Abstract
A critical step in skeletal morphogenesis is the formation of synovial joints, which define the relative size of discrete skeletal elements and are required for the mobility of vertebrates. We have found that several Wnt genes, including Wnt4, Wnt14, and Wnt16, were expressed in overlapping and complementary patterns in the developing synovial joints, where β-catenin protein levels and transcription activity were up-regulated. Removal of β-catenin early in mesenchymal progenitor cells promoted chondrocyte differentiation and blocked the activity of Wnt14 in joint formation. Ectopic expression of an activated form of β-catenin or Wnt14 in early differentiating chondrocytes induced ectopic joint formation both morphologically and molecularly. In contrast, genetic removal of β-catenin in chondrocytes led to joint fusion. These results demonstrate that the Wnt/β-catenin signaling pathway is necessary and sufficient to induce early steps of synovial joint formation. Wnt4, Wnt14, and Wnt16 may play redundant roles in synovial joint induction by signaling through the β-catenin-mediated canonical Wnt pathway.
Keywords: Wnt, β-catenin, joint formation, skeletal development
Formation of synovial joints between different skeletal elements is essential for the mobility of vertebrates. The number and position of joints also determine characteristic skeletal patterns in each vertebrate species by defining the size and shape of skeletal elements. As alterations of early patterning signals often lead to changes in the position and number of joints in the developing limb (Dahn and Fallon 2000; Suzuki et al. 2004), understanding the regulation of joint formation in the limb will also provide critical insights into how early-limb patterning is linked to later skeletal morphogenesis at the molecular level.
In the developing limb, studies of descriptive embryology have shown that skeletal elements form through temporally and spatially regulated processes that include mesenchymal condensation, elongation, branching, and/or segmentation (Shubin and Alberch 1986). Most of the synovial joints in the limb form through segmentation of a pre-existing cartilage rod. For instance, in the developing forelimb, the initial de novo mesenchymal condensation forms the cartilage anlagen of the humerus, the growth and branching of which then produce a Y-shaped bifurcation. It is the segmentation of this Y-shaped cartilage primordium that forms the elbow joint that separates the radius and ulna from the humerus (Shubin and Alberch 1986).
Synovial joint formation starts from the differentiation of newly differentiated chondrocytes into flattened and densely packed interzone cells (for review, see Archer et al. 2003), which express joint-specific markers such as Gdf5 and lose the expression of chondrocyte-specific markers such as ColII (Craig et al. 1987; Nalin et al. 1995; Morrison et al. 1996; Storm and Kingsley 1996). Later in development, the interzone cells differentiate and form three layers. The middle layer eventually cavitates to form a fluid-filled joint space. Synthesis of hyaluronan (HA), expression of its principle receptor CD44 in the interzone cells, and movement of the embryo have all been suggested to play an essential role in joint cavitation (Pitsillides et al. 1999). Meanwhile, the two lateral layers of the interzone participate in the formation of articular cartilage of the two opposing skeletal elements. In the mature joint, the opposing articular cartilages are wrapped in the joint capsule, which is enforced by ligaments and tendons outside and lined by synovial membrane inside (Archer et al. 2003). In adult life, the mature joint structures need to be properly maintained, as disruption of articular cartilage leads to pathological conditions such as rheumatoid arthritis (RA) and osteoarthritis (OA), which are common diseases.
The molecular mechanisms regulating joint formation are just beginning to be elucidated. It has been shown that cell-cell signaling mediated by Gdf5, Noggin, and Wnt14 plays a critical role in controlling synovial joint formation (Storm et al. 1994; Brunet et al. 1998; Hartmann and Tabin 2001). However, none of these signaling molecules are both necessary and sufficient for synovial joint induction. First, bone morphogenetic protein (Bmp) family members Gdf5 and Gdf6 are essential for joint formation in certain regions of the limb, but they are not sufficient for inducing joint formation. Gdf5 and Gdf6 null mutant mice exhibit joint development defects in digits, wrists, and ankles (Storm and Kingsley 1996; Settle et al. 2003). Yet, overexpression of Gdf5 in both chick and mouse does not induce joint formation (Francis-West et al. 1999a; Merino et al. 1999; Storm and Kingsley 1999; Tsumaki et al. 1999). In contrast, Gdf5 overexpression resulted in extensive cartilage overgrowth and complete absence of joints (Tsumaki et al. 1999). Second, joint formation is inhibited in the Noggin mutant mice (Brunet et al. 1998), but overexpression of Noggin only inhibited cartilage formation (Capdevila and Johnson 1998; Pathi et al. 1999; Pizette and Niswander 2000). Lastly, ectopic expression of Wnt14 in the chick limb is sufficient to induce joint formation (Hartmann and Tabin 2001), but it is not clear whether Wnt14 is also required in the mouse for joint formation. Furthermore, it was unknown prior to this study how Wnt14 transduces its signal in joint induction.
There are 19 Wnt family members and they can transduce their signals through several different pathways (Veeman et al. 2003; Yang 2003; Nelson and Nusse 2004 and references therein). Among them, the canonical Wnt pathway plays pivotal roles in controlling cell proliferation and cell-fate determination during many embryonic development processes. Central to the canonical Wnt pathway is the stabilization and nuclear translocation of β-catenin after Wnt ligands bind to their receptors Frizzled and LRP5/6. β-Catenin then activates downstream gene expression through binding to the LEF/TCF transcription factors.
Here, we found that Wnt4, Wnt 14, and Wnt16 were expressed in the forming joints in overlapping and complementary patterns. These Wnts can transduce their signals through the canonical Wnt pathway both in vitro and in vivo. The activity of Wnt14 was blocked when β-catenin was inactivated in the forming cartilage. Overexpression of a constitutively active β-catenin or Wnt14 in early differentiating chondrocytes induced ectopic joint formation both morphologically and molecularly. Conversely, genetic ablation of β-catenin in early differentiating chondrocytes led to fusion of skeletal elements. Our data demonstrate that the canonical Wnt signaling is not only sufficient, but also necessary for inducing at least early steps of synovial joint formation. Our results also indicate that Wnt4, Wnt 14, and Wnt16 may play redundant roles in inducing joint formation by signaling through the canonical Wnt pathway.
Results
Expression of Wnt4, Wnt14, Wnt16 and accumulation of β-catenin protein in the developing synovial joints
To test the role of Wnt signaling in synovial joint induction, we first examined the expression of Wnt genes in the forming synovial joints of the developing mouse limb. We found that Wnt4, Wnt14, and Wnt16 were already expressed in the future joint region, as indicated by Gdf5 expression, at the same time when chondrogenic mesenchymal condensations form at 11.5 days postcoitum (dpc) (Fig. 1A,B). However, the expression patterns of the Wnts were distinct. Wnt14 was expressed at higher levels in the mesenchyme surrounding the cartilage primordium than in the presumptive joint (Fig. 1A,B). Interestingly, Wnt14 was also expressed in the presumptive joint region between the scapula and humerus (Fig. 1A) that does not form by segmentation of a pre-existing chondrogenic condensation. Wnt4 was also expressed in the future elbow joint, but its expression in the flanking mesenchyme, which will give rise to the joint capsule, was stronger (Fig. 1B,D). Wnt16 was initially expressed as a stripe that marks the future metatarsophalangeal (MTP) area (Fig. 1A). At 11.5 dpc, Gdf5 expression was broad and Wnt4 expression was stronger than Wnt14 (Fig. 1B). At 12.5 dpc, Gdf5 expression marks future joints in the digit rays. Wnt14 expression was still observed around the forming cartilage with stronger expression in the future joint (Fig. 1A). Interestingly, the initial stripe-like expression pattern of Wnt16 was expanded at 12.5 dpc. Wnt16 was expressed almost exclusively in the MTP joints and the proximal interdigital area (Fig. 1A). At 13.5 dpc, Wnt4, Wnt14, and Wnt16 were all expressed in the developing digit joints, but Wnt4 expression was the strongest (Fig. 1C). In the MTP joint region, however, Wnt16 expression was the strongest (Fig. 1C). In the elbow region, where the joint was more mature and Gdf5 was expressed as a narrow stripe, Wnt14 expression was stronger than Wnt4 in the joint interzone. Wnt14 was also expressed strongly in the mesenchyme surrounding the cartilage and in the cells that will form tendons (Fig. 1D). Wnt4 was expressed at much higher levels in the mesenchyme that will form the joint capsule (Fig. 1D). Taken together, the dynamic expression of Wnt4, Wnt14, and Wnt16 in the forming joints of the limb was both overlapping and complementary. These Wnt genes were also expressed in noncartilaginous mesenchymal cells that will form fibrous tissues, such as the joint capsule, ligaments, and tendons.
Figure 1.
