Abstract
Vibrio parahaemolyticus, a gram-negative marine bacterium, is a worldwide cause of food-borne gastroenteritis. Recent genome sequencing of the clinical V. parahaemolyticus strain RIMD2210633 identified two sets of genes for the type III secretion system (TTSS), TTSS1 and TTSS2. Here, we constructed a series of mutant strains from RIMD2210633 to determine whether the two putative TTSS apparatus are functional. The cytotoxic activity of mutant strains having a deletion in one of the TTSS1 genes was significantly decreased compared with that of the parent and TTSS2-related mutant strains. In an enterotoxicity assay with the rabbit ileal loop test, intestinal fluid accumulation was diminished by deletion of the TTSS2-related genes while TTSS1-related mutants caused a level of fluid accumulation similar to that of the parent. VopD, a protein encoded in the proximity of the TTSS1 region and a homologue of the Yersinia YopD, was secreted in a TTSS1-dependent manner. In contrast, VopP, which is encoded by a pathogenicity island on chromosome 2 and is homologous to the Yersinia YopP, was secreted via the TTSS2 pathway. These results provide evidence that V. parahaemolyticus TTSSs function as secretion systems and may have a role in the pathogenicity of the organism. This is the first report of functional TTSSs in Vibrio species. The presence of TTSS apparatus gene homologues was demonstrated in other vibrios, such as Vibrio alginolyticus, Vibrio harveyi, and Vibrio tubiashii, suggesting that some other vibrios also contain TTSS and that the TTSS has a role in protein secretion in those organisms during interaction with eukaryotic cells.
Vibrio parahaemolyticus, one of the human-pathogenic vibrios, is a gram-negative halophilic bacterium that naturally inhabits marine and estuarine environments. The organism causes three major types of clinical illness: gastroenteritis (the most common illness), wound infections, and septicemia (4, 7, 10, 16). Almost all of the clinical V. parahaemolyticus isolates from diarrheal patients show β-type hemolysis on Wagatsuma agar (22), a specialized blood agar medium. This hemolysis has been called the Kanagawa phenomenon (KP), and it is considered to be a good marker of pathogenic strains. The thermostable direct hemolysin (TDH) is responsible for the KP (10). TDH is a protein toxin composed of 165 amino acid residues, and it displays several biological activities, i.e., hemolytic activity, enterotoxicity, cytotoxicity, and cardiotoxicity (10, 17, 23, 25). Thus, TDH has been considered a major virulence factor of the organism. The overall mechanism of pathogenesis by V. parahaemolyticus, however, has not yet been elucidated.
The type III secretion system (TTSS) is an apparatus used by several gram-negative pathogenic bacteria to secrete and translocate virulence factor proteins into the cytosol of eukaryotic cells (11). The TTSS apparatus is well conserved among these bacteria, whereas the specific properties of the effectors and, hence, the resulting symptomatic effects on the host organism vary widely (11).
Recently, a consortium including our laboratory completed sequencing of the genome of the TDH-producing (KP-positive) V. parahaemolyticus strain RIMD2210633 (14). The sequencing revealed the presence of two sets of genes for the TTSS on chromosomes 1 and 2 (TTSS1 and TTSS2). TTSS1 was found in all of the V. parahaemolyticus strains tested while TTSS2 was present only in the KP-positive strains (14). In this study, to determine the function of the two TTSSs, we constructed a series of mutant V. parahaemolyticus RIMD2210633 strains with a deletion in the TTSS-related genes. The contribution of the two TTSSs to the pathogenicity of this bacterium was also investigated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All of the V. parahaemolyticus and Vibrio species strains used in this study were obtained from the Laboratory for Culture Collection, Research Institute for Microbial Diseases, Osaka University. V. parahaemolyticus strain RIMD2210633 (KP-positive, serotype O3:K6) was used for the construction of deletion mutants and for functional studies. Escherichia coli DH5α and SM10λpir (15) strains were used for the general manipulation of plasmids and the mobilization of plasmids into V. parahaemolyticus, respectively. The bacteria were cultured at 37°C with shaking in Luria-Bertani (LB) medium (for E. coli) and LB supplemented with 3% NaCl (for V. parahaemolyticus). TCBS agar (Nissui, Tokyo, Japan) was used for the screening of mutant strains. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 5 μg/ml.
Construction of deletion mutant strains.
