (50 mM Tris-HCl at pH 8.0, 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate), and twice with 10 mM Tris-HCl (pH 7.6) containing 10 mM EDTA. Samples were treated with proteinase K for 4 h at 42°C and cross-linking reversed by overnight incubation at 65°C. After phenol-chloroform extraction and ethanol precipitation, the DNA was resuspended in 20 μL of water. Five percent of the total DNA was used in quantitative PCR analysis as described above (“RNA analysis”). The difference in threshold cycles obtained when amplifying Pol II immunoprecipitated DNA and control DNA precipitated using nonspecific (HA) or no antibody was at least 10 cycles. Control experiments demonstrated that less than twofold differences in the density of RNA Pol II on a gene were readily detectable with our protocol.
Acknowledgments
We thank members of the Zamore lab for support and for comments on the manuscript, Tamal Raha and Michael Green for help with Pol II ChIP, Janet Partridge and Robin Allshire for the chp1Δ, clr4Δ, and swi6Δ strains, and Shiv Grewal and Danesh Moazed for the tas3Δ strain.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1218004.
Corresponding authors.
References
- Aravin A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11: 1017-1027. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie III, A., Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943-951. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Azzalin, G., Macino, G., and Cogoni, C. 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes & Dev. 16: 790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C., Pallotta, M., ReFalo, P., Sachs, M.S., Vayssie, L., Macino, G., and Cogoni, C. 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell Biol. 24: 2536-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti L., Mian, N., and Bateman, A. 2000. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25: 481-482. [DOI] [PubMed] [Google Scholar]
- Cogoni C. and Macino, G. 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. 94: 10233-10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166-169. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543-553. [DOI] [PubMed] [Google Scholar]
- Elgin S.C. and Grewal, S.I. 2003. Heterochromatin: Silence is golden. Curr. Biol. 13: R895-R898. [DOI] [PubMed] [Google Scholar]
- Fagard M., Boutet, S., Morel, J.-B., Bellini, C., and Vaucheret, H. 2000. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. 97: 11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23-34. [DOI] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana, G.D., Noma, K.-I., Ayoub, N., Cohen, A., and Grewal, S.I.S. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232-2237. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950-952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293-296. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146-1150. [DOI] [PubMed] [Google Scholar]
- Hutvágner G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056-2060. [DOI] [PubMed] [Google Scholar]
- Kennerdell J.R., Yamaguchi, S., and Carthew, R.W. 2002. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes & Dev. 16: 1884-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862-864. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69-81. [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.-J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science (in press). [Epub ahead of print July 29, 2004; 10.1126/science.1102513.] [DOI] [PubMed]
- Livak K.J. and Schmittgen, T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402-408. [DOI] [PubMed] [Google Scholar]
- Lyne R., Burns, G., Mata, J., Penkett, C.J., Rustici, G., Chen, D., Langford, C., Vetrie, D., and Bahler, J. 2003. Whole-genome microarrays of fission yeast: Characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens H., Novotny, J., Oberstrass, J., Steck, T.L., Postlethwait, P., and Nellen, W. 2002. RNAi in Dictyostelium: The role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell 13: 445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska, A., Urlaub, H., Lührmann, R., and Tuschl, T. 2002. Single stranded antisense siRNA guide target RNA cleavage in RNAi. Cell 110: 563-574. [DOI] [PubMed] [Google Scholar]
- Maundrell K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127-130. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185-197. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Fine, N.A., Fujisawa, T., and Gorovsky, M.A. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689-699. [DOI] [PubMed] [Google Scholar]
- Morel J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in posttranscriptional gene silencing and virus resistance. Plant Cell 14: 629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar, A., and Nurse, P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795-823. [DOI] [PubMed] [Google Scholar]
- Nykänen A., Haley, B., and Zamore, P.D. 2001. ATP Requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309-321. [DOI] [PubMed] [Google Scholar]
- Okamura K., Ishizuka, A., Siomi, H., and Siomi, M.C. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 18: 1655-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M., Bhadra, U., and Birchler, J.A. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9: 315-327. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Leibovitch, B.A., Gandhi, S.G., Rao, M., Bhadra, U., Birchler, J.A., and Elgin, S.C. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669-672. [DOI] [PubMed] [Google Scholar]
- Partridge J.F., Borgstrom, B., and Allshire, R.C. 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes & Dev. 14: 783-791. [PMC free article] [PubMed] [Google Scholar]
- Raponi M. and Arndt, G.M. 2003. Double-stranded RNA-mediated gene silencing in fission yeast. Nucleic Acids Res. 31: 4481-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V. and Allshire, R. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069-1074. [DOI] [PubMed] [Google Scholar]
- Shi H., Djikeng, A., Tschudi, C., and Ullu, E. 2004. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: Control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell Biol. 24: 420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A., Spoerke, J., Stacey, S., Klein, M., Mackin, N., and Maine, E. 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10: 169-178. [DOI] [PubMed] [Google Scholar]
- Smith N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407: 319-320. [DOI] [PubMed] [Google Scholar]
- Song J.-J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science (in press). [Epub ahead of print July 29, 2004; 10.1126/science.1102514.] [DOI] [PubMed]
- Tabara H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123-132. [DOI] [PubMed] [Google Scholar]
- Tabara H., Yigit, E., Siomi, H., and Mello, C.C. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DexH-box helicase to direct RNAi in C. elegans. Cell 109: 861-871. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Saitoh, S., and Yanagida, M. 2000. Application of the chromatin immunoprecipitation method to identify in vivo protein-DNA associations in fission yeast. Science STKE 2000: PL1. [DOI] [PubMed] [Google Scholar]
- Tijsterman M., Okihara, K.L., Thijssen, K., and Plasterk, R.H. 2002. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr. Biol. 12: 1535-1540. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du, T., Haley, B., Schwarz, D.S., Bennett, R., Cook, H.A., Koppetsch, B.S., Theurkauf, W.E., and Zamore, P.D. 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831-841. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez, F., Crete, P., and Bartel, D.P. 2004. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Dev. 18: 1187-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I.S., and Martienssen, R.A. 2002. Regulation of heterchromatic silencing and Histone H3 Lysine-9 methylation by RNAi. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Williams R.W. and Rubin, G.M. 2002. Argonaute1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. 99: 6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25-33. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao, X., and Jacobsen, S.E. 2003. Argonaute4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716-719. [DOI] [PubMed] [Google Scholar]