Expression of Wnt genes correlates with the expression of the joint marker Gdf5, up-regulation of β-catenin protein, and down-regulation of Sox9 expression. (A) Whole-mount in situ hybridizations. At 11.5 dpc, Wnt14 and Gdf5 were expressed in the future shoulder and elbow joints (arrows). Gdf5 was also expressed in the distal limb bud, whereas Wnt14 was also expressed in the mesenchymal cells surrounding the forming cartilage. Wnt16 was expressed in the future MTP joints (arrow). At 12.5 dpc, Wnt14 was still expressed around the cartilage, but stronger expression was detected in the forming joints (arrows). Gdf5 was expressed in all developing joints (arrow) and the interdigital area. The MTP region was expanded and Wnt16 was expressed in the forming MTP joints (arrow). Joint-specific Wnt4 expression detected by whole-mount in situ hybridization was obscure at 11.5 or 12.5 dpc due to its expression in the surface ectoderm and tissues around the cartilage. (B) A limb bud section at 11.5 dpc hybridized with a 35S-labeled Gdf5 probe is shown as a bright-field image. The boxed elbow region was enlarged, and expression of Gdf5, Wnt14, and Wnt4 (arrowheads) was detected in consecutive sections. Wnt14 and Wnt4 were also expressed at stronger levels in the mesenchyme surrounding the forming cartilage (arrows) than in the presumptive joint (arrowheads). Immunohistochemistry of β-catenin and Sox9 was performed on a consecutive section. β-Catenin was up-regulated in the future joint region (arrowhead). The boxed region was enlarged in an inset to show a cell with nuclear localized β-catenin protein (arrow). Sox9 was detected in the forming cartilage (arrow), but down-regulated in the forming joint (arrowhead). (H) Humerus; (R) radius; (U) ulna. (C) Expression of Gdf5, Wnt14, Wnt4, and Wnt16 was all detected in the developing joints between phalanges (P) at 13.5 dpc (arrows). Expression of Wnt16 in the MTP joints was much stronger than Wnt14 and Wnt4. (D) Only Gdf5 and Wnt14 expression was detected in the elbow joint at 13.5 dpc (arrows). Gdf5 and Wnt4 were also expressed in the future joint capsule region (arrowhead). Wnt14 was also expressed in the future tendons (arrowhead). Complementary patterns of β-catenin and Sox9 protein levels in the joint were similar to those in B.
As the first attempt to identify the pathway through which these Wnts transduce their signaling in the developing joint, we examined β-catenin protein levels. Up-regulation of β-catenin protein levels due to stabilization of β-catenin protein is an important outcome of the canonical Wnt signaling activation. We found that β-catenin protein levels were low in newly differentiated chondrocytes that express Sox9 at 11.5 dpc (Fig. 1B). However, in the developing joint region, β-catenin protein levels were up-regulated and nuclear localization of β-catenin protein was observed (Fig. 1B). In the same region, Sox9 expression was down-regulated (Fig. 1B). This reciprocal relationship between β-catenin and Sox9 protein levels was also observed later in development at 13.5 dpc in more mature joints (Fig. 1D). The correlation between Wnt gene expression and β-catenin protein accumulation during synovial joint formation suggested that at least some of the Wnts expressed in the forming synovial joint may signal through the canonical Wnt pathway to increase β-catenin protein levels and its subsequent nuclear localization.
The canonical Wnt signaling pathway was up-regulated in the developing joint and activated by Wnt4/14/16
We next tested whether the canonical Wnt signaling activity was activated in the developing synovial joints using the TOPGAL mouse embryo (DasGupta and Fuchs 1999; Topol et al. 2003), in which LacZ expression is under the control of LEF/TCF-binding sites and can be activated by β-catenin. Consistent with the increased β-catenin protein levels, the canonical Wnt signaling activity was up-regulated in the developing joints in vivo, indicated by the LacZ-expressing cells (Fig. 2B). To further test whether Wnt4, Wnt14, and Wnt16 signal through the canonical Wnt pathway, Wnt 4, Wnt14, and Wnt16 were expressed in a rat chondrocyte cell line RCS (Mukhopadhyay et al. 1995). In the in vitro LEF/TCF reporter (TOPFLASH) assay for the canonical Wnt activity (Korinek et al. 1997), expression of Wnt4, Wnt14 and Wnt16, like Wnt3a or ΔNβ-catenin, which encodes a consistitutively active β-catenin, led to the up-regulation of TOPFLASH activity (Fig. 2C). In addition, we performed micromass culture with the limb mesenchymal cells from the TOPGAL mouse embryo to mimic the in vivo process of cartilage formation in vitro. We found that Gdf5, Wnt 4, Wnt14, and Wnt16 were expressed around the cartilage nodule in the micromass culture (data not shown). The LacZ staining was found only in the cells that surrounded the cartilage nodules, but not within the nodules (Fig. 2D). Ectopic expression of Wnt14, by infecting micromass cultures with a Wnt14-adenovirus, completely inhibited cartilage nodule formation (Fig. 2E), consistent with the previous observation in the chick micromass culture infected by a RCAS-Wnt-14 virus (Hartmann and Tabin 2001). Importantly, we found that LacZ expression was activated to a higher level throughout the micromass infected with the Wnt14-adenovirus (Fig. 2E). All these data indicate that Wnt14 transduces its signal through β-catenin in regulating downstream gene expression.
Figure 2.
The canonical Wnt pathway was activated by Wnt4/14/16 and ectopic Wnt14 signaling inhibited chondrogenesis. (A) Whole-mount in situ hybridization showing that Gdf5 expression was detected in the forming digit joints of the right foot of a TOPGAL embryo at 14.5 dpc. (B) LacZ staining showing that the canonical Wnt signaling activity was up-regulated in the forming digit joint and joint capsule (arrows) of the left foot of the same embryo shown in A. Skin was peeled off in most areas to facilitate LacZ staining of the cartilage. (C). Expression of Wnt4, Wnt14, and Wnt16, like Wnt3a and ΔNβ-catenin, activated TOPFLASH reporter (blue bars), but not the control FOPFLASH reporter (black bars), in RCS cells. (D,E) Micromass cultures using the limb mesenchymal cells from the TOPGAL mice. A comparable representative area was enlarged and shown in the inset. (D) LacZ staining was absent in cartilage nodules (brown color, arrow). (E) After infected by a Wnt14-adenovirus, LacZ staining was up-regulated and cartilage nodule formation was completely inhibited.
Wnt14 activity is blocked by inactivation of β-catenin or β-catenin transcription activity
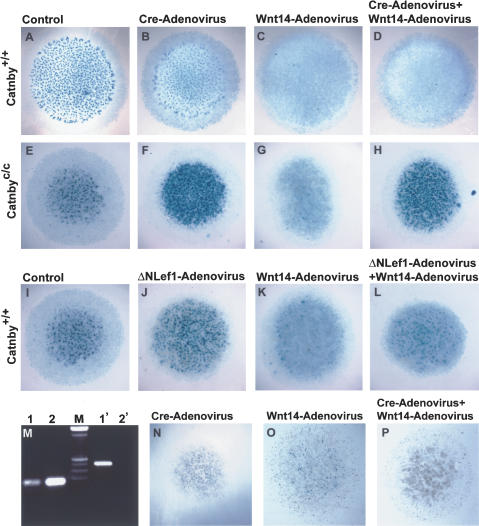
We further tested whether the activity of Wnt14 in inducing joint formation requires β-catenin by performing micromass assays where the Wnt14-adenovirus was added to limb mesenchymal cells, in which β-catenin was inactivated by the Cre recombinase. We generated a conditional targeted allele of β-catenin, Catnbyc/c (Supplementary Fig. S1). Because the joint-inducing activity of Wnt14 is reflected in part in its ability to inhibit chondrocyte differentiation, we performed micromass cultures with the mesenchymal cells from the Catnbyc/c limb bud at 12.5 dpc to determine whether Wnt14-mediated inhibition of chondrogenesis requires β-catenin. The dissociated limb mesenchymal cells were first infected with a Cre-adenovirus before plated at high density to achieve efficient virus infection. We found that chondrogenesis was unaffected or slightly reduced by the Cre-adenovirus in the Catnby+/+ micromass culture (Fig. 3A,B) and Cre-mediated deletion of β-catenin occurred in most cells of the Catnbyc/c culture (Fig. 3M). However, when β-catenin was inactivated in the Catnbyc/c culture by the Cre-adenovirus infection, cartilage nodule formation was greatly enhanced and the nodules were bigger in size when compared with the uninfected Catnbyc/c micromass culture (Fig. 3E,F). Wnt14-adenovirus blocked chondrogenesis completely in both Catnby+/+ and Catnbyc/c micromass cultures when β-catenin was functional (Fig. 3C,G). Significantly, only when the Catnbyc/c micromass culture (Fig. 3H), not the Catnby+/+ one (Fig. 3D), was sequentially infected by the Cre-adenovirus and the Wnt14-adenovirus, cartilage nodule formation was not blocked. Cartilage nodule formation between the Wnt14-adenovirus-infected and noninfected micromass cultures was similar to each other when β-catenin was inactivated (Fig. 3F,H). In addition, Wnt14-adenovirus was not able to up-regulate LacZ expression in the Cre-adenovirus infected Catnbyc/c;TOPGAL micromass cultures (Fig. 3O,P). These results demonstrated that the activity of Wnt14 in inhibiting chondrogenesis requires β-catenin. We further found that expression of a dominant-negative Lef1 (ΔNLef1), which blocks the transcription activity of β-catenin (Niemann et al. 2002) resulted in increased cartilage nodule formation (Fig. 3J) and blocked Wnt14-mediated inhibition of chondrogenesis (Fig. 3L) in the Catnby+/+ micromass culture. Because β-catenin is involved in both cell adhesion and activation of downstream gene transcription in the canonical Wnt pathway, these data indicate that activation of LEF/TCF-mediated transcription by β-catenin is required for Wnt14 to execute chondrogenic inhibitory effect.
