Abstract
Mycobacterium marinum causes a systemic tuberculosis-like disease in a large number of poikilothermic animals and is used as a model for mycobacterial pathogenesis. In the present study, we infected zebra fish (Danio rerio) with different strains of M. marinum to determine the variation in pathogenicity. Depending on the M. marinum isolate, the fish developed an acute or chronic disease. Acute disease was characterized by uncontrolled growth of the pathogen and death of all animals within 16 days, whereas chronic disease was characterized by granuloma formation in different organs and survival of the animals for at least 4 to 8 weeks. Genetic analysis of the isolates by amplified fragment length polymorphism showed that M. marinum strains could be divided in two clusters. Cluster I contained predominantly strains isolated from humans with fish tank granuloma, whereas the majority of the cluster II strains were isolated from poikilothermic species. Acute disease progression was noted only with strains belonging to cluster I, whereas all chronic-disease-causing isolates belonged to cluster II. This difference in virulence was also observed in vitro: cluster I isolate Mma20 was able to infect and survive more efficiently in the human macrophage THP-1 and the carp leukocyte CLC cell lines than was the cluster II isolate Mma11. We conclude that strain characteristics play an important role in the pathogenicity of M. marinum. In addition, the correlation between genetic variation and host origin suggests that cluster I isolates are more pathogenic for humans.
Bacterial strains belonging to the Mycobacterium tuberculosis complex show an unusually high degree of conservation, not only in housekeeping genes (19) but also in genes encoding (putative) targets of the immune system (13). In fact, most genes in strains isolated worldwide show a negligible variation. These data are best explained by assuming that contemporary strains have all originated from a recent common ancestor, which would have lived 10,000 to 20,000 years ago. However, genetic analysis by DNA fingerprinting has revealed that there is genetic variation in M. tuberculosis, and recently it was shown that this diversity correlates markedly with pathogenicity (11). The M. tuberculosis Beijing strain, which is highly prevalent in Asia and the former USSR, causes a significantly higher and earlier mortality in mice compared to the prototype H37Rv strain. This effect was correlated with a nonprotective immune response. These results show that also for highly homogeneous strains, genetic variation plays a role in disease development.
In the past years, Mycobacterium marinum, the causative agent of fish tuberculosis, was adopted as a model to study mycobacterial infections. There are good reasons for this approach: M. marinum is the mycobacterial species most closely related to members of the M. tuberculosis complex (17), it has a relatively short generation time of 4 to 6 h (compared to 20 h for M. tuberculosis), and it grows optimally at 30°C and hardly at all at 37°C (4, 2). Because of this optimal growth temperature, M. marinum infections of humans are found almost exclusively as superficial lesions on the extremities (3, 5, 6). The histopathology of these M. marinum infections, generally called fish tank granuloma or swimming pool granuloma, shows the formation of granulomas that resemble those associated with tuberculosis (6, 9, 23). Another advantage of M. marinum is that this bacterium is a natural pathogen of poikilothermic species, which provides the opportunity to study infection in a natural host (4). Different infection models that use the leopard frog (Rana pipiens) (16), the goldfish (Carassius auratus) (21), and recently also the genetically tractable zebra fish (Danio rerio) (7, 14, 22) as a host have been described. Those studies used either the M. marinum M strain, originally isolated from an infected patient, or M. marinum ATCC 927, isolated from fish. In the present study, we analyzed the genetic variation between different isolates of M. marinum and their pathogenicities for zebra fish. We observed that strains of M. marinum can be grouped into two clusters based on genetic analysis (amplified fragment length polymorphism [AFLP]). Interestingly, representative strains of the first cluster, which consists almost exclusively of M. marinum strains isolated from humans, induced an acutely lethal disease in the zebra fish, whereas strains of the second cluster induced a chronic progressive disease characterized by granuloma formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type M. marinum isolates included in this study (Table 1) were obtained from the collections of the National Institute of Public Health and the Environment (RIVM) (Bilthoven, The Netherlands), the Institute of Aquaculture (Stirling, United Kingdom), and the Institute for Animal Science and Health (CIDC) (Lelystad, The Netherlands) and were assigned to the species M. marinum by 16S sequencing and growth characteristics, such as photochromogenic behavior. We also included reference strain M, a human isolate (21). Bacteria were grown at 30°C in Middlebrook 7H9 medium supplemented with Middlebrook oleic acid-albumin-dextrose-catalase (BD Biosciences), 0.2% Tween and 1 mg of d-arabinose per ml (1) to decrease clumping of cells. Prior to inoculation in zebra fish, bacteria were washed three times with phosphate-buffered saline (PBS); bacterial numbers were determined by measuring the optical density at 600 nm and by plating and CFU determination.