PCR-amplified DNA fragments used for constructing the in-frame deletion mutation of vcrD1 were generated by overlap PCR with the PCR primers VP1662A (5′-ACGACACGTTGGATCCTGCTT-3′), 1662B (5′-ATCTTTTTCCTTTTGGATGTTCACGATGAT-3′), 1662C (5′-ATCATCGTGAACATCCAAAAGGAAAAAGAT-3′), and 1662D (5′-GTTGATCTGCAGAGTGAGTTC-3′). Two DNA fragments were amplified by PCR with V. parahaemolyticus strain RIMD2210633 chromosomal DNA as a template with primer pairs VP1662A and 1662B and 1662C and 1662D, respectively. Primers 1662B and 1662C included a complementary 15-bp sequence at their 3′ and 5′ ends, respectively. These two fragments were used as templates in a second PCR with primers 1662A and 1662D, resulting in the construction of a fragment with a deletion in the vcrD1 gene. The fragment containing the deletion was purified and cloned into the pT7Blue T-vector (Novagen, Inc.). This fragment was then removed from the pT7Blue T-vector by digestion with BamHI and PstI and cloned into an R6K-ori suicide vector, pYAK1 (13), which contains the sacB gene conferring sensitivity to sucrose. This plasmid was introduced into E. coli SM10 λpir and then mated with V. parahaemolyticus strain RIMD2210633. TCBS agar containing chloramphenicol at a concentration of 5 μg/ml was used to screen vcrD1 deletion mutants, which were then selected on LB plates supplemented with 10% sucrose.
The following primers were used to create vscC1, vscN1, vopD, vcrD2, vscC2, vscN2, and vopP mutants in a similar manner to that described for the construction of the vcrD1 deletion mutant. For the vscC1 mutant, primers VP1696-1 (5′-AACAACTTTGGATCCAACTAT-3′), 1696-2 (5′-AATGTTGTTGAGCTGGCTCTCTAACGCTTC-3′), 1696-3 (5′-GAAGCGTTAGAGAGCCAGCTCAACAACATT-3′), and 1696-4 (5′-CTTCTACTGCAGCATCCGATA-3′) were used. For the vscN1 mutant, primers VP1668-1 (5′-ATTGGTGGATCCTCTCTTCCG-3′), 1668-2 (5′-CATCACTCGACTTGCTTTGATGCATCGGTC-3′), 1668-3 (5′-GACCGATGCATCAAAGCAAGTCGAGTGATG-3′), and 1668-4 (5′-GTTCTGCTTTCTGCAGCTCTG-3′) were used. For the vopD mutant, primers VPA1656-1 (5′-GGACATGGGATCCAAAGCTGC-3′), 1656-2 (5′-CGCGCCACGCTTGGTCGCGACCCGTTCCAC-3′), 1656-3 (5′-GTGGAACGGGTCGCGACCAAGCGTGGCGCG-3′), and 1656-4 (5′-GTGACACCTGCAGTACCTATG-3′) were used. For the vcrD2 mutant, primers VP1355-1 (5′-AATTGAGGATCCCAAAAGAGG-3′), 1355-2 (5′-TTTAATACGTGTCAAAACAAACTGACCAAC-3′), 1355-3 (5′-GTTGGTCAGTTTGTTTTGACACGTATTAAA-3′), and 1355-4 (5′-GACAACTGCAGGTAGTCCAAG-3′) were used. For the vscC2 mutant, primers VP1339-1 (5′-AAGTTAAAGGGGATCCTGGAT-3′), 1339-2 (5′-CGCCTGAACCTTCGTCCCCTTCATCTCCGA-3′), 1339-3(5′-TCGGAGATGAAGGGGACGAAGGTTCAGGCG-3′), and 1339-4 (5′-AACGTCAATGCTGCAGTTTGC-3′) were used. For the vscN2 mutant, VPA1338-1 (5′-GTAACAGGATCCAAGTTGGAG-3′), 1338-2 (5′-AATCGCAGGGTAACGATTGTTGGCCATAAT-3′), 1338-3 (5′-ATTATGGCCAACAATCGTTACCCTGCGATT-3′), and 1338-4 (5′-GTTCTCTTTTTCTGCAGTTTA-3′) were used. For the vopP mutant, primers VPA1346-1 (5′-GAATCAAAGGGATCCACTTTG-3′), 1346-2 (5′-GGCGTTGTTGGCGCGTCCGTAAAAACTTCG-3′), 1346-3 (5′-CGAAGTTTTTACGGACGCGCCAACAACGCC-3′), and 1346-4 (5′-CAACTGTGTCTGCAGTGTTTC-3′) were used. We have compared the growth rates of the parent and mutant strains. Both in LB medium with 3% NaCl and Dulbecco's modified Eagle's medium (DMEM), we could not observe a significant difference in growth rate between the parent and the mutants.
