Abstract
Matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis identified enolase as a cell surface component of Streptococcus mutans, which was confirmed by enzyme-linked immunosorbent assay, Western blotting, and transmission electron microscopy. Surface enolase was demonstrated to bind to human plasminogen and salivary mucin MG2. The results suggested a role for enolase in S. mutans attachment, clearance, or breach of the bloodstream barrier.
Streptococcus mutans is an alpha-hemolytic and nongroupable streptococcus, which causes dental caries and occasionally bacterial endocarditis (BE) (20). Some surface proteins of S. mutans are virulence factors with functions critical to viability, such as adhesion (2). Adhesion is one of the most important steps in the pathogenicity of microorganisms, and bacterial cell surface-mediated interactions with salivary proteins may be the first step in S. mutans attachment to a tooth surface coated with salivary pellicle or saliva-coated plaque (11). Liu et al. reported that S. mutans binds to salivary mucins, but the specific molecule responsible for this binding was unknown (18). Salivary mucin MG2 is a 180-kDa glycoprotein (22). Mucin MG2 binds to several oral microbes, including S. mutans, Actinobacillus actinomycetemcomitans, and Candida albicans, and forms heterotypic complexes with salivary proteins, including secretory immunoglobulin A (SIgA) and lactoferrin (4, 15, 18, 19, 29).
Enolase is a 47-kDa cytoplasmic enzyme in the glycolytic pathway (21). There are three distinct isoforms (α, β, and γ) of enolase, each with the same molecular mass. α-Enolase is the isoform typically found in bacteria. Surface α-enolase has been recently identified on Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus pneumonia, Streptococcus oralis, and A. actinomycetemcomitans (3, 13, 16, 25, 32). Surface enolase of S. pyogenes binds and activates human plasminogen, a molecule which plays a crucial role in fibrinolysis, homeostasis, and the degradation of extracellular matrix (8). Proteins, such as enolase, with exposed carboxyl-terminal lysines on the cell surface can bind and promote plasminogen activation (33).
In the present study, we identified α-enolase from a cell surface protein preparation of S. mutans A32-2 by two-dimensional polyacrylamide gel electrophoresis (PAGE) and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis. The results of enzyme-linked immunosorbent assay (ELISA), Western blot, and transmission electron microscopy (TEM) confirmed α-enolase on the cell surface as well as in the cytoplasm of S. mutans. Results of Western blot, TEM, and ELISA experiments demonstrate that S. mutans surface enolase binds to human plasminogen and human salivary mucin MG2.
S. mutans A32-2 was isolated from a highly-caries-active patient and expresses significantly more surface proteins than isolates from caries-free subjects (26). Surface protein, cell wall, and cytoplasmic samples were purified according to methods described previously (17, 24, 26). Briefly, S. mutans was harvested, washed in surface protein buffer (10 mM phosphate-buffered saline, 1 mM CaCl2, pH 7.2, 1% phenylmethylsulfonyl fluoride), and sheared in a Waring blender for three 1-min cycles at high speed. The unbroken cells and debris were removed by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant containing surface proteins was centrifuged at 16,000 × g for 15 min at 4°C, transferred to a fresh tube, and centrifuged again at 110,000 × g for 2.5 h at 4°C. The pellet containing the surface proteins was resuspended in surface protein buffer and stored at −80°C. Cytoplasmic and cell wall fractions were collected from disrupted cells by a sucrose gradient. Protein concentrations of all preparations were measured by a QuantiPro bicinchoninic acid assay kit (Sigma).