TABLE 1.
Sources and origins of the M. marinum strains used in this study
| Strain |
Source | Reference or source | |
|---|---|---|---|
| Original name | Name in present study | ||
| E6 | Sciaenops ocellatus (red drum) | 15 | |
| E7 | Mma7 | Chaetodon fasciatus (butterfly fish) | 15 |
| E11 | Mma11 | Dicentrarchus labrax (sea bass) | 15 |
| E12 | Dicentrarchus labrax (sea bass) | 15 | |
| E15 | Siganus rivulatus (marbled spinefoot) | 15 | |
| E16 | Dicentrarchus labrax (sea bass) | 15 | |
| 420472-4 | Mma42 | Snake (unknown sp.) | CIDCa |
| 551962 | Shinisaurus crocodiluris (crocodile lizard) | CIDC | |
| M strain | Human | 21 | |
| Mis 14 | Human | RIVMb | |
| 14641 | Human | RIVM | |
| 18347 | Human | RIVM | |
| 9800607 | Mma98 | Human | RIVM |
| 9801756 | Human | RIVM | |
| 9900036 | Human | RIVM | |
| 2000-01053 | Mma20 | Human | RIVM |
| 2001-00796 | Mma21 | Human | RIVM |
Collection of the Institute for Animal Science and Health, Lelystad, The Netherlands.
Collection of the National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
Animals.
Male zebra fish (D. rerio) (approximately 1 g and 1 year old) were chosen from our breeding facilities, acclimated for 1 week to their new environment in the infection room, and kept at 28°C on a 14-h/10-h light/dark cycle. The infected zebra fish were housed in 10-liter separate tanks with separate pumps and filter systems (Ecco; Eheim).
Infection of zebra fish.
The zebra fish were anesthetized in a 0.02% aqueous solution of ethyl-3-aminobenzoate methanesulfonate salt (MS-222) (Sigma) and inoculated intraperitoneally with 10 μl of M. marinum suspension in PBS. Ten zebra fish per group were inoculated with 104 CFU of M. marinum strains Mma98 and Mma7, 8 zebra fish per group were inoculated with Mma21 and Mma42, and 15 zebra fish per group were inoculated with Mma20 and Mma11. In addition, 10 control fish were injected with 10 μl of PBS. Viable bacterial counts present in the livers and kidneys of three fish inoculated with Mma11 and Mma20 were determined by plating serial 10-fold dilutions of organ homogenates, decontaminated with BBL MycoPrep (BD), on Middlebrook 7H10 agar. The colonies were identified as mycobacterial species by morphology and photochromogenic behavior. Counts were performed at 1 day postinfection (dpi), 1 week postinfection (wpi), 4 wpi, and 8 wpi. Two fish inoculated with Mma20, Mma11, or Mma7 were used for histological examination and Ziehl-Neelsen (ZN) staining at 10 dpi (Mma20), 4 wpi, and 8 wpi. All animal experiments were approved by the local Animal Welfare Committee, under protocol number MM 01-02 and MM 03-02.
Zebra fish pathology and tissue processing.
The zebra fish were observed for gross signs of infection and were sacrificed when they exhibited moribund behavior. For histological examination, zebra fish were sacrificed by incubation in an overdose of MS-222 (A-5040; Sigma), fixed in Bouin (5 ml of 40% formaldehyde, 15 ml of water-saturated picric acid, 1 ml of acetic acid), and processed for paraffin embedding. Frontal sections (4 to 7 μm) were stained with hematoxylin and eosin or according to the ZN method and observed under a Zeiss Axioskop light microscope. Photographs were taken with a Nikon Coolpix 900 camera and processed by using Adobe Photoshop software, version 6.0.
AFLP analysis.
Bacterial strains were incubated with protein K for 60 min at room temperature prior to DNA isolation with the DNeasy tissue kit (Qiagen). AFLP was performed essentially as described previously, with EcoRI and MseI as restriction enzymes and the primers EcoA and MseC (10, 18, 24). AFLP fragments were separated on an ABI Prism Genetic Analyzer 3100 (Applied Biosystems). Data were analyzed by Pearson correlation and clustered by unweighted pair group matrix analysis with Bionumerics software, version 3.0 (Applied Maths).
crtB sequence analysis.