For vcrD1 complementation studies, the vcrD1 gene with the 376-bp upstream sequence containing a promoter region was amplified by PCR by using V. parahaemolyticus strain RIMD2210633 chromosomal DNA as a template and the primers 5′-ACAACTGGATCCGCGTTACCT-3′ and 5′-CGTCACTTGGATCCCCGTTTG-3′ (boldface indicates the restriction site of the specific enzyme). PCR products were cloned into the pT7Blue T-vector, removed from the construct by digestion with BamHI, and cloned into pSA19CP-MCS (18). In the vcrD2 complementation, the promoter of the tdhA gene was used for expression, since vcrD2 is a part of an operon. The tdhA promoter region (the 188-bp upstream sequence of tdhA) was cloned into the PstI and BamHI sites of pSA19CP-MCS. The vcrD2 gene was amplified by PCR and cloned downstream the tdhA promoter in pSA19CP-MCS by insertion into the BamHI and EcoRI sites. The following primers were used to create a vcrD2 complementation plasmid: for the tdhA promoter, 5′-GTTACCGCTGCAGGAATCACA-3′ and 5′-TTTTGCAAAGGATCCGTACTT-3′; for the vcrD2 promoter, 5′-AAGAGGATTGGATCCGATATG-3′ and 5′-GTTCAGTGAATTCTTCTGCCA-3′. The plasmids were introduced into the vcrD1 or vcrD2 mutant strain by electroporation with a Gene Pulse apparatus (Bio-Rad) as previously described (6).
Cytotoxicity assays.
For cytotoxicity assays, HeLa cells were grown at 37°C in 5% CO2 in DMEM (Sigma) supplemented with 10% fetal bovine serum (Sigma). Before infection, HeLa cells were washed with phosphate-buffered saline (PBS, pH 7.2) and incubated further with DMEM without phenol red. The release of lactate dehydrogenase (LDH) into the medium was assayed by using the CytoTox96 nonradioactive cytotoxicity kit (Promega) according to the manufacturer's instructions. At 5 h after infection, the supernatants were collected and the release of LDH was quantified. The LDH release (percent cytotoxicity) was calculated with the following equation: (optical density at 490 nm [OD490] of experimental release − OD490 of spontaneous release)/(OD490 of maximum release − OD490 of spontaneous release) × 100. The spontaneous release is the amount of LDH released from the cytoplasm of uninfected HeLa cells, whereas the maximum release is the amount released by total lysis of uninfected HeLa cells.
To observe the morphological change of the cells, HeLa cells were seeded into six-well plates containing coverslips at a density of 2 × 105 cells per well. At 5 h after infection with a bacterial multiplicity of infection of 1, cells were washed twice in PBS and fixed in 4% paraformaldehyde for 15 min at room temperature. After being washed twice in PBS, the cells were stained with 1 μg of Hoechst/ml and observed by microscopy.
Rabbit ileal loop test and histology.
To analyze the enterotoxicity of the parent and TTSS-related mutants, we examined the induction of fluid accumulation in rabbit ileal loops, as previously described (26). Rabbit ileal loop tests were performed with 1.5-kg female New Zealand White rabbits. The bacterial cells (108 CFU) were injected into the ligated ileal loops of rabbits, and then the fluid accumulation in each loop was measured 15 h after challenge. We dissected pieces of the small intestine from the ileal loops, washed them with cold PBS (pH 7.2), and fixed them with 4% paraformaldehyde in PBS (pH 7.2). The samples were embedded in paraffin, sectioned at 4 μm, stained with hematoxylin and eosin, and then observed by microscopy.
Protein purification and antibody production.
PCR-amplified DNA harboring the RIMD2210633 vopD or vopP gene was cloned into pET-30a (+) under the control of a T7lac promoter by using the following oligonucleotide primers: for VopD, 5′-TTAAGGAGCGGATCCATGTTGGAT-3′ and 5′-AGTACTAAAAGTCGACCGCAATTA-3′; for VopP, 5′-GCCAGATTTGAATTCAATATGGGG-3′ and 5′-TAAGATTCAATGTCGACAAATTTA-3′. His tag-containing VopD and VopP proteins were expressed in E. coli BL21, purified over Talon resin columns (Clontech), and dialyzed with PBS (pH 8.0). Polyclonal antibodies to VopD or VopP were raised by immunizing female New Zealand White rabbits, as described previously (2).