Figure 3.
β-catenin transcription activity is required for the function of Wnt14 in inhibiting chondrocyte differentiation and activating TOPGAL expression. Micromass cultures were stained by Alcian blue to show cartilage nodules. (A-D) Wild-type (Catnby+/+) micromass cultures. (B) The Cre-adenovirus had no or little effect on cartilage nodule formation. (C) The Wnt14-adenovirus completely inhibited cartilage nodule formation. (D) The Cre-adenovirus infection did not block the chondrogenic inhibitory effect of Wnt14. (E-H) Homozygous conditional (Catnbyc/c) micromass cultures. The Cre-adenovirus significantly increased cartilage nodule formation (F) compared with the uninfected Catnbyc/c control culture (E). The Wnt14-adenovirus completely inhibited cartilage nodule formation (G), but such inhibition was completely blocked by the Cre-adenovirus, which inactivates β-catenin (H). (I-L) Wild-type (Catnby+/+) micromass cultures. The ΔNLef1-adenovirus increased cartilage nodule formation (J) compared with the uninfected control culture (I). The Wnt14-adenovirus completely inhibited cartilage nodule formation (K), but such inhibition was significantly blocked by the ΔNLef1-adenovirus (L). (M) Genotyping the cells in the micromass cultures for the conditional allele (lanes 1,2) and the null allele (lanes 1′,2′). (Lanes 1,1′) Cre-adenovirus-infected Catnbyc/c micromass cultures. Most Catnbyc/c cells infected by the Cre-adenovirus had undergone deletion of the β-catenin gene (cf. lanes 1 and 1′). (Lanes 2,2′) Catnbyc/c micromass cultures without virus infection. (Lane 2′) No deletion occurred in the absence of Cre. (N-P) Micromass cultures performed using limb mesenchymal cells from the homozygous conditional embryos that also carry the TOPGAL transgene (Catnbyc/c; TOPGAL). (O) The Wnt14-adenovirus inhibited cartilage formation and up-regulated LacZ expression. (P) Both effects of the Wnt14-adenovirus were blocked by the Cre-adenovirus.
Ectopic expression of a constitutively active β-catenin or Wnt14-induced ectopic joint formation
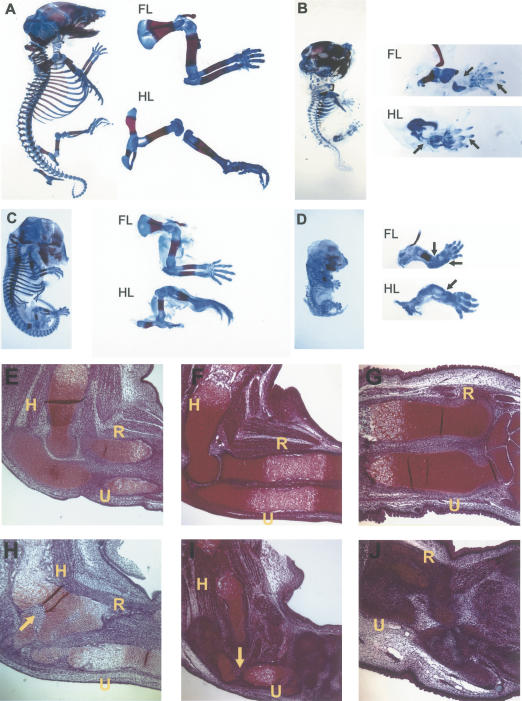
To test whether the β-catenin-mediated canonical Wnt signaling plays a pivotal role in inducing joint formation in vivo, we generated Col2a1-ΔNβ-catenin transgenic mice, in which a constitutively active, N-terminally truncated form of β-catenin (ΔNβ-catenin) (Topol et al. 2003) was expressed in chondrocytes under the Col2a1 promoter/enhancer (Yang et al. 2003). We also generated Col2a1-Wnt14 transgenic mice to confirm that Wnt14 signals through the canonical Wnt pathway. The Col2a1-ΔNβ-catenin mouse was perinatal lethal and the Col2a1-Wnt14 mouse embryos died around 16.5 dpc. Both the Col2a1-ΔNβ-catenin and the Col2a1-Wnt14 mouse embryos had dome-shaped heads and shorter limbs, tails, and snouts as compared with their wild-type litter mates (Fig. 4B,D). When skeletal preparations were examined, we found that cartilage formation in both transgenic mice was greatly reduced, as indicated by much weaker Alcian blue staining (Fig. 4B,D). In addition, endochondral ossification, but not intramembranous ossification, was significantly reduced in both transgenic mice (Fig. 4B,D), possibly caused by the impaired cartilage formation. We also observed joint fusions in both Col2a1-ΔNβ-catenin and the Col2a1-Wnt14 transgenic mice (Fig. 4D,H). The similar phenotypes in the Col2a1-ΔNβ-catenin and the Col2a1-Wnt14 mice supported our conclusion that Wnt14 signals through stabilizing β-catenin in controlling skeletal development.
Figure 4.
Expression of a constitutively active β-catenin or Wnt14 under the control of the Col2a1 promoter and enhancer-disrupted skeletal morphogenesis. (A-D) Skeletal preparations in which cartilage was stained by Alcian blue and ossified bone was stained by Alizarin red. Forelimbs (FL) and hindlimbs (HL) are shown in higher magnification. (A) Wild-type embryo at 18.5 dpc. (B) Col2a1-ΔNβ-catenin transgenic littermate. Cartilage and endochondral bone formation was greatly reduced (arrows). (C) Wild-type embryo at 15.5 dpc. (D) Col2a1-Wnt14 transgenic littermate. Cartilage and endochondral bone formation was also greatly reduced and the joint between the humerus and radius was fused (arrows). (E-J) A 5-μm section of the limb stained according to the Weigert-Safranin staining procedure. Chondrocytes were stained red by Safranin O. (H) Humerus; (R) radius; (U) ulna. (E,F) Wild-type limb at 15.5 dpc. (G) Wild-type limb at 18.5 dpc. (H) Limb from Col2a1-ΔNβ-catenin transgenic littermate of E. Some areas in the cartilage had lost cartilage-specific staining (arrow). The joint between humerus and radius was fused. (I) Limb from the Col2a1-Wnt14 transgenic littermate of F. Cartilage-specific staining was greatly reduced, and some areas had lost cartilage-specific staining (arrow). (J) Limb from the Col2a1-ΔNβ-catenin transgenic littermate of G. Loss of cartilage specific staining was more extensive.
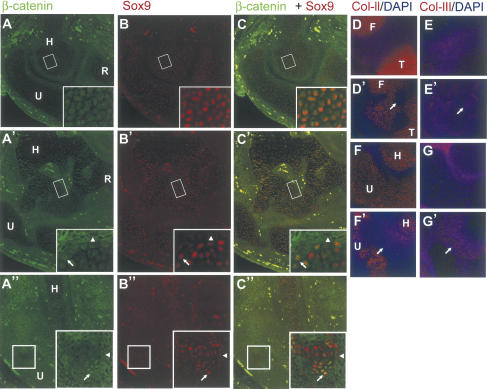
To analyze chondrocyte differentiation and synovial joint formation in the Col2a1-ΔNβ-catenin and Col2a1-Wnt14 transgenic mouse embryos in more detail, we first examined histological sections of the developing limb at 15.5 and 18.5 dpc. We found that in many areas of the cartilage, chondrocyte-specific staining was lost. For instance, we observed regions inside the cartilage where chondrocyte-specific staining was greatly diminished in both Col2a1-ΔNβ-catenin and Col2a1-Wnt14 mice (Fig. 4H,I). Loss of cartilage tissue was more severe at 18.5 dpc than at 15.5 dpc (Fig. 4J). We concluded that chondrocytes had lost their characteristic cellular phenotypes and may have adopted other cell fates. To test this, we examined alterations of cell morphology and gene expression by immunohistochemistry and in situ hybridization. Chondrocytes have a characteristic small, cobble stone-like shape, whereas other cells, including the joint-forming interzone cells, osteoblasts, and cells that will form the tendon and ligament are elongated. At the molecular level, chondrocytes express very low levels of β-catenin, but high levels of Sox9 (Fig. 1B,D). We found that Wnt14 ectopic expression leads to strong β-catenin accumulation and nuclear translocation (Fig. 5A″), again confirming that Wnt14 signals through stabilizing β-catenin protein. In addition, we found that ectopic expression of ΔNβ-catenin or Wnt14 leads to loss of Sox9 expression and the cell morphology change from cobble stone-like to elongated (Fig. 5, cf. insets in A′,B′,C′,A″,B″,C″ and A,B,C). Chondrocytes also express specific extracellular matrix protein such as ColII (Fig. 5D,F), but not ColIII (Craig et al. 1987; Nalin et al. 1995; Fig. 5E,G). In both Col2a1-ΔNβ-catenin and Col2a1-Wnt14 embryos, ColIII was ectopically expressed (Fig. 5E′,G′) at the expense of ColII expression (Fig. 5D′,F′). As the Col2a1 promoter and enhancer are directly activated by Sox9, which is expressed in differentiating chondrogenic mesenchymal condensations (Wright et al. 1995; Lefebvre et al. 1996), our results indicate that Wnt14 signals though β-catenin in differentiating chondrocytes in vivo to reverse chondrocyte differentiation, the first step in synovial joint formation.