In order to determine the nucleotide sequence of the crtB gene, the gene was first amplified by PCR on the various chromosomal DNA preparations with crtB-specific primers (wbcrtBF, TCGACCTGAAAGCACAGTTG; wbcrtBR, AGTCTTCAATCGGGATGTCG). Subsequently, the PCR product was purified with a PCR purification column (Qiagen) and used in a sequence reaction with one of these primers. The different elongation products were separated on an ABI Prism Genetic Analyzer 3100 (Applied Biosystems).
Cell lines and culture conditions.
The human acute monocytic leukemia cell line THP-1 was cultured in RPMI 1640 medium (GIBCO BRL) with 10% fetal calf serum (FCS) at 37°C with 5% CO2. The adherent carp monocyte/macrophage cell line (CLC) was obtained through the European Collection of Cell Culture, Salisbury, United Kingdom (ECACC no. 95070628;) and was maintained at 28°C with 5% CO2 in RPMI 1640 medium supplemented with 10% FCS.
Intracellular survival assays.
Cellular infection assays were carried out in 24-well tissue culture plates (Costar) as previously described (8, 12). To differentiate the THP-1 cells into macrophage-like cells, the cells were treated with phorbol myristate acetate (Sigma). THP-1 cells were harvested by centrifugation for 9 min at 200 × g and the pellet was suspended in 1 ng of phorbol myristate acetate per ml-RPMI 1640-10% FCS to a density of approximately 106 THP-1 cells/ml. One milliliter of cell suspension was added to each well of a 24-well plate. The plate was incubated for 24 h at 37°C with 5% CO2. The medium was removed from each well, and adherent cells were washed once with RPMI 1640-10% FCS, refreshed with new RPMI 1640-10% FCS, and incubated for an additional 24 h. CLC cells were seeded at a density of 106 cells per well 24 h prior to use. Immediately before infection, cells were washed once with fresh RPMI 1640-10% FCS. Bacteria were harvested by centrifugation for 5 min at 3,000 × g and washed twice with RPMI 1640-10% FCS medium. The bacteria were suspended in RPMI 1640-10% FCS at a concentration to achieve a multiplicity of infection of 10 for Mma11 and 1 for Mma20. Bacteria and cells were incubated for 1 h at 33°C for THP-1 and at 28°C for CLC. Cells were then washed twice with RPMI 1640-10% FCS to remove free bacteria and incubated in fresh medium with amikacin (200 μg/ml; Sigma Chemical) at the appropriate temperature. After 2 h, the medium was replaced by medium with 30 μg of amikacin per ml. The cells were incubated at the appropriate temperature and then lysed at different time points with 1 ml of 0.1% (vol/vol) Triton X-100 in PBS. One well was processed immediately (time zero) for determination of initial bacterial counts. Each lysate was diluted as necessary, and portions were plated on 7H10 agar plates. Survival was expressed as the percentage of CFU at each time point, with the number of CFU at time zero as the reference.
RESULTS
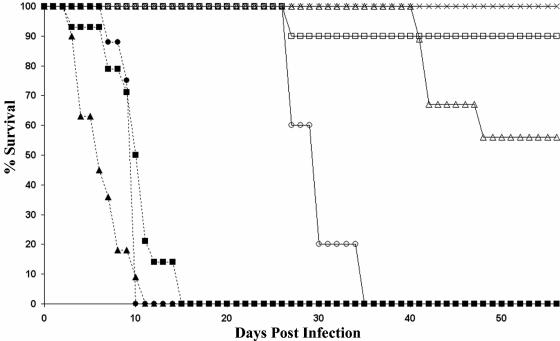
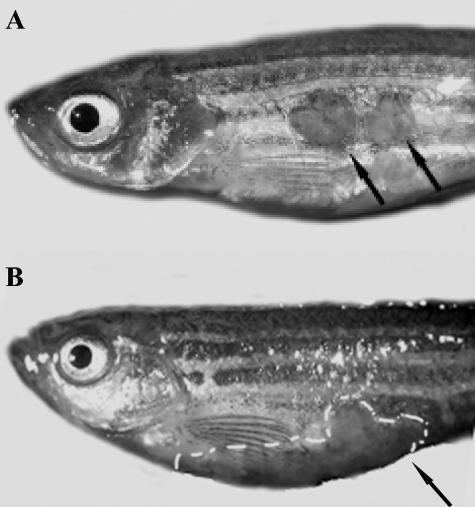
Different M. marinum strains cause different forms of disease. To study possible differences in pathogenicity between different M. marinum strains in zebra fish, groups of 10 or 15 zebra fish were inoculated intraperitoneally with 104 CFU of M. marinum. Four different strains were used: Mma7, Mma11, Mma20, and Mma98. The last two strains were originally isolated from skin lesions of patients with fish tank granuloma, whereas Mma11 and Mma7 were isolated from infected fish. Most zebra fish inoculated with Mma7 or Mma11 survived up to the end of the experiment, at 56 days postinfection (Fig. 1). However, differences in external signs of disease were observed. The zebra fish inoculated with Mma7 showed no signs of infection for 8 weeks, while three of the zebra fish infected with Mma11 showed skin lesions (ulcerations) after approximately 45 days (Fig. 2). Notably, these skin lesions were not at the site of the injection, which shows that the infection was disseminated. Furthermore, these overtly ill animals also showed buoyancy problems for a short period of time (1 to 2 days) before they were sacrificed.