Analysis of secreted proteins.
Secreted proteins from the parent and mutant strains were isolated from the supernatants of 3 ml of bacterial cell cultures grown for 5 or 16 h at 37°C in heart infusion broth or LB medium. Bacterial cells were removed from the cultures by centrifugation. Supernatants were passed through a 0.2-μm-pore-size syringe filter and precipitated by the addition of trichloroacetic acid to a final concentration of 10% (vol/vol). The proteins were collected by centrifugation at 17,500 × g for 30 min at 4°C. Pellets were washed in 1.5 ml of ice-cold acetone, dried, and suspended in 30 μl of sodium dodecyl sulfate (SDS) sample buffer prior to analysis by SDS-polyacrylamide gel electrophoresis with 10% polyacrylamide.
Western blot analysis.
Secreted proteins were loaded onto an SDS-10% polyacrylamide gel. After electrophoresis, proteins were electrophoretically transferred to an Immobilon-P transfer membrane (Millipore). The membrane was blocked with 5% skim milk in Tris-buffered saline (20 mM Tris, 137 mM NaCl [pH 7.6]) containing 0.1% Tween 20 and probed with anti-VopD or anti-VopP for 30 min at room temperature. The secondary antibody was anti-rabbit immunoglobulin conjugated to horseradish peroxidase (Amersham Life Sciences). The blots were developed by using the ECL Western blotting kit (Amersham) according to the manufacturer's instructions.
RESULTS
Organization of TTSS genes on chromosomes 1 and 2.
From the completed genome sequence of TDH-producing V. parahaemolyticus strain RIMD2210633, two sets of genes for TTSS were identified (14). One set of genes is located at positions 1.77 to 1.81 Mb from the replication origin of chromosome 1. In this region, about 30 open reading frames were identified to be homologous to the TTSS-related genes of other gram-negative bacteria. We designated the TTSS apparatus genes on chromosome 1 as TTSS1. As shown in Fig. 1, the organization of the V. parahaemolyticus TTSS1 are similar to those of Yersinia spp. Therefore, each gene was named based on the homology to the Yersinia genes. For example, the largest protein encoded on TTSS1, which is homologous to the Yersinia YscC (identity 48%), was named VscC1, following the uniform nomenclature for the TTSS genes proposed by Hueck (11). There were some differences between the TTSS1 and Yersinia TTSS genes. First, 12 hypothetical genes existed between vscU1 and vscL1 in V. parahaemolyticus TTSS1 (Fig. 1). These genes are not found in Yersinia spp. or other species. Second, a total of 13 genes (comprising vscA1 to vscL1, virG1, and virF1) were encoded in the direction opposite to those of Yersinia species. The average G+C content in the TTSS1 region was similar to that of the rest of the genome. Another set of genes for the TTSS were identified within the pathogenicity island (PAI) region of strain RIMD2210633 (14). The PAI exists at positions 1.38 to 1.47 Mb from the replication origin on chromosome 2 (14). The G+C content of the region was 39.8%, obviously lower than that of the rest genome (45.4%), suggesting that this region was obtained by recent lateral transfer. In this region, two copies of genes for TDH were present. Within the PAI, we could identify at least nine genes that are homologous to the TTSS apparatus genes of Yersinia spp. and other gram-negative bacteria. The TTSS gene on chromosome 2 was designated TTSS2, and each gene was named by attaching the number 2, as in vscC2. The nine TTSS apparatus genes were transcribed in the same direction. Within the TTSS2 region, there existed a number of hypothetical genes. Within or in the proximity of the TTSS2 region, there existed several virulence and effector gene candidates such as homologues of the Yersinia YopJ (YopP in Y. enterocolitica, 36% identity), E. coli cytotoxic necrotizing factor (38% identity), Pseudomonas exoenzyme T (45% identity), and Shigella OspB (24% identity).
FIG. 1.
Organization of the V. parahaemolyticus TTSS1 locus. Numbers above the genes of V. parahaemolyticus TTSS1 are percent identities at the amino acid level to the corresponding proteins of Yersinia TTSS.
Construction of deletion mutant strains of vcrD, vscC, or vscN in TTSS1 and TTSS2.