S. mutans subcellular samples or saliva components were separated by sodium dodecyl sulfate (SDS)-PAGE (7.5% polyacrylamide). MALDI-TOF MS was performed on trypsin-digested, Coomassie stained spots excised from surface proteins separated on two-dimensional PAGE gels (32) at the Biochemistry Biotechnology Facility (Dept. of Biochemistry, Indiana University School of Medicine). About 160 spots from the gel were visualized by computer-generated imaging, and 96 spots were analyzed by MALDI-TOF MS with Profound software (34) (http://129.85.19.192/profound_bin/WebProFound.exe), based on Z value and coverage percentage (data not shown). Two spots with molecular masses of 47 and 60 kDa and pIs of pIs 4.7 and 6.0, respectively, were identified as enolase. Enolase is predicted to be encoded by a single-copy gene designated SMU.1247 (1). Around 67% of the smaller enolase peptide mass matched the computed enolase mass using the top 50 MALDI-TOF peptides, and the Z value was 2.36. Around 38% of the larger measured enolase peptide mass matched the computed enolase mass using the top 50 MALDI-TOF peptides, and the Z value was 1.65. Modification of each protein was also predicted based on the MALDI-TOF data. The enolase peptide 57SRYGGLGTQK66 was predicted to be phosphorylated in both spots, and the computed mass for the phosphorylated peptide was 1,305.455, while the measured mass was 1,304.812. Serine, threonine, or tyrosine in this peptide may be potentially phosphorylated. The peptides 11EVLDSR16, 426SFYNLK431, and 393TGSLSR398 may also be potentially phosphorylated in the larger spot. Posttranslational modification may alter the binding properties of enolase as well as the molecular weight. Our analysis of the predicted protein sequence of enolase of S. mutans reveals that this protein lacks the hexameric motif (LPXTGX) typical for anchoring proteins of gram-positive bacteria to the cell wall (9). It is not yet understood how enolase is transported to and attaches to the surface of the bacterium (23). Human enolase can be posttranslationally acylated or phosphorylated in vivo and in vitro (5, 7). Our analysis of the enolase peptide sequence of S. mutans suggests the possibility of phosphorylation sites based on the MALDI-TOF data. Phosphorylation can affect protein-protein interactions (6), and phosphorylated enolase may bind to the receptor on the cell surface.
Bacterial samples for immunogold bead staining were prepared for TEM (http://www.nanoprobes.com/Inf2016.html) to study the presence of surface enolase on S. mutans and its binding ability to human plasminogen. The presence of gold beads at the outer edge of S. mutans cells demonstrated anti-α-enolase antibody (1:200; Laboratory of Bacterial Pathogenesis and Immunology, Rockefeller University, New York, N.Y.) reacting with a protein on the cell wall of S. mutans (results not shown). Control cells without anti-α-enolase antibody did not stain. These results supported the MALDI-TOF finding of enolase on the surface of S. mutans. Incubation of S. mutans with human plasminogen (0.02 mg/ml; ICN Biomedicals, Aurora, Ohio) followed by incubation with goat anti-plasminogen antibody (1:200; ICN) and anti-goat antibody conjugated to gold beads (1:200; Nanoprobes, Yaphank, N.Y.) demonstrated the ability of human plasminogen to bind to the surface of S. mutans. Control cells without human plasminogen did not stain. The results suggested that S. mutans surface proteins bound to human plasminogen.
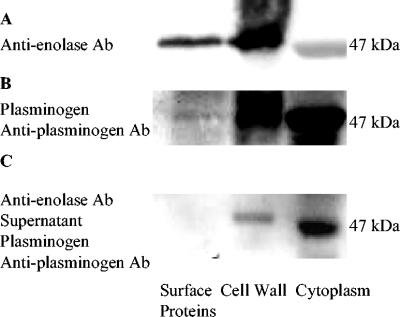
Far-Western blot analysis (14) was used to study the binding ability of enolase to human plasminogen and mucin MG2. Enolase in the cell surface, cell wall, and cytoplasmic samples reacted with anti-α-enolase antibody (1:300) in Western blots (Fig. 1A). Enolase (47 kDa) in the cell surface, cell wall, and cytoplasmic fractions binds plasminogen (0.02 mg/ml; Fig. 1B). Cytoplasmic enolase binding with plasminogen was most intense in Fig. 1B, while cell wall enolase reacting with antienolase antibody was most intense in Fig. 1A. Since antienolase antibody is polyclonal, it can recognize multiple domains of the enolase molecule and may recognize domains of cell wall enolase molecules with high efficiency. Plasminogen recognizes only C-terminal lysines (8), and since the concentration of enolase in cytoplasm was higher than that in the other samples, this produced a more intense band. The binding of enolase to plasminogen can be blocked by antienolase antibody (Fig. 1C). Negative controls were not incubated with plasminogen or antiplasminogen antibody, and there was no band in the negative controls.
FIG. 1.
Plasminogen binding to S. mutans enolase. (A) Electrophoretically separated cell surface, cell wall, and cytoplasmic samples were incubated with rabbit anti-enolase antibody (Ab) (1:300) and anti-rabbit IgG-peroxidase (1:500). (B) Separated surface protein, cell wall, and cytoplasm samples were incubated with plasminogen (0.02 mg/ml), goat anti-plasminogen, and anti-goat IgG-peroxidase (1:500). (C) Surface protein, cell wall, and cytoplasm samples were incubated with blocking rabbit anti-enolase antibody (1:300) and centrifuged. The supernatants of those samples were isolated by SDS-PAGE and transferred to a PVDF membrane, which was incubated with plasminogen (0.02 mg/ml), goat anti-plasminogen, and anti-goat IgG-peroxidase (1:500). The binding activity of enolase with plasminogen was decreased by blocking rabbit anti-enolase antibody.