FIG. 1.
Survival curves for zebra fish infected intraperitoneally with 104 CFU of Mma7 (□), Mma11 (▵), Mma98 (▴), Mma20 (▪), Mma21 (○), and Mma42 (•) or treated with PBS (×).
FIG. 2.
Macroscopic characteristics of M. marinum chronic (A) and acute (B) infections. (A) Intraperitoneal infection with strain Mma11 induces skin ulcerations at 7 wpi (arrows). These ulcerations were usually not located at the site of infection. (B) Intraperitoneal infection with strain Mma20 causes a swelling of the abdomen and severe hemorrhages within 2 wpi (arrow and white dashed line). No ulcerations were observed in control animals.
All of the zebra fish inoculated with 104 Mma20 and Mma98 bacteria developed acute symptoms, including hemorrhages and inflammation at the site of infection, and all infected fish died or were sacrificed in a moribund state at between 5 to 16 days postinfection (Fig. 1). To examine whether this effect was dose dependent, six zebra fish were infected with 102 CFU of Mma20. Half of these zebra fish also displayed acute disease symptoms and had to be sacrificed within 16 days (results not shown), which might indicate that the acute disease symptoms are not highly dose dependent. Together, these results show that M. marinum causes an acute or chronic infection in a strain-dependent manner.
M. marinum recovery from zebra fish organs.
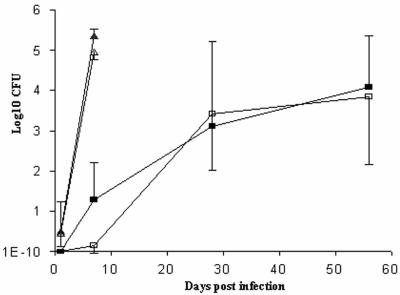
To assess the ability of the different M. marinum strains to persist and replicate in host tissue, fish were inoculated with 104 CFU of Mma11 or Mma20. Three fish were sacrificed at 1 and 7 dpi and at 4 and 8 wpi, and the livers and the posterior kidneys were collected for bacteriological examination. From 7 dpi onwards, all tested organs of both groups were colonized, which showed that the bacteria were disseminated. However, a significant difference in the bacterial load between organs derived from zebra fish infected with Mma20 and Mma11 was observed. Whereas livers from zebra fish inoculated with Mma11 contained only a small number of bacteria at 7 dpi, livers from zebra fish inoculated with Mma20 contained as much as 105 CFU (Fig. 3). Since the growth rates of the two strains in vitro are similar, these data suggest that Mma20 is able to survive and/or replicate better in zebra fish than Mma11. Probably the high bacterial load caused the early death of the zebra fish. Upon prolonged infection, the number of Mma11 bacteria increased steadily and reached numbers of the same order of magnitude as the numbers of Mma20 bacteria at 7 dpi (Fig. 3). As can be seen in Fig. 3 the bacterial numbers recovered from fish infected with Mma11 varied markedly, but the numbers of CFU recovered from the liver and posterior kidney of the same fish were always comparable. This probably means that the onset and progression of disease in Mma11 were highly variable. We observed the same phenomenon upon prolonged incubation (6 months) of zebra fish infected with Mma11. The onset of overt signs of disease varied between 7 weeks and 6 months.
FIG. 3.
Total CFU counts per homogenized liver and posterior kidney from zebra fish inoculated with 104 bacilli of M. marinum strain Mma11 or strain Mma20. The mean for three samples per time point per group is shown. The error bars represent the standard errors of the means. Bacterial numbers isolated from Mma20 liver (▴), Mma20 posterior kidney (▵), Mma11 liver (▪), and Mma11 posterior kidney (□) are shown.
Histology of M. marinum infection.