Figure 5.
Ectopic Wnt14/β-catenin signaling in chondrocytes resulted in alteration of cell morphology and gene expression. Forelimb sections at the elbow region of a wild-type embryo (A,B), a Col2a1-ΔNβ-catenin transgenic embryo (A′,B′) and a Col2a1-Wnt14 transgenic embryo (A″,B″) at 15.5 dpc were costained for β-catenin and Sox9. Merged images of A and B, A′ and B′, and A″ and B″ are shown in C, C′, and C″, respectively. Boxed regions inside Humerus or Ulna were enlarged and shown in insets. Cells with higher levels of β-catenin and low levels of Sox9 were elongated, similar to fibroblasts (arrowheads). Chondrocytes containing both nuclear β-catenin and Sox9 were indicated by arrows. (D,D′,E,E′) Adjacent sections of a limb at the knee joint region at 15.5 dpc stained for ColII or ColIII. The nucleus was stained by DAPI. (D,E) Wild-type controls. (D′,E′) Sections of the Col2a1-ΔNβ-catenin transgenic embryo. (F,F′,G,G′) Adjacent sections of a limb at the elbow joint region at 15.5 dpc stained for ColII or ColIII. The nucleus was stained by DAPI. (F,G) Wild-type controls. (F′,G′) Sections of the Col2a1-Wnt14 transgenic embryo at 15.5 dpc. Arrows indicate the area where ColIII was ectopically expressed and ColII expression was absent. (H) Humerus; (R) radius; (U) ulna; (F) femur; (T) tibia
We next tested whether up-regulation of the Wnt14/β-catenin signaling specifically leads to ectopic joint formation by examining the expression of joint-specific markers. We found that Gdf5 expression was ectopically activated in the cartilage by ectopic expression of ΔNβ-catenin or Wnt14, indicating that ectopic joint formation was induced by the canonical Wnt signaling (Fig. 6A,B). The domain of ectopic Wnt14 expression overlaps with that of Gdf5 induction (Fig. 6A,B). But, in the Col2a1-ΔNβ-catenin mice, ectopic Gdf5 expression was detected at the periphery of the regions where chondrocyte phenotypes were absent (Fig. 6A). In the adjacent section of the same Col2a1-ΔNβ-catenin embryo (Fig. 5A',B′,C′), nuclear β-catenin colocalized with Sox9 in the peripheral cells of the same region where chondrocyte phenotypes were absent, and it is likely that these peripheral cells ectopically expressed Gdf5. Because the Col2a1 promoter was activated by Sox9, absence of Gdf5 expression in the cells that had lost chondrocyte phenotypes and Sox9 expression in the Col2a1-ΔNβ-catenin embryo (Fig. 6A) suggested that Wnt/β-catenin not only activates Gdf5 expression, it is also required to maintain it. The expression of other joint markers such as Chordin and Fgf18 was also found to be ectopically expressed in the Col2a1-ΔNβ-catenin and the Col2a1-Wnt14 mouse embryos (Fig. 6A,B; data not shown). As Fgf18 was not expressed in early condensed mesenchyme (Maruoka et al. 1998), the Wnt/β-catenin signaling did not simply block chondrocyte differentiation or reverse the differentiating chondrocytes to the earlier mesenchymal condensation stage, it ectopically activated early joint formation process in what was previously cartilage proper.
Figure 6.
Induction of joint markers by ectopic expression of ΔNβ-catenin and Wnt14 in chondrocytes. In situ hybridization with 35S-labeled riboprobes was performed on adjacent limb sections. (A) In the elbow region, ectopic expression of Gdf5, ColI, and Chordin (arrows) was induced at 15.5 dpc in the Col2a1-ΔNβ-catenin transgenic mouse embryo (same embryo as in Fig. 5A′) and correlated with ectopic β-catenin expression shown in Figure 5A′. Ectopic expression of Gdf5, ColI (arrows) in the Col2a1-Wnt14 embryos at 15.5 dpc correlated with the ectopic expression of Wnt14 (arrows). (B) In the digit region of a Col2a1-Wnt14 transgenic embryo at 15.5 dpc, ectopic expression of Wnt14 correlated with ectopic expression of Gdf5, Chordin, and Fgf18 (arrows). Noggin expression was suppressed (arrow). No ectopic expression was detected for ColX, Cbfa1, and Cd44. Ectopic expression was detected for Autotaxin and Scleraxis in the cartilage (arrows) in a pattern similar to Gdf5 ectopic expression, but stronger expression was detected in the cells around the cartilage (arrowheads). In the wild-type (WT) samples, expression of Autotaxin and Scleraxis in ligaments were indicated (arrowheads).
To further examine the loss of chondrocyte phenotype in the Col2a1-ΔNβ-catenin and the Col2a1-Wnt14 mouse embryo, we examined the expression of additional markers for chondrocyte differentiation including Noggin and ColX in the Col2a1-ΔNβ-catenin or Col2a1-Wnt14 embryos. Noggin was expressed in early proliferative chondrocytes and its expression was significantly decreased in both Col2a1-ΔNβ-catenin and Col2a1-Wnt14 embryos (Fig. 6B; data not shown). In addition, ColX was expressed in hypertrophic chondrocytes that did not express ColII. In both Col2a1-ΔNβ-catenin and Col2a1-Wnt14 embryos, ColX expression was delayed, because its expression was absent in the digit (Fig. 6B; Supplementary Fig. S2B), but normal in the humerus (Supplementary Fig. S2A). These data indicate that loss of early chondrocyte characteristics occurs before chondrocyte hypertrophy in the Col2a1-ΔNβ-catenin and Col2a1-Wnt14 embryos.
We then examined the differentiation of other cell types such as osteoblasts and cells that form the tendons and ligaments, because these cells express ColI and/or ColIII (Niederreither et al. 1995; Rossert et al. 1995), both of which were ectopically expressed in the Col2a1-ΔNβ-catenin and Col2a1-Wnt14 embryos (Figs. 6A, 5E′,G′). However, neither ΔNβ-catenin nor Wnt14 induced ectopic osteoblast formation, as Cbfa1 expression was not ectopically activated (Fig. 6B; Supplementary Fig. S2B). It has been reported that many genes that are expressed in the interzone are also expressed in cells that form the tendons and ligaments. For instance, Cd44 and Autotaxin are expressed in the developing joints (Edwards et al. 1994; Noonan et al. 1998; Bachner et al. 1999). However, both are also strongly expressed in some of the connective tissue sheets (Noonan et al. 1996; Bachner et al. 1999), where Scleraxis is highly expressed (Cserjesi et al. 1995; Schweitzer et al. 2001). Scleraxis was also expressed in the joints at a weaker level compared with its expression in the tendon and ligament (Fig. 6B; Supplementary Fig. S2A,B). We found that in the early stages of Wnt14 ectopic expression in the digit cartilage, ectopic expression of Autotaxin and Scleraxis was detected in the cartilage, but stronger ectopic expression of these genes was only detected at the peripheral of the digits where ligament and tendon tissues normally form (Fig. 6B). These results suggested that ectopic Wnt/β-catenin signaling did not transdifferentiate chondrocytes into ligaments or tendons, as the ectopically activated Autotaxin and Scleraxis expression in cartilage was at the level typical of its normal expression in the joints, weaker than its expression in the normal ligament and tendons.
We also found that Cd44 expression was not activated at the same time as Autotaxin or Scleraxis in the digits at 15.5 dpc in the Col2a1-Wnt14 embryo (Fig. 6B). However, after Wnt14 or ΔNβ-catenin had been expressed for a longer period of time, for example, in the humerus at 15.5 dpc, ectopic expression of Cd44 in the cartilage was detected (Supplementary Fig. S2A). This is consistent with the normal temporal expression order of Autotaxin, Scleraxis, and Cd44 in the developing joint and the previous studies that showed that CD44 acts at later stages of joint development to promote cavitation (Pitsillides 2003). During normal synovial joint development, Autotaxin and Scleraxis expression was strong, but the expression of Cd44 was weak in the newly formed digit joints at 15.5 dpc (Fig. 6B). In contrast, in more mature joints, for instance, the elbow joint at 15.5 dpc or the digit joint at 18.5 dpc, Autotaxin and Scleraxis expression was weaker and Cd44 expression was stronger (Supplementary Fig. S2A,B). It appears that the joint markers were ectopically induced by the Wnt/β-catenin signaling in the same temporal order as that of the endogenous one.