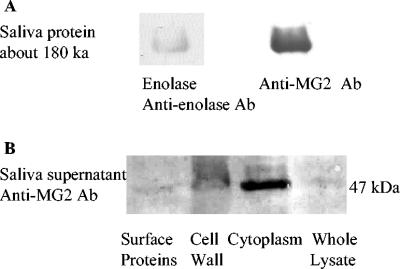
To investigate the binding of salivary proteins to enolase, electrophoretically separated salivary proteins (saliva supernatant from laboratory volunteers; Institutional Review Board approval) were transferred to the polyvinylidene difluoride (PVDF) membrane and incubated with purified enolase (0.05 mg/ml; Sigma) followed by incubation with antienolase antibody (1:200). Enolase interacted with only one protein, with a molecular mass of 180 kDa (Fig. 2A). Schiff's staining of the salivary proteins indicated the 180-kDa protein was a highly glycosylated protein (results not shown), and the Western blots of salivary proteins incubated with anti-mucin MG2 antibody (1:1,000; Laboratory of Biochemistry, Boston University) demonstrated that the 180-kDa protein was mucin MG2 (Fig. 2A). Separated subcellular proteins, when incubated with salivary supernatant (containing mucins MG1 and MG2) followed by incubation with anti-mucin MG2 antibody, confirmed enolase in subcellular samples binding to salivary mucin MG2 (Fig. 2B). Negative controls were not incubated with saliva supernatant or anti-mucin MG2 antibody, and there was no band in the negative controls. The interaction was confirmed by coimmunoprecipitation (results not shown). The complex, formed by mixing the S. mutans subcellular samples with salivary proteins, was purified with anti-γ-enolase antibody (1:200) and protein A (10 μl; Sigma). Mucin MG2 in the complex was confirmed with anti-mucin MG2 antibody (1:1,000) in Western blot analysis.
FIG. 2.
Enolase binding to salivary mucin MG2. (A) Saliva supernatant was separated by SDS-PAGE and transferred to a PVDF membrane. The left lane contained a salivary protein of about 180 kb, which interacted with purified enolase (0.05 mg/ml), rabbit anti-enolase antibody (Ab) (1:200), and anti-rabbit IgG conjugated with peroxidase (1:500). The right lane contained a salivary protein of about 180 kb, which interacted with rabbit anti-mucin MG2 (1:1,000) and anti-rabbit IgG conjugated with peroxidase (1:500). (B) Subcellular S. mutans samples were separated by SDS-PAGE and transferred to a PVDF membrane. Enolase (47 kDa) in surface protein, cell wall, cytoplasm, and whole-cell lysate samples was incubated with saliva supernatant (as a source of mucin MG2), rabbit anti-mucin MG2 antibody (1:1,000), and anti-rabbit IgG-peroxidase (1:500).
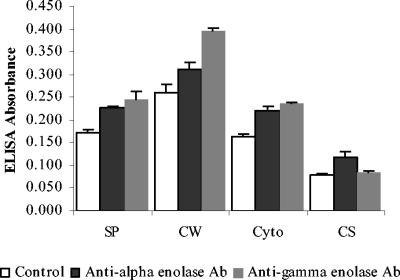
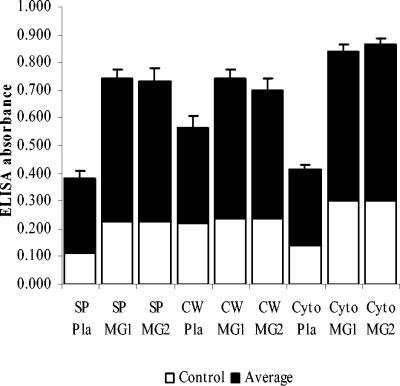
ELISA was used to assess enolase localization and the ability of enolase to bind human plasminogen and mucins MG1 and MG2. ELISA confirmed that not only was enolase present in the cytoplasmic fraction as expected, it was also observed in the cell surface fraction (Fig. 3). As expected, α-enolase reacted with anti-α-enolase (1:500) and anti-γ-enolase antibodies (1:500; Sigma), while the no-antibody negative controls did not. The data were analyzed by Student's t test. The version for independent samples was used. The variances in the groups were assumed to be equal, and the test was two sided. The P values of all groups were <0.05, which means there are significant differences between experimental groups and control groups. The presence of enolase in the culture supernatant suggested either an active secretion of enolase or the presence of lysed bacterial cells. ELISA plates were coated with anti-α enolase antibody (1:500), which bound enolase in surface, cell wall, and cytoplasm samples. Enolase subsequently reacted with human plasminogen (0.02 mg/ml), which was detected by incubation with goat anti-human plasminogen antibody (1:1,000) followed by peroxidase-conjugated anti-goat IgG antibody (1:1,000; Sigma), while the no-plasminogen controls were negative. The data were analyzed by t test as described before, and the P values of all groups were <0.05. ELISA results supported the conclusion that S. mutans surface enolase can bind to human plasminogen (Fig. 4). In a similar experiment, the plate was coated with S. mutans subcellular samples, incubated with saliva supernatant, and reacted with anti-mucin MG1 or anti-mucin MG2 antibodies (1:1,000). The data were analyzed by t test, and the P values were <0.05. ELISA results indicated that proteins in the subcellular samples reacted with both mucins MG1 and MG2 (Fig. 4). Combining the ELISA data with the Western blot in Fig. 2 confirmed that enolase in the subcellular samples reacted with salivary mucin MG2.