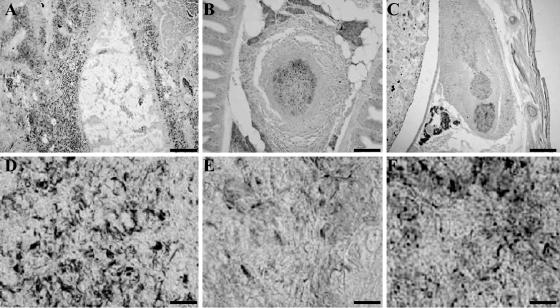
To determine the nature of the pathological response to M. marinum, zebra fish infected with Mma20, Mma11, and Mma7 were sacrificed and fixed in Bouin. Paraffin sections were made, and slides were alternately stained with ZN stain or hematoxylin and eosin. Microscopic examination of the sections revealed that moribund fish infected with Mma20 contained high numbers of mycobacteria at 5 dpi (Fig. 4A and D). The peritoneum and the surrounding tissues showed severe necrosis, loss of structural organization, and the influx of large numbers of inflammatory cells. This finding also suggests that the early death of the zebra fish was caused by the outgrowth of bacteria to very high numbers, which caused severe peritonitis. Also, high numbers of bacteria other than mycobacteria could be seen in infected fish tissue. In contrast to Mma20-infected fish, zebra fish infected with Mma11 contained well-organized granulomas in the liver, pancreas, kidney, and intestines at 4 wpi. Sometimes granulomas were located in the connective tissues. Histological examination showed that most granulomas consisted of an outer layer of tightly packed epithelial cells and a central region containing mainly macrophages and possibly some granulocytes and lymphocytes. In addition, the central regions of most granulomas showed necrosis and the presence of mycobacteria. The number of granulomas and their size seemed to increase over time, as observed at 8 wpi (Fig. 4B and E). In contrast to the case for Mma11-infected fish, only one of the two zebra fish infected with Mma7 displayed granulomas at 4 and 8 wpi (Fig. 4C and E). The morphology of the granulomas was slightly different from that of the granulomas in Mma11-infected zebra fish. Mma7 induced fewer and less organized granulomas; sometimes they contained an outer layer of epithelial cells and a necrotic center. In addition, single M. marinum cells of Mma7 were observed outside granulomas, free in the tissues (Fig. 4C). Neither bacteria nor granulomas were observed in sections of the second zebra fish infected with Mma7 at 4 wpi, but sections of the zebra fish at 8 wpi contained individual mycobacteria (i.e., not grouped in clusters).
FIG. 4.
ZN-stained sections of zebra fish infected with Mma20 (A and D), Mma11 (B and E), or Mma7 (C and F). (A and D) Section of Mma20-infected zebra fish at 5 dpi. (A) Considerable tissue damage and many mycobacteria can be observed. The mycobacteria are not organized in granulomas. (D) A 1,000× magnification showing the individual mycobacteria. (B and E) Section of Mma11-infected zebra fish at 8 wpi. (B) A well-organized granuloma with a necrotic center can be observed in the pancreas. (E) At high magnification, some mycobacteria in the center of the granuloma can be seen. (C and F) Section of Mma7-infected zebra fish at 8 wpi. (C) Less-well-organized granulomas and mycobacteria not organized in granulomas can be seen. (F) At high magnification, mycobacteria are found in the centers of the granulomas. (A, B, and C) Bars represent 100 μm; (D, E, and F) bars represent 10 μm.
AFLP analysis of M. marinum: two large clusters correlated with pathogenic properties.
Large differences in disease progression were observed for the four M. marinum strains analyzed. To determine the genetic relationships between the strains causing acute and chronic disease, DNA fingerprinting by AFLP was performed. For this analysis we used chromosomal DNA of 17 different M. marinum isolates, including the strains used in the infection experiments (Table 1; Fig. 5). Following analysis by use of the Pearson correlation coefficient and unweighted pair group matrix analyses, the 17 strains were grouped in two AFLP clusters at a delineation level of 60% (Fig. 5). Interestingly, the compositions of these two clusters roughly corresponded to the origins of isolation of the M. marinum strains; i.e., cluster I contained seven of the nine human isolates, whereas cluster II contained six of the eight isolates from poikilothermic animals. This could indicate that cluster I isolates are more pathogenic for humans. To substantiate this result, the AFLP analysis was extended with an extra eight human isolates, which all clustered in cluster I (results not shown). Both of the M. marinum strains responsible for acute disease in zebra fish, i.e., Mma20 and Mma98, also fell in cluster I, whereas both chronic disease-causing strains fell in cluster II. This could mean that M. marinum strains belonging to cluster I provoke an acute disease in zebra fish or that M. marinum strains that cause disease in humans are responsible for this. To test these assumptions, zebra fish were inoculated with Mma21, a strain that was isolated from a human infection case but fell in cluster II upon AFLP fingerprinting, and with Mma42, a strain that clustered in group I but was isolated from a reptile. Infection with Mma21 resulted in a more chronic infection, while Mma42 caused an acute infection (Fig. 1), which suggests that M. marinum strains belonging to cluster I, irrespective of their origin, give rise to acute lethal infections in zebra fish.