Loss of β-catenin in the early differentiating chondrocytes leads to joint fusion
To test whether the β-catenin mediated Wnt signaling is also required for synovial joint formation, we crossed mice containing the β-catenin conditional allele Catnbyc/c with the prion-Cre mice (Scheel et al. 2003) and Col2a1-Cre mice (Ovchinnikov et al. 2000) to inactivate β-catenin in the early differentiated chondrocytes. We found that the Catnbyc/-;Col2a1-Cre and Catnbyc/c; Col2a1-Cre embryos exhibited similar skeletal development defects and they died shortly after birth. The limbs were shortened and the heads were dome shaped (Y. Yang, unpubl.). When synovial joint formation in the limb was examined, we found that some joints between the future tarsal bones in the ankle region were either missing or incompletely formed (Fig. 7A′). The calcaneus was fused to the cuboid and the navicular was partially fused to the intermedial cuneiforms at 15.5 dpc (Fig. 7A′). The joints between these tarsal bones had just formed at 15.5 dpc in the Catnbyc/+;Col2a1-Cre mouse embryos (Fig. 7A). Furthermore, we found that lack of joint formation at 15.5 dpc was not due to the delay in joint formation, because no joint between the fused tarsal bones was found at 18.5 dpc in the mutant embryo (Fig. 7B′). We then examined gene expression in these fused joints. We found that ColII expression was not decreased (Fig. 7D′) and Gdf5 expression was missing or greatly reduced in the fused or partially fused joints, respectively (Fig. 7C′). BrdU labeling showed that chondrocyte proliferation was reduced in the fused joint as compared with the normal joint (data not shown). These data indicate that joint fusion was not a result of extensive cell proliferation and that β-catenin activity is required for inducing synovial joint formation.
Figure 7.
Loss of β-catenin in the early cartilage caused joint fusions. (A,B,A′,B′) Limb sections in the ankle region were stained according to the Weigert-Safranin staining procedure. (A,B) Catnbyc/+;Col2a1-Cre embryo. (A′,B′) The conditional mutant embryo (Catnbyc/c;Col2a1-Cre). The joint between the calcaneus (1) and cuboid (2) was fused (arrow), and the joint between the navicular (3) and the intermediate cuneiform (4) was partially fused (arrowhead) in A′, and completely fused in B′ in the ankle region of the conditional mutant embryo. (C,C′) At 15.5 dpc, Gdf5 expression was missing in the Catnbyc/c; Col2a1-Cre embryo in the fused joint and reduced in the partially fused joint (arrowhead). (D,D′) Immunohistochemistry showing that at 15.5 dpc in the Catnbyc/c;Col2a1-Cre embryo, ColII expression was not down-regulated in the fused joint (arrow), partially down-regulated in the partially fused joint (arrowhead). (E,F) Forelimb (E) and hindlimb (F) of a Catnbyc/+; Dermo1-Cre embryo at 15.5 dpc. (E′,F″) Forelimb (E′) and hindlimb (F′) of a Catnbyc/c;Dermo1-Cre littermate. The joints in the elbow, pelvic, and knee regions were fused (arrow). (G,G′,H,H′) Weigert-Safranin staining procedure (G,G′) or in situ hybridization showing the expression of Gdf5 (H,H′) were performed on 5-μm hind limb sections of a Catnbyc/+; Dermo1-Cre embryo (G,H) and a Catnbyc/c;Dermo1-Cre embryo (G′,H′) at 15.5 dpc. The knee joint (arrows) between femur (F) and tibia (T) was fused (G′) and Gdf5 expression was missing in the fused knee joint in the Catnbyc/c; Dermo1-Cre embryo (H′).
The lack of joint phenotypes in most synovial joints of the Catnbyc/c;Col2a1-Cre embryos may be due to the timing of Cre expression. As Col2a1 promoter is inactivated in the interzone that forms soon after chondrocyte differentiation, the joint interzone cells only express Cre briefly in the Col2a1-Cre mice. The brief expression of Cre driven by the Col2a1 promoter may not be enough to inactivate β-catenin to stop the progress of joint formation in most joints. Because joint formation in the ankle region is completed relatively late (at 15.5 dpc), we reasoned that the time window of Cre expression may be wide enough only in the ankle region to affect the joints between the tarsal bones. To test whether expressing Cre earlier in skeletal formation would result in more efficient β-catenin deletion and lead to more profound joint formation, we used the Dermo1-Cre mice (Yu et al. 2003) to inactivate β-catenin in early mesenchymal condensations. Joint fusions were observed in the elbow, hip, knee, ankle, and digit regions of the Catnbyc/c;Dermo1-Cre embryos (Fig. 7E′,F′G′; data not shown). In addition, in the fused knee joint of a Catnbyc/c;Dermo1-Cre embryo (Fig. 7G′), Gdf5 expression was missing (Fig. 7H′), demonstrating that β-catenin is required for joint formation.
Discussion
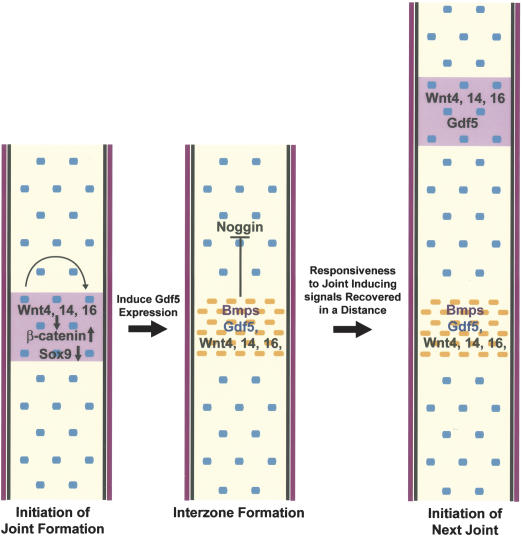
Here, we report the identification of the β-catenin-mediated canonical Wnt pathway as the first signaling pathway that is both sufficient and necessary for the induction of synovial joints in the limb. This indicates that the Wnt/β-catenin may act on the top of the regulatory hierarchy in inducing synovial joint formation. We also showed that Wnt4, Wnt14, and Wnt16 were expressed in the forming joint in overlapping and complementary patterns, and they may play redundant roles in activating β-catenin and joint formation. Taken together, we propose a model for the molecular control of synovial joint induction (Fig. 8). Expression of Wnt genes such as Wnt4, Wnt14, andWnt16 in the presumptive joint results in up-regulation of β-catenin protein levels and inhibition of Sox9 expression. This leads to the induction of synovial joint formation, the first step of which is the formation of interzone cells by inhibiting or reversing chondrocyte differentiation and inducing Gdf5 expression (Fig. 8). Our study is consistent with the previous studies in chick that show that Wnt14 induces early steps of joint formation and that β-catenin protein levels decrease dramatically upon chondrocyte differentiation, and that ectopic accumulation of β-catenin-reversed chondrocyte differentiation (Hartmann and Tabin 2001; Ryu et al. 2002). Our results further indicate that the canonical Wnt signaling pathway has at least two physiological functions in synovial joint formation, suppression of chondrocyte differentiation and the induction of Gdf5 expression. Reversion of chondrocyte differentiation itself is not sufficient for joint formation. Chondrocyte differentiation is reversed in Sox9c/c;Col2a1-Cre mice, but Gdf5 expression is not up-regulated or ectopically expressed (Akiyama et al. 2002). Conversely, induction of Gdf5 alone by the Wnt/β-catenin signaling cannot reverse chondrocyte differentiation, because overexpression of Gdf5 instead caused excessive chondrocyte differentiation and cartilage overgrowth (Francis-West et al. 1999a; Tsumaki et al. 1999).
Figure 8.
Model for the induction of joint formation by the Wnt/β-catenin signaling. Expression of Wnt4, Wnt14, and Wnt16 (shown in pink) in the presumptive joint region of the newly formed cartilage may be the first step in initiation of joint formation. These Wnts signal through the canonical pathway, leading to up-regulation of β-catenin protein level and down-regulation of Sox9 protein in the presumptive joint region, the result of which is the induction of Gdf5 expression and interzone formation. Wnt signaling at this stage suppressed Noggin expression, whereas Gdf5 and other Bmp family members may antagonize Noggin protein function by sequestering it. As Noggin is required for the chondrocytes to sense the joint-inducing signaling, new joint would only form in a distance where active Noggin protein concentration recovers from the inhibitory signals (Wnts, Gdf5, and Bmps) from the previously formed interzone.
We have observed joint fusions when ectopic joint formation was detected in both the Col2a1-ΔN β-catenin and Col2a1-Wnt14 transgenic embryos (Fig. 4D,H). This is consistent with what has been shown in the chick limb bud that ectopically expresses Wnt14, and confirms that the spacing of joint can be regulated by a previously formed joint (Hartmann and Tabin 2001). We speculate that Noggin expressed in chondrocytes may play a critical role in regulating joint spacing by regulating the responsiveness of the differentiating chondrocytes to joint-inducing signals such as Wnt4, Wnt14, and Wnt16 (Fig. 8). It has been proposed that Chordin might act downstream of Wnt14 signaling in regulating joint spacing (Hartmann and Tabin 2001) on the basis of the observation that exogenous application of Noggin in the interdigit area resulted in alteration of Wnt14 expression and decrease in the number of phalanges in the altered digit (Dahn and Fallon 2000; Hartmann and Tabin 2001). However, in the developing joint region, both Chordin and Bmp family members including Bmp2 and Bmp4 are expressed (Francis-West et al. 1999b). Moreover, there is no direct genetic evidence for the function of Chordin in regulating joint spacing. In contrast, Noggin is required for joint formation (Brunet et al. 1998) and Noggin expression was down-regulated by Wnt/β-catenin signaling. In addition, Noggin protein activity might be neutralized by Bmp family members including Gdf5 when joint formation is induced, because GDF5 can also bind Noggin (Merino et al. 1999). Therefore, one mechanism of regulating joint spacing could be through regulating the active Noggin protein level, which is low in a chondrogenic condensation near a developing joint. It is possible that a new joint could only form at a distance from the previously formed joint where the accumulation of active Noggin protein above a certain threshold could occur. Thus, the positions of synovial joints may be regulated at two levels. First, the early patterning signals including Bmps determine the expression pattern of key inductive signaling molecules, which may include Wnt4, Wnt14, and Wnt16 at the presumptive joint region at the beginning of skeletal development. Second, later in the developing skeletal system, Wnt/β-catenin signaling regulates the expression of Noggin and Gdf5 in the developing cartilage to further modify the program of joint spacing.