FIG. 3.
Localization of enolase in the subcellular fractions of S. mutans by ELISA. ELISA plates were coated with surface protein (SP), cell wall (CW), cytoplasm (Cyto), and cultural supernatant (CS) samples and incubated with rabbit anti-α-enolase or anti-γ-enolase antibody (1:500) and peroxidase-conjugated anti-rabbit IgG antibody (1:1,000). Following color development, the absorbance of each well was measured at 490 nm. The data were analyzed by Student's t test (independent samples with equal variances and two sides), and the P values of all groups were <0.05.
FIG. 4.
Ability of S. mutans enolase to bind to plasminogen and salivary mucins MG1 and MG2. The ability of enolase in surface protein (SP), cell wall (CW), and cytoplasm (Cyto) samples to bind to human plasminogen was detected by ELISA. An ELISA plate was coated with rabbit anti-enolase antibody (1:500) and incubated with sequentially subcellular samples of plasminogen (Pla; 0.02 mg/ml), goat anti-plasminogen antibody (1:1,000), and anti-goat IgG conjugated with peroxidase (1:1,000). In a similar experiment, proteins in the subcellular samples able to bind to human salivary mucins MG1 and MG2 were detected by ELISA. An ELISA plate was coated with subcellular samples (surface protein, cell wall, and cytoplasm samples), incubated sequentially with saliva supernatant (containing salivary mucins MG1 and MG2), rabbit anti-mucin MG1 (1:1,000) or anti-mucin MG2 antibodies (1:1,000), and anti-rabbit IgG conjugated with peroxidase (1:1,000). Following color development, the absorbance of each well was measured at 490 nm. The data were analyzed by Student's t test (independent samples with equal variances and two sides), and the P values of all groups were <0.05.
Wistedt et al. (33) found that proteins presenting exposed carboxyl-terminal lysines on the streptococcal cell surface can promote plasminogen activation. S. pyogenes surface enolase binds to the Kringle region of plasminogen, promotes plasminogen activation, and protects plasmin from its inhibitor, α2 antiplasmin (27, 33). Preliminary analysis of the S. mutans enolase amino acid sequence reveals two C-terminal lysines, which bind to plasminogen (our laboratory's unpublished data). Surface enolase may help S. mutans disseminate through oral tissues to enter the bloodstream, where S. mutans may cause BE (12). The reported range of cases of BE caused by S. mutans is 1.7 to 4.8%. This may be greatly underreported, however, due to S. mutans present in the less-specific diagnosis of BE due to viridans group streptococcal (48% of all BE cases) infections (30). Enolase may serve to potentiate S. mutans entering the bloodstream by activating plasminogen. Enolase is highly conserved in the genus Streptococcus (23). The enolase protein sequences of S. pneumonia, S. agalactiae, Streptococcus sobrinus, and S. mutans have been compared and have greater than 90% identity (31). In addition, there is similarity between streptococcal enolase and human enolase (49% identity). Because of the similarity between human and streptococcal enolase, antienolase antibodies reactive with human enolase are produced after S. pyogenes infection (10). Furthermore, in some autoimmune diseases, antienolase antibodies have been identified (10, 28). Similarly, cross-reactivity of human enolase with antibodies specific for S. mutans surface enolase may play a role in pathophysiological processes.