FIG. 5.
AFLP-DNA fingerprint. Numerical analysis of normalized AFLP band patterns generated from chromosomal DNAs of M. marinum isolates and M. tuberculosis H37Rv as outgroup was performed. M. marinum strains isolated from humans are indicated in boldface italic. The dendrogram was constructed by using unweighted pair group matrix analyses. The clusters representing the human isolates (cluster I) and the poikilothermic animal isolates (cluster II) were defined at a delineation level of 60%. The grey error flags at each branch show the standard deviation of the average similarity at this position.
By multilocus sequence typing it has been shown previously (20) that M. marinum strains can be divided into two clusters. The same study also showed that one of these clusters is in fact closely linked to the human pathogen Mycobacterium ulcerans, the causative agent of Buruli ulcer. To determine whether the observed subdivision of our set of M. marinum strains, based on AFLP analysis, is consistent with that of Stinear et al. (20), we sequenced the crtB gene. This gene is the most differentiating gene in the multilocus sequence typing. We analyzed 10 strains and found consistent clustering of the strains by AFLP analysis and by crtB sequence analysis. Furthermore, AFLP cluster I, which contained most human isolates, was identical to the cluster closely linked with M. ulcerans in the study by Stinear et al. (20).
Cell culture.
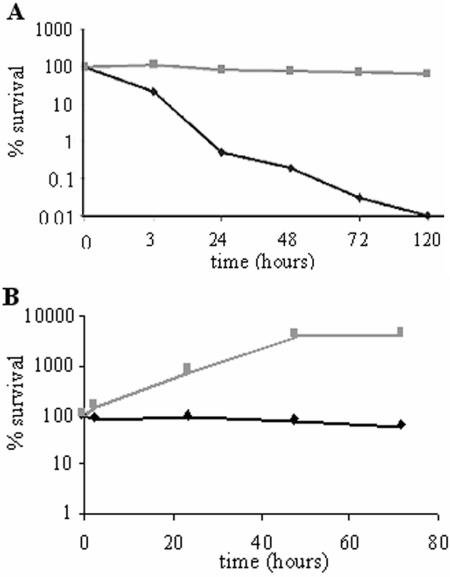
From the in vivo infection studies it is clear that at 7 dpi there is a major difference between the bacterial loads of fish infected with Mma20 or Mma11 (Fig. 3). To test whether this might be attributed to differences in intracellular survival and/or persistence of the M. marinum strains, the human THP-1 cell line was infected with Mma20 or Mma11 and the intracellular survival of the bacteria was monitored over time (Fig. 6). Already at 4 h of incubation with THP-1 cells, a decrease in Mma11 CFU was observed. This number showed a steady decline, and at 120 h postinfection hardly any Mma11 could be cultured from these cells. In contrast, bacteria of strain Mma20 were able to survive intracellularly for a long time in THP-1 cells, since the number of cells did not change substantially over 120 h. M. marinum strain M, which is used to determine the genome sequence, belongs to cluster I (Fig. 5) and is also able to maintain itself in the human macrophage cell line THP-1, similar to the case for Mma20 (results not shown). In a second experiment, the in vitro growth of these two M. marinum strains in carp leukocytes (CLC line) was determined. This cell line can be maintained at 28°C, the optimal growth temperature of M. marinum, and has been shown previously to be a useful model for M. marinum intracellular growth and survival (10). In contrast to the results obtained with the THP-1 cells, Mma11 was able to maintain itself in these CLC cells (Fig. 6). However, in this experiment also there was a clear difference in outgrowth of Mma20 compared to Mma11 (Fig. 6). This showed that strain Mma20 is able to survive and/or replicate more efficiently in macrophages, which might explain the increased virulence of this strain.
FIG. 6.
Survival of Mma11 (♦) and Mma20 (░⃞) in the human macrophage cell line THP-1 (A) and the carp leukocyte cell line CLC (B). Bacteria and cells were incubated at 33°C for THP-1 and at 28°C for CLC. After a 1-h coincubation of cells and bacteria, the cells were washed, treated with 200 μg of amikacin per ml, and incubated in medium with 30 μg of amikacin per ml at the appropriate temperature. At different time points, samples were taken for CFU determination. Survival was expressed as the percentage of CFU at each time point, with the number of CFU at time zero as a reference. The means from three experiments are shown.