The β-catenin-mediated canonical Wnt pathway plays a pivotal role in synovial joint formation
In our study, we found that ectopic expression of a constitutively active β-catenin (ΔNβ-catenin) or Wnt14 in newly differentiated chondrocytes under the control of Col2a1 promoter and enhancer both induced early steps of joint formation morphologically and molecularly. Just as in endogenous joint formation, chondrocyte characteristics, such as the expression of the master controlling transcription factor Sox9 were lost, whereas joint markers were ectopically induced in both Col2a1-ΔN β-catenin and Col2a1-Wnt14 transgenic embryos. In addition, the induced joint marker expression has the same temporal order as the endogenous one, which indicates that there are at least two types of genes associated with joint induction. The ones that were induced early such as Gdf5, Fgf18, Autotaxin, and Scleraxis are candidates for being direct targets of Wnt signaling and are likely to act more upstream in the regulatory hierarchy of joint or even tendon formation. Interestingly, Fgf18 has been reported to be a direct transcription target of the canonical Wnt signaling in colon cancer cells (Shimokawa et al. 2003) and joint fusions between the tarsal bones have been observed in the mice in which Fgf receptor 2 (FgfR2) is inactivated in the early condensing mesenchyme (Yu et al. 2003). The joint markers that were induced later, such as Cd44, are likely to be expressed as a result of the differentiation of interzone cells, and they may act at later stages of joint formation. Indeed, CD44-hyaluronan signaling has been implicated in synovial joint cavitation (Pitsillides 2003), a later step in joint morphogenesis. Therefore, both the Wnt14 and β-catenin signaling activate not only the early regulatory gene expression, but also expression of genes important in later joint maturation.
Prior to this work, it was unknown how Wnt14 transduces its signal in joint induction (Hartmann and Tabin 2001). Here, we found that Wnt14 activated LEF/TCF-mediated transcription in vivo and in vitro. In addition, Wnt14 and the activated form of β-catenin not only had the same activity in inducing joint formation, β-catenin was also required for the activity of Wnt14 and joint formation, demonstrating that Wnt14 transduces its signal through the canonical Wnt pathway in joint induction. Because Wnt4, Wnt14, and Wnt16 are expressed in the developing joint region, and they all can activate β-catenin transcription activity, it is not surprising that the function of each individual Wnt ligand is redundant when they all signal through the same canonical pathway in inducing joint formation. Interestingly, we found that Wnt14 was also expressed in the developing joint between the future scapular and humerus, even though this joint does not form through segmenting a pre-existing chondrogenic condensation. It is likely that Wnt14/β-catenin signaling regulates the first step in different kinds of joint formation by either preventing chondrocyte differentiation from mesenchymal precursors or reversing chondrocyte differentiation that has just started.
Relationship of Wnt/β-catenin and Bmp signaling in skeletal morphogenesis
Unlike the β-catenin-mediated canonical Wnt signaling pathway, previously identified signaling molecules such as Gdf5, Noggin, and Wnt14 are either necessary or sufficient, but not both, in joint formation. Gdf5 appears to be a transcription target of the Wnt/β-catenin signaling in our study, it is likely that the Gdf5 signaling pathway is one of those immediately downstream of Wnt/β-catenin signaling, and these pathways need to act together in joint induction. As a potent antagonist of Bmp signaling, Noggin plays a critical role in regulating the strength of Bmp activity. Because joint formation was inhibited in the Noggin mutant mouse limb or in the chick limb that misexpresses an activated Bmp receptor IB (BmpRIB) (Zou et al. 1997), up-regulation of Bmp signaling generates a similar phenotype to that of the loss of the canonical Wnt signaling activity in synovial joint formation. This raises an interesting question as to whether Wnt and BMP signalings are antagonistic to each other, and that their relative signaling strength may determine a particular biological outcome such as joint formation. Interestingly, mutually exclusive expression patterns and functional antagonism between wingless (Wg) and decapentaplegic (dpp), the Drosophila orthologs of vertebrate Wnt and Bmps, respectively, have been observed in several embryonic developmental processes in Drosophila (Jiang and Struhl 1996; Lee and Treisman 2001). Testing whether Wnt4, Wnt14, and Wnt16 are still expressed, and whether ectopic Wnt14/β-catenin signaling can still induce joint formation in the Noggin mutant embryo, will provide more insight into the functional relationship of Wnt and Bmps in joint formation.
Apart from their expression in the forming joints, Wnt4, Wnt14, and Wnt16 were also expressed in mesenchymal cells that surround the newly formed cartilage (Fig. 1). This pattern of expression may lead to inhibition of cartilage appositional growth to limit cartilage lateral expansion, which is critical for maintaining the spacing between each individual skeletal element. Because Wnt antagonists such as SFRP-2 and SFRP-3 are expressed in the prechondrogenic mesenchymal condensations (Lescher et al. 1998; Duprez et al. 1999), it is likely that the complementary expression patterns of Wnt antagonists and Wnt ligands in chondrogenic mesenchymal condensations and their surrounding mesenchyme are functionally important in defining the position and spacing of parallel skeletal elements such as the digit rays in the limb. Indeed, implanting cell pellets that ectopically express SFRP-2 in the interdigit area induced ectopic cartilage formation (Topol et al. 2003). Moreover, we have observed partial fusion of digit 4 with digit 5 in the Catnbyc/c;Dermo1-Cre embryo (Y. Yang, unpubl.). Interestingly, formed cartilages expanded laterally, fused with each other, and interdigital tissue was lost in the limb of Noggin-/- embryos. This phenotype further indicates that the Wnt and BMP signalings exhibit opposite activities not only in maintaining chondrocyte phenotype and inducing joint formation, but also in recruiting chondrocytes from mesenchymal progenitors. This is supported by our observation that deletion of β-catenin resulted in significantly more cartilage nodule formation in micromass culture (Fig. 3F), a similar phenotype has been observed when exogenous GDF5 or BMP4 proteins were applied to wild-type micromass cultures (Hatakeyama et al. 2004).
Maintenance of joint cartilage
After the formation of a mature joint, the two opposing articular cartilages must be properly maintained. Because the Wnt/β-catenin signaling can reverse chondrocyte differentiation, it would be important to keep the Wnt/β-catenin signaling at an appropriate level in the articular chondrocytes. Abnormal expression of Wnts in articular chondrocytes or the fibroblast-like synoviocytes that cover the articular cartilage would be likely to disrupt normal joint structure in pathological conditions such as rheumatoid arthritis and osteoarthritis. It appears that Wnt signaling level is kept low at least in part through the expression of Wnt antagonists. SFRP-3 is expressed in the articular chondrocytes and SFRP-2 is expressed in the perichondrium including the articular surface (Y. Yang, unpubl.). In addition, it was recently shown that Sox9 in differentiated chondrocytes promotes the degradation of β-catenin (Akiyama et al. 2004). Because constitutive activation of the canonical Wnt signaling pathway has been found in the fibroblast-like synoviocytes in rheumatoid arthritis and up-regulated Wnt signaling is associated with fibronectin and pro-matrix metalloproteinase 3 expression (Sen et al. 2002), further understanding of the Wnt/β-catenin signaling in joint formation will be important for elucidating a fundamental developmental process and identifying therapeutic targets for treating cartilage and joint diseases.
Materials and methods
Generation of transgenic and targeted mouse lines
The Col2a1 transgenic vector used to generate the Col2a1-ΔNβ-catenin and Col2a1-Wnt14 mice has been described (Yang et al. 2003). Transgenic mice were generated by pronuclear injection and G0 embryos at 15.5 and 18.5 dpc were analyzed. All transgenic embryos that expressed the transgenes showed phenotypes and the severity of phenotypes correlated with the level of transgene expression. The results on representative ones (more than 50% of the transgenic embryos) were shown. Genomic DNA prepared from embryonic liver was genotyped by PCR. Oligos pWnt14 5′-GGTTCACTGCCTGTTAGC-3′ and pColII 5′-GCAACGTGCTGGTTGTTGTG-3′ were used to genotype Col2a1-Wnt14 mice. Oligos pcatnb 5′-CAGCATCAAACTGT GTAGATG-3′ and pColII were used to genotype the Col2a1- ΔNβ-catenin mice. Col2a1-Cre transgenic mice that have been described before were kindly provided by Dr. Richard Behringer (Department of Molecular Genetics, M.D. Anderson Cancer Center, Houston, TX) (Ovchinnikov et al. 2000).