Mucins MG1 and MG2 are highly glycosylated proteins, which can form heterotypic complexes with other salivary proteins in nonglycosylated regions (22). Mucin MG1 can bind to amylase, proline-rich proteins, statherin, and histatins, and mucin MG2 can bind to SIgA and lactoferrin (4, 29). Mucin MG2 protects lactoferrin from proteolytic attack, and the interaction of mucin MG2 with oral bacteria exhibits bactericidal activity (29). Mucin MG2 was observed to bind to some unknown surface components of S. mutans (18). Our results indicate that surface enolase is one ligand responsible for the binding of salivary mucin MG2 to S. mutans. Soluble mucin MG2 may serve as a bridge between S. mutans and other salivary proteins. The interaction of surface enolase with mucin MG2 may either lead to bacterial attachment to oral tissues or facilitate the removal of S. mutans from the oral cavity.
This paper describes the identification of enolase as a component of S. mutans surface proteins and demonstrates the binding of surface enolase to human plasminogen. Additionally, this is the first report that a bacterial surface enolase interacts with the human salivary component mucin MG2. Further studies will be needed to define the role of enolase in the virulence of S. mutans.
Acknowledgments
We thank Vijay Pancholi for the gift of the anti-S. pyogenes enolase antibodies and Rodrigo V. Soares for rabbit anti-mucin MG1 and MG2 antibodies.
Editor: J. B. Bliska
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Joe, and J. J. Ferretti. 2002. Genomic sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banas, A. J. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9:1267-1277. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasminogen-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 4.Biesbrock, A. R., M. S. Reddy, and M. J. Levine. 1991. Interaction of a salivary mucin-secretory immunoglobulin A complex with mucosal pathogens. Infect. Immun. 59:3492-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottalico, L. A., N. C. Kendrick, A. Keller, Y. Li, and I. Tabas. 1993. Cholesteryl ester loading of mouse peritoneal macrophages is associated with changes in expression or modification of specific cellular proteins, including increase in an α-enolase isoform. Arterioscl. Thromb. 13:264-275. [DOI] [PubMed] [Google Scholar]
- 6.Cong, M., S. J. Perry, F. T. Lin, I. D. Fraser, L. A. Hu, W. Chen, J. A. Pitcher, J. D. Scoot, and R. J. Lefkowitz. 2001. Regulation of membrane targeting of the G protein-coupled receptor kinase 2 by protein kinase A and its anchoring protein AKAP79. J. Biol. Chem. 276:15192-15199. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, J. A., F. S. Esch, S. S. Taylor, and T. Hunter. 1984. Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine kinases in vivo and in vitro. J. Biol. Chem. 259:7835-7841. [PubMed] [Google Scholar]
- 8.Derbise, A., Y. P. Song, S. Parikh, V. A. Fischetti, and V. Pancholi. 2004. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect. Immun. 72:94-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from Gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 10.Fontan, P. A., V. Pancholi, M. M. Nociari, and V. A. Fischetti. 2000. Antibodies to streptococcal surface enolase react with human α-enolase: implications in poststreptococcal sequelae. J. Infect. Dis. 182:1712-1721. [DOI] [PubMed] [Google Scholar]
- 11.Fontana, M., L. E. Gfell, and R. L. Gregory. 1995. Characterization of preparations enriched for Streptococcus mutans fimbriae: salivary immunoglobulin A antibodies in caries-free and caries-active subjects. Clin. Diagn. Lab. Immunol. 2:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauduchon, V., L. Chalabreysse, J. Etienne, M. Celard, Y. Benito, H. Lepidi, F. Thivolet-Bëjui, and F. Vandenesch. 2003. Molecular diagnosis of infective endocarditis by PCR amplification and direct sequencing of DNA from valve tissue. J. Clin. Microbiol. 41:763-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara, H., H. Ohta, T. Inoue, T. Ohashi, S. Takashiba, Y. Murayama, and K. Fukui. 2000. Cell surface associated enolase in Actinobacillus actinomycetemcomitans. Microbiol. Immunol. 44:349-356. [DOI] [PubMed] [Google Scholar]