DISCUSSION
Since the 1920s, M. marinum has been known as the causative agent of fish tuberculosis in poikilothermic species and of fish tank granuloma or swimming pool granuloma in humans. Strain variation with respect to virulence, however, was never investigated. In the present study we demonstrate that different M. marinum strains show marked differences in pathogenicity. These differences correlated with the origin of the isolates (from infected humans or infected fish) and with genetic clustering. Indeed, by AFLP fingerprinting, two clusters, designated clusters I and II, could be recognized, with human isolates falling predominantly in cluster I and isolates from poikilothermic species falling predominantly in cluster II. In addition, we confirmed that the zebra fish is a useful animal model to study mycobacterial infection (14).
Depending on the M. marinum strain used, the zebra fish developed an acute or a chronic infection. The acute symptoms were not expected, since the relative inoculum was known to result in a chronic disease in goldfish (21). Zebra fish with an acute disease suffered from loss of equilibrium, swelling, and hemorrhage at the site of infection, hung at the bottom of the tank, and did not eat. Histological examination showed a massive amount of acid-fast rods at the site of infection and a severe peritonitis. The fish with chronic infections showed signs typical of fish tuberculosis, i.e., a systemic spread of the infection, granuloma formation in different organs, shedding of scales, and skin lesions. The differences in disease progression that we observed in fish inoculated with strain Mma20 (a human isolate) compared to those inoculated with strain Mma11 (a fish isolate) were not merely an effect of inoculum size, since strain Mma20 also caused acute infection in 50% of the fish when a 100-times-lower inoculum was used (102 CFU versus 104 CFU) (data not shown). Since the zebra fish infected with the human isolates had to be sacrificed earlier than originally planned, due to the unexpectedly fast progression of the infection, we could isolate organs of these zebra fish only at 1 and 7 dpi. At day 7 only the fish infected with the human isolate Mma20 showed a large increase in bacterial numbers in the liver and the posterior kidney. These results were substantiated in experiments with both human THP-1 and carp CLC cells: cluster I isolate Mma20 infected and survived better in macrophages than cluster II strain Mma11. The striking difference in disease characteristics between the different M. marinum isolates correlated with genetic differences, as was determined by AFLP analysis. This analysis showed that all isolates grouped in two clusters, I and II. All strains causing acute disease belonged to cluster I. Surprisingly, most strains of this cluster were human isolates, which raised the hypothesis of whether passage through a human host would result in an increase in poikilothermic virulence. However, the cluster I snake isolate Mma42 also induced an acute lethal disease, which indicates that the genetic background is important and not the human passage.
Of course, human infection is caused by isolates that are transmitted to humans from fish or other poikilothermic species, but our results suggests that strains that have the potential to induce infection also in humans differ genetically from strains that cause infection only in fish. The human isolates were collected during the last decade from patients admitted to Dutch hospitals; therefore, these strains were isolated from patients with severe infections in need of medical attention. The finding that these human isolates are strongly overrepresented in cluster I indicates that strains of cluster I more frequently give rise to more severe and persistent human infections. This observation, combined with the evidence that these strains show enhanced survival in both human and fish cell lines, suggests that cluster I forms a subspecies of M. marinum with increased pathogenicity for humans and zebra fish. Cluster I is genetically more closely related to the human pathogen M. ulcerans than cluster II (20). However, the implication of this relationship is at present unclear. The observed differences in M. marinum virulence are in contrast with previous studies, which mention briefly that there were no differences in disease outcome when different strains of M. marinum were used to infect the leopard frog or the goldfish (16, 21). The difference between those studies and our report is probably not due to the use of different M. marinum strains, since those studies also report the use of several strains, derived both from humans and from fish and frogs. The observed difference might be due to the choice of host organism; the leopard frog has been shown to be relatively resistant to M. marinum, with stable bacterial loads and noncaseous granulomas (16). On the other hand, the link with the human isolates indicates that the difference in virulence can be seen in widely different species.
The finding that M. marinum strains cluster in two major groups, with one cluster containing strains pathogenic for humans and zebra fish, allows us to use the zebra fish model to identify mycobacterial virulence factors that are important for survival and persistence in fish and in humans.
Acknowledgments
We thank Monique Raats and Wim Schouten for their expert technical assistance, Ben Appelmelk for helpful discussions, and Sandrine Florquin (AMC, Amsterdam, The Netherlands) for her assistance with the interpretation of the histopathology. We thank Peter Willemsen (CIDC, Lelystad, The Netherlands), Kim Thompson (The Institute of Aquaculture, Stirling, United Kingdom), and Dick van Soolingen and Tridia van der Laan (RIVM, Bilthoven, The Netherlands) for the field isolates of M. marinum.