For the conditional β-catenin allele, a complete description of the targeting vector construct, chimera production, and allele identification is provided in the Supplemental Material.
LacZ staining, histology, in situ hybridization, and immunohistochemistry
Wild-type and mutant embryos were dissected in PBS. For LacZ staining, embryos were fixed in 0.5% formaldehyde and 0.5% glutaraldehyde for 10 min at room temperature. LacZ staining was performed as previously described (Topol et al. 2003). For in situ hybridizations, embryos were fixed in 4% paraformaldehyde at 4°C overnight. Some fixed samples were embedded in paraffin and sectioned at 5-μm thickness. Histological analysis, BrdU labeling, immunohistochemistry, whole mount and radioactive 35S RNA in situ hybridization were performed as described before (Yang et al. 2003). Primary antibodies included anti-Sox9 (Santa Cruz) at 1:100, anti-ColII (Santa Cruz) at 1:200, Anti-ColIII (Santa Cruz) at 1:50, and anti-β-catenin (Transduction laboratories) at 1:50. Signals were detected using Alexa Fluor-conjugated secondary antibodies (Molecular Probes). Full-length cDNAs of Wnt14, Wnt16, and Fgf18 were generated by PCR and subcloned into the pBluescript vector. The cDNAs were verified by sequencing and used to generate probes for in situ hybridization. EST clones for Autotaxin (Clone ID: 5043391), Scleraxis (Clone ID: 5056 300), Noggin (UI-M-AM0-adr-a-03-0-UI), Cd44 (Clone ID: 4911674), and Chordin (UI-M-BH3-bsf-u-05-0-UI) were ordered from Invitrogen and used to generate RNA probes. Lef1 cDNA was ordered from ATCC. Other RNA probes have been described previously: ColX, ColII, and Cbfa1 (Yang et al. 2003); Wnt4 (Parr et al. 1993); and Gdf5 (Storm and Kingsley 1999).
Skeletal analysis
Embryos at 15.5 dpc or 18.5 dpc were dissected in PBS. The embryos were then skinned, eviscerated, and fixed in 95% ethanol. Skeletal preparations were performed as described (McLeod 1980).
Plasmids and virus
The coding regions of Wnt4, Wnt14, and Wnt16 were inserted into pCS-2, pIRES-hrGFP-1a expression vector (Stratagene), and pCDNA3 (Invitrogen), respectively. The Cre-adenenovirus was provided by Dr. Francis Collins (National Genome Research Institute, National Institutes of Health, Bethesda, MD), the Wnt14 and ΔNLef1-adenovirus were generated using a kit purchased from Clontech according to manufacturer's instruction.
Cell culture, transfection, and reporter analysis
Rat chondrosarcoma cells were obtained from Dr. Yoshi Yamada (National Institutes of Health, Bethesda, MD). Cells were seeded the day before transfection and electroporation was done with the Nucleofector technology (Amaxa GmbH). The TOPFLASH assay was performed as described before (Topol et al. 2003). Luciferase activity was measured 24 h after transfection according to the Dual-Luciferase Reporter Assay System (Promega). The results are shown as relative luciferase activity. The histograms are presented as the average ± S.D. from three independent transfections.
Micromass cultures
Micromass cultures were performed according to a procedure described previously (Akiyama et al. 2002). The Cre-adenovirus was added to the limb mesenchymal cell suspension (∼2 × 106 cells/mL) at 5 × 109 pfu/mL, and the cell suspension was gently rocked at 4°C for 2 h. Then, the cell suspension was precipitated and resuspended to 2 × 107 cells/mL with medium (DMEM, 10% FCS) containing the Cre-adenovirus (5 × 109 pfu/mL) and 20-μL drops were plated. The Wnt14-adenovirus at 1 × 108 pfu/mL was added to the micromass culture 24 h after the cells were plated in high density.
Acknowledgments
We are very grateful to Dr. David Ornitz for providing us the Dermo1-Cre mice and Drs. Francis Collins and Peter Scacheri for providing us the Cre-Adenovirus. We thank Drs. Lee Niswander, Robert Nussbaum, and Leslie Biesecker for critical reading of the manuscript; members of the Yang lab for stimulating discussion; and Mike Cichanowski for help with figure preparation.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1230704.
References
- Akiyama H., Chaboissier, M.C., Martin, J.F., Schedl, A., and de Crombrugghe, B. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes & Dev. 16: 2813-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H., Lyons, J.P., Mori-Akiyama, Y., Yang, X., Zhang, R., Zhang, Z., Deng, J.M., Taketo, M.M., Nakamura, T., Behringer, R.R., et al. 2004. Interactions between Sox9 and β-catenin control chondrocyte differentiation. Genes & Dev. 18: 1072-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer C.W., Dowthwaite, G.P., and Francis-West, P. 2003. Development of synovial joints. Birth Defects Res. Part C Embryo Today 69: 144-155. [DOI] [PubMed] [Google Scholar]
- Bachner D., Ahrens, M., Betat, N., Schroder, D., and Gross, G. 1999. Developmental expression analysis of murine autotaxin (ATX). Mech. Dev. 84: 121-125. [DOI] [PubMed] [Google Scholar]
- Brunet L.J., McMahon, J.A., McMahon, A.P., and Harland, R.M. 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455-1457. [DOI] [PubMed] [Google Scholar]
- Capdevila J. and Johnson, R.L. 1998. Endogenous and ectopic expression of noggin suggests a conserved mechanism for regulation of BMP function during limb and somite patterning. Dev. Biol. 197: 205-217. [DOI] [PubMed] [Google Scholar]
- Craig F.M., Bentley, G., and Archer, C.W. 1987. The spatial and temporal pattern of collagens I and II and keratan sulphate in the developing chick metatarsophalangeal joint. Development 99: 383-391. [DOI] [PubMed] [Google Scholar]
- Cserjesi P., Brown, D., Ligon, K.L., Lyons, G.E., Copeland, N.G., Gilbert, D.J., Jenkins, N.A., and Olson, E.N. 1995. Scleraxis: A basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 121: 1099-1110. [DOI] [PubMed] [Google Scholar]
- Dahn R.D. and Fallon, J.F. 2000. Interdigital regulation of digit identity and homeotic transformation by modulated BMP signaling. Science 289: 438-441. [DOI] [PubMed] [Google Scholar]
- DasGupta R. and Fuchs, E. 1999. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126: 4557-4568. [DOI] [PubMed] [Google Scholar]
- Duprez D., Leyns, L., Bonnin, M.A., Lapointe, F., Etchevers, H., De Robertis, E.M., and Le Douarin, N. 1999. Expression of Frzb-1 during chick development. Mech. Dev. 89: 179-183. [DOI] [PubMed] [Google Scholar]
- Edwards J.C., Wilkinson, L.S., Jones, H.M., Soothill, P., Henderson, K.J., Worrall, J.G., and Pitsillides, A.A. 1994. The formation of human synovial joint cavities: A possible role for hyaluronan and CD44 in altered interzone cohesion. J. Anat. 185: 355-367. [PMC free article] [PubMed] [Google Scholar]
- Francis-West P.H., Abdelfattah, A., Chen, P., Allen, C., Parish, J., Ladher, R., Allen, S., MacPherson, S., Luyten, F.P., and Archer, C.W. 1999a. Mechanisms of GDF-5 action during skeletal development. Development 126: 1305-1315. [DOI] [PubMed] [Google Scholar]
- Francis-West P.H., Parish, J., Lee, K., and Archer, C.W. 1999b. BMP/GDF-signalling interactions during synovial joint development. Cell Tissue Res. 296: 111-119. [DOI] [PubMed] [Google Scholar]
- Hartmann C. and Tabin, C.J. 2001. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell 104: 341-351. [DOI] [PubMed] [Google Scholar]
- Hatakeyama Y., Tuan, R.S., and Shum, L. 2004. Distinct functions of BMP4 and GDF5 in the regulation of chondrogenesis. J. Cell. Biochem. 91: 1204-1217. [DOI] [PubMed] [Google Scholar]
- Jiang J. and Struhl, G. 1996. Complementary and mutually exclusive activities of decapentaplegic and wingless organize axial patterning during Drosophila leg development. Cell 86: 401-409. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker, N., Morin, P.J., van Wichen, D., de Weger, R., Kinzler, K.W., Vogelstein, B., and Clevers, H. 1997. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC-/- colon carcinoma. Science 275: 1784-1787. [DOI] [PubMed] [Google Scholar]
- Lee J.D. and Treisman, J.E. 2001. The role of Wingless signaling in establishing the anteroposterior and dorsoventral axes of the eye disc. Development 128: 1519-1529. [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Zhou, G., Mukhopadhyay, K., Smith, C.N., Zhang, Z., Eberspaecher, H., Zhou, X., Sinha, S., Maity, S.N., and de Crombrugghe, B. 1996. An 18-base-pair sequence in the mouse proα1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol. Cell. Biol. 16: 4512-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescher B., Haenig, B., and Kispert, A. 1998. sFRP-2 is a target of the Wnt-4 signaling pathway in the developing metanephric kidney. Dev. Dyn. 213: 440-451. [DOI] [PubMed] [Google Scholar]