- 14.Hoeffler, J. P., J. W. Lustbader, and C. Y. Chen. 1991. Identification of multiple nuclear factors that interact with cyclic adenosine 3′, 5′-monophosphated response element-binding protein and activating transcription factor-2 by protein-protein interaction. Mol. Endocrinol. 5:256-266. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman, M. P., and C. G. Haidaris. 1993. Analysis of Candida albicans adhesion to salivary mucin. Infect. Immun. 61:1940-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes, M. J., J. C. Moore, J. D. Lane, R. Wilson, P. K. Pribul, Z. N. Younes, R. J. Dobson, P. Everest, A. J. Reason, J. M. Redfern, F. M. Greer, T. Paxton, M. Panico, H. R. Morris, R. G. Feldman, and J. D. Santagelo. 2002. Identification of major outer surface proteins of Streptococcus agalactiae. Infect. Immun. 70:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kling, D. E., L. C. Madoff, and J. L. Michel. 1999. Subcellular fractionation of group B streptococcus. BioTechniques 27:24-28. [DOI] [PubMed] [Google Scholar]
- 18.Liu, B., S. Rayment, F. G. Oppenheim, and R. F. Troxler. 1999. Isolation of human salivary mucin MG2 by a novel method and characterization of its interactions with oral bacteria. Arch. Biochem. Biophys. 364:286-293. [DOI] [PubMed] [Google Scholar]
- 19.Liu, B., S. Rayment, R. V. Soares, F. G. Oppenheim, G. D. Offner, P. Fives-Taylor, and R. F. Troxler. 2002. Interaction of salivary mucin MG2, its recombinant N-terminal region and a synthetic peptide with Actinobacillus actinomycetemcomitans. J. Periodontol. Res. 37:416-424. [DOI] [PubMed] [Google Scholar]
- 20.Loesche, W. J., and L. H. Straffon. 1979. Longitudinal investigation of the role of Streptococcus mutans in human fissure decay. Infect. Immun. 26:498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohman, K., and O. Meyerhof. 1934. Enzymatic transformation of phosphoglyceric acid into pyruvic and phosphoric acid. Biochem. Z. 273:60-72. [Google Scholar]
- 22.Loomis, R. E., S. A. Prakobphol, M. J. Levine, M. S. Reddy, and P. C. Jones. 1987. Biochemical and biophysical comparison of two mucins from human submandibular-sublingual saliva. Arch. Biochem. Biophys. 258:452-464. [DOI] [PubMed] [Google Scholar]
- 23.Pancholi, V. 2001. Multifunctional α-enolase: its role in diseases. Cell. Mol. Life. Sci. 58:902-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pancholi, V., and V. A. Fischetti. 1988. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J. Bacteriol. 170:2618-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasminogen binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 26.Perrone, M., L. E. Gfell, M. Fontana, and R. L. Gregory. 1997. Antigenic characterization of fimbria preparations from Streptococcus mutans isolates from caries-free and caries-susceptible subjects. Clin. Diagn. Lab. Immunol. 4:291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponting, C. P., J. M. Marshall, and S. A. Cederholm-Williams. 1992. Plasminogen: a structural review. Blood Coagul. Fibrinolysis 3:606-614. [PubMed] [Google Scholar]
- 28.Pratesi, F., S. Moscato, A. Sabbatini, D. Chimenti, S. Bombardieri, and P. Migliorini. 2000. Autoantibodies specific for α-enolase in systemic autoimmune disorders. J. Rheumatol. 27:109-115. [PubMed] [Google Scholar]
- 29.Soares, R. V., C. C. Siqueira, L. S. Brono, F. G. Oppenheim, G. D. Offner, and R. F. Troxler. 2003. MG2 and lactoferrin form a heterotypic complex in salivary secretions. J. Dent. Res. 82:471-475. [DOI] [PubMed] [Google Scholar]
- 30.Ullman, R. F., S. J. Miller, M. J. Strampfer, and B. A. Cunha. 1988. Streptococcus mutans endocarditis: report of three cases and review of the literature. Heart Lung 17:209-212. [PubMed] [Google Scholar]
- 31.Veiga-Malta, I., M. Duarte, M. Dinis, D. Tavares, A. Videira, and P. Ferreira. 2004. Enolase from Streptococcus sobrinus is an immunosuppressive protein. Cell. Microbiol. 6:79-88. [DOI] [PubMed] [Google Scholar]
- 32.Wilkins, J. C., K. A. Homer, and D. Beighton. 2002. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl. Environ. Microbiol. 68:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wistedt, A. C., U. Ringdahl, W. Mjller-Esterl, and U. Sjobring. 1995. Identification of a plasminogen-binding motif in PAM, a bacterial surface protein. Mol. Microbiol. 18:569-578. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]