This research was supported in part by grant 050-71-001 from The Netherlands Genomics Initiative (NROG).
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Anton, V., P. Rouge, and M. Daffe. 1996. Identification of the sugars involved in mycobacterial cell aggregation. FEMS Microbiol. Lett. 144:167-170. [DOI] [PubMed] [Google Scholar]
- 2.Aronson, J. D. 1926. Spontaneous tuberculosis in saltwater fish. J. Infect. Dis. 39:315-320. [Google Scholar]
- 3.Bailey, J. P., S. J. Stevens, W. M. Bell, H. G. Mealing, D. H. Loebl, and E. H. Cook. 1982. Mycobacterium marinum infection—a fishy story. JAMA 247:1314. [PubMed] [Google Scholar]
- 4.Clark, H. F., and C. C. Chepard. 1963. Effect of environmental temperatures on infection with Mycobacterium marinum (Balnei) of mice and a number of poikilothermic species. J. Bacteriol. 86:1057-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, R. B., H. Spector, D. M. Friedman, K. J. Oldrati, C. L. Young, and S. C. Nelson. 1990. Osteomyelitis and synovitis produced by Mycobacterium marinum in a fisherman. J. Clin. Microbiol. 28:2570-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, C. H., J. M. Grange, W. C. Noble, and M. D. Yates. 1985. Mycobacterium marinum infections in man. J. Hyg. 94:135-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, J. M., H. Clay, J. L. Lewis, N. Ghori, P. Herbomel, and L. Ramakrishnan. 2002. Real-time visualization of Mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17:693-702. [DOI] [PubMed] [Google Scholar]
- 8.El Etr, S. H., L. Yan, and J. D. Cirillo. 2001. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect. Immun. 69:7310-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huminer, D., S. D. Pitlik, C. Block, L. Kaufman, S. Amit, and J. B. Rosenfeld. 1986. Aquarium-borne Mycobacterium marinum skin infection—report of a case and review of the literature. Arch. Dermatol. 122:698-703. [PubMed] [Google Scholar]
- 10.Koeleman, J. G. M., J. Stoof, D. J. Biesman, P. H. M. Savelkoul, and C. M. J. E. Vandenbroucke-Grauls. 1998. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J. Clin. Microbiol. 36:2522-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez, B., D. Aguilar, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, R. Hernandez-Pando, K. Huygen, and D. Van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, B. H., and T. M. Shinnick. 2000. Evaluation of Mycobacterium tuberculosis genes involved in resistance to killing by human macrophages. Infect. Immun. 68:387-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musser, J. M., Al Aminb, and S. Ramaswamy. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure genetics. Genetics 155:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prouty, M. G., N. E. Correa, L. P. Barker, P. Jagadeeswaran, and K. E. Klose. 2003. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol. Lett. 225:177-182. [DOI] [PubMed] [Google Scholar]
- 15.Puttinaowarat, S., K. Thompson, J. Lilley, and A. Adams. 1999. Characterization of Mycobacterium spp isolated from fish by pyrolysis mass spectrometry (PyMS) analysis. AAHRI Newslett. 8:4-8. [Google Scholar]
- 16.Ramakrishnan, L., R. H. Valdivia, J. H. McKerrow, and S. Falkow. 1997. Mycobacterium marinum causes both long-term subclinical infection and acute disease in the leopard frog (Rana pipiens). Infect. Immun. 65:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogall, T., J. Wolters, T. Flohr, and E. C. Bottger. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 18.Savelkoul, P. H. M., H. J. M. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. Wrademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shreevatsan, S., P. Escalante, X. Pan, K. E. Stockbauer, N. D. Conell, B. N. Kreiwirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stinear, T. P., G. A. Jenkin, P. D. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talaat, A. M., R. Reimschuessel, S. S. Wasserman, and M. Trucksis. 1998. Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect. Immun. 66:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Sar, A. M., B. J. Appelmelk, C. M. J. E. Vandenbroucke-Grauls, and W. Bitter. 2004. A star with stripes: zebrafish as an infection model. Trends Microbiol. 12:451-457. [DOI] [PubMed]
- 23.Vanduijn, C. 1981. Tuberculosis in fishes. J. Small Anim. Pract. 22:391-411. [DOI] [PubMed] [Google Scholar]
- 24.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Vandelee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP—a new technique for DNA-fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]