- Maruoka Y., Ohbayashi, N., Hoshikawa, M., Itoh, N., Hogan, B.L., and Furuta, Y. 1998. Comparison of the expression of three highly related genes, Fgf8, Fgf17 and Fgf18, in the mouse embryo. Mech. Dev. 74: 175-177. [DOI] [PubMed] [Google Scholar]
- McLeod M.J. 1980. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology 22: 299-301. [DOI] [PubMed] [Google Scholar]
- Merino R., Macias, D., Ganan, Y., Economides, A.N., Wang, X., Wu, Q., Stahl, N., Sampath, K.T., Varona, P., and Hurle, J.M. 1999. Expression and function of Gdf-5 during digit skeletogenesis in the embryonic chick leg bud. Dev. Biol. 206: 33-45. [DOI] [PubMed] [Google Scholar]
- Morrison E.H., Ferguson, M.W., Bayliss, M.T., and Archer, C.W. 1996. The development of articular cartilage: I. The spatial and temporal patterns of collagen types. J. Anat. 189: 9-22. [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay K., Lefebvre, V., Zhou, G., Garofalo, S., Kimura, J.H., and de Crombrugghe, B. 1995. Use of a new rat chondrosarcoma cell line to delineate a 119-base pair chondrocyte-specific enhancer element and to define active promoter segments in the mouse pro-α 1(II) collagen gene. J. Biol. Chem. 270: 27711-27719. [DOI] [PubMed] [Google Scholar]
- Nalin A.M., Greenlee Jr., T.K., and Sandell, L.J. 1995. Collagen gene expression during development of avian synovial joints: Transient expression of types II and XI collagen genes in the joint capsule. Dev. Dyn. 203: 352-362. [DOI] [PubMed] [Google Scholar]
- Nelson W.J. and Nusse, R. 2004. Convergence of Wnt, β-catenin, and cadherin pathways. Science 303: 1483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K., D'Souza, R., Metsaranta, M., Eberspaecher, H., Toman, P.D., Vuorio, E., and De Crombrugghe, B. 1995. Coordinate patterns of expression of type I and III collagens during mouse development. Matrix Biol. 14: 705-713. [DOI] [PubMed] [Google Scholar]
- Niemann C., Owens, D.M., Hulsken, J., Birchmeier, W., and Watt, F.M. 2002. Expression of ΔNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development 129: 95-109. [DOI] [PubMed] [Google Scholar]
- Noonan K.J., Stevens, J.W., Tammi, R., Tammi, M., Hernandez, J.A., and Midura, R.J. 1996. Spatial distribution of CD44 and hyaluronan in the proximal tibia of the growing rat. J. Orthop. Res. 14: 573-581. [DOI] [PubMed] [Google Scholar]
- Noonan K.J., Reiter, R.S., Kurriger, G.L., Martin, J.A., Maynard, J.A., and Stevens, J.W. 1998. Spatial and temporal expression of CD44 isoforms in the developing and growing joints of the rat limb. J. Orthop. Res. 16: 100-103. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D.A., Deng, J.M., Ogunrinu, G., and Behringer, R.R. 2000. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26: 145-146. [PubMed] [Google Scholar]
- Parr B.A., Shea, M.J., Vassileva, G., and McMahon, A.P. 1993. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development 119: 247-261. [DOI] [PubMed] [Google Scholar]
- Pathi S., Rutenberg, J.B., Johnson, R.L., and Vortkamp, A. 1999. Interaction of Ihh and BMP/Noggin signaling during cartilage differentiation. Dev. Biol. 209: 239-253. [DOI] [PubMed] [Google Scholar]
- Pitsillides A.A. 2003. Identifying and characterizing the joint cavity-forming cell. Cell. Biochem. Funct. 21: 235-240. [DOI] [PubMed] [Google Scholar]
- Pitsillides A.A., Skerry, T.M., and Edwards, J.C. 1999. Joint immobilization reduces synovial fluid hyaluronan concentration and is accompanied by changes in the synovial intimal cell populations. Rheumatology (Oxford) 38: 1108-1112. [DOI] [PubMed] [Google Scholar]
- Pizette S. and Niswander, L. 2000. BMPs are required at two steps of limb chondrogenesis: Formation of prechondrogenic condensations and their differentiation into chondrocytes. Dev. Biol. 219: 237-249. [DOI] [PubMed] [Google Scholar]
- Rossert J., Eberspaecher, H., and de Crombrugghe, B. 1995. Separate cis-acting DNA elements of the mouse pro-α 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell. Biol. 129: 1421-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.H., Kim, S.J., Kim, S.H., Oh, C.D., Hwang, S.G., Chun, C.H., Oh, S.H., Seong, J.K., Huh, T.L., and Chun, J.S. 2002. Regulation of the chondrocyte phenotype by β-catenin. Development 129: 5541-5550. [DOI] [PubMed] [Google Scholar]
- Scheel J.R., Garrett, L.J., Allen, D.M., Carter, T.A., Randolph-Moore, L., Gambello, M.J., Gage, F.H., Wynshaw-Boris, A., and Barlow, C. 2003. An inbred 129SvEv GFPCre transgenic mouse that deletes loxP-flanked genes in all tissues. Nucleic Acids Res. 31: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R., Chyung, J.H., Murtaugh, L.C., Brent, A.E., Rosen, V., Olson, E.N., Lassar, A., and Tabin, C.J. 2001. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128: 3855-3866. [DOI] [PubMed] [Google Scholar]
- Sen M., Reifert, J., Lauterbach, K., Wolf, V., Rubin, J.S., Corr, M., and Carson, D.A. 2002. Regulation of fibronectin and metalloproteinase expression by Wnt signaling in rheumatoid arthritis synoviocytes. Arthritis Rheum. 46: 2867-2877. [DOI] [PubMed] [Google Scholar]
- Settle S.H., Rountree, R.B., Sinha, A., Thacker, A., Higgins, K., and Kingsley, D.M. 2003. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 254: 116-130. [DOI] [PubMed] [Google Scholar]
- Shimokawa T., Furukawa, Y., Sakai, M., Li, M., Miwa, N., Lin, Y.M., and Nakamura, Y. 2003. Involvement of the FGF18 gene in colorectal carcinogenesis, as a novel downstream target of the β-catenin/T-cell factor complex. Cancer Res. 63: 6116-6120. [PubMed] [Google Scholar]
- Shubin N.H. and Alberch, P. 1986. A morphogenetic approach to the origin and basic organization of the tetrapod limb. Evol. Biol. 20: 319-387. [Google Scholar]
- Storm E.E. and Kingsley, D.M. 1996. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development 122: 3969-3979. [DOI] [PubMed] [Google Scholar]
- ____. 1999. GDF5 coordinates bone and joint formation during digit development. Dev. Biol. 209: 11-27. [DOI] [PubMed] [Google Scholar]
- Storm E.E., Huynh, T.V., Copeland, N.G., Jenkins, N.A., Kingsley, D.M., and Lee, S.J. 1994. Limb alterations in brachypodism mice due to mutations in a new member of the TGF β-superfamily. Nature 368: 639-643. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Takeuchi, J., Koshiba-Takeuchi, K., and Ogura, T. 2004. Tbx genes specify posterior digit identity through Shh and BMP signaling. Dev. Cell 6: 43-53. [DOI] [PubMed] [Google Scholar]
- Topol L., Jiang, X., Choi, H., Garrett-Beal, L., Carolan, P.J., and Yang, Y. 2003. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent β-catenin degradation. J. Cell. Biol. 162: 899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumaki N., Tanaka, K., Arikawa-Hirasawa, E., Nakase, T., Kimura, T., Thomas, J.T., Ochi, T., Luyten, F.P., and Yamada, Y. 1999. Role of CDMP-1 in skeletal morphogenesis: Promotion of mesenchymal cell recruitment and chondrocyte differentiation. J. Cell. Biol. 144: 161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M.T., Axelrod, J.D., and Moon, R.T. 2003. A second canon. Functions and mechanisms of β-catenin-independent Wnt signaling. Dev. Cell 5: 367-377. [DOI] [PubMed] [Google Scholar]
- Wright E., Hargrave, M.R., Christiansen, J., Cooper, L., Kun, J., Evans, T., Gangadharan, U., Greenfield, A., and Koopman, P. 1995. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat. Genet. 9: 15-20. [DOI] [PubMed] [Google Scholar]
- Yang Y. 2003. Wnts and wing: Wnt signaling in vertebrate limb development and musculoskeletal morphogenesis. Birth Defects Res. Part C Embryo Today 69: 305-317. [DOI] [PubMed] [Google Scholar]
- Yang Y., Topol, L., Lee, H., and Wu, J. 2003. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development 130: 1003-1015. [DOI] [PubMed] [Google Scholar]
- Yu K., Xu, J., Liu, Z., Sosic, D., Shao, J., Olson, E.N., Towler, D.A., and Ornitz, D.M. 2003. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130: 3063-3074. [DOI] [PubMed] [Google Scholar]
- Zou H., Wieser, R., Massague, J., and Niswander, L. 1997. Distinct roles of type I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes & Dev. 11: 2191-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]