TDH has been considered to be a major virulence factor of V. parahaemolyticus. TDH shows several biological activities, such as hemolytic activity, enterotoxicity, and cytotoxicity (10, 17, 23, 25). Previously, we constructed deletion mutants from the RIMD2210633 strain, which had both of the tdh genes (tdhA and tdhS) deleted (19). The tdhAS double deletion mutant, POR-1, completely lost the hemolytic activity, but the deletion did not affect its cytotoxicity in HeLa cells (19). Enterotoxicity, assayed by the rabbit ileal loop test, was decreased by the tdh deletion, but POR-1 still showed partial but apparent fluid accumulation in the rabbit intestine (19). In this study, to exclude the effect of TDH, we used the strain POR-1 as the parent strain for construction of further deletion mutants in the following experiments. To examine the role of two putative TTSSs of TTSS1 and TTSS2, vcrD1 and vcrD2, each encoding an inner membrane protein, vscC1 and vscC2, encoding an outer membrane protein, or vscN1 and vscN2, encoding a cytoplasmic protein of the TTSS apparatus, were disrupted by an in-frame deletion with homologous recombination by using the positive selection suicide vector pYAK1 (13). Deletion of the target genes was confirmed by PCR and Southern hybridization analysis (data not shown). The growth curve of the mutant strains was identical to that of the parent strain under in vitro culture conditions such as in LB medium (data not shown).
Cytotoxicity of TTSS1- or TTSS2-deficient strains.
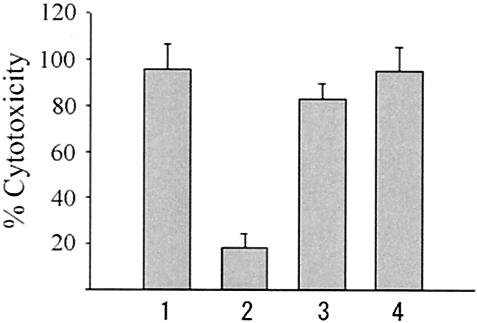
The cytotoxicity of the constructed mutant strains in HeLa cells was assessed by the release of cytosolic LDH. The strains were analyzed for their cytotoxic effect on HeLa cells 5 h after infection. The cytotoxicity of the vcrD2 deletion mutant was identical to that of the parent strain while the vcrD1 deletion mutant had little effect on HeLa cells. The decrease in cytotoxicity was fully restored by an in trans complementation of vcrD1 (Fig. 2). To observe morphological changes by parent and mutant strain infection, HeLa cells were incubated with parent or mutant strains for 5 h, fixed, and stained with Hoechst, a DNA-binding fluorophore. Cells challenged by the parent or vcrD2 mutant strain for 5 h displayed rounding with shrunken cytoplasm and nuclei, as illustrated by the intense areas of Hoechst staining (Fig. 3). In contrast, the vcrD1 deletion mutant caused little change in the HeLa cell morphology compared with the negative control uninfected cells (Fig. 3). The cytotoxicities of other vscC1 (17% ± 3%) and vscN1 (21% ± 5%) mutant strains were phenotypically similar to that of vcrD1 (18% ± 6%), whereas vscC2 (85% ± 10%) and vscN2 (82% ± 11%) mutant strains were phenotypically similar to the vcrD2 (83% ± 7%) mutant strain. These data suggest that TTSS1, but not TTSS2, is involved in the cytotoxic activity of the parent strain.
FIG. 2.
Cytotoxicity of parent and TTSS-related deletion mutant strains to HeLa cells. HeLa cells were infected with bacteria at a multiplicity of infection of 20. Five hours after infection, cytotoxic activity was assayed by measuring total cellular LDH release into the culture supernatant. The percent cytotoxicity was calculated by using the following equation: (OD490 of experimental release − OD490 of spontaneous release)/(OD490 of maximum release − OD490 of spontaneous release) × 100. Bars: 1, parent strain; 2, vcrD1 deletion mutant; 3, vcrD2 deletion mutant; 4, vcrD1 complementation into vcrD1 deletion mutant. Results are representative of at least three independent experiments.
FIG. 3.
Morphological changes induced by V. parahaemolyticus infection. HeLa cells were infected with the following strains: parent strain (A, B), vcrD1 deletion mutant (C, D), vcrD2 deletion mutant (E, F). Uninfected cells were used as a control (G, H). Five hours after infection, cells were fixed, stained, and observed by phase-contrast microscopy (left) and Hoechst staining (right).
Enterotoxicity of TTSS1- or TTSS2-deficient strains.
To investigate the contribution of TTSS1 and TTSS2 to the enterotoxicity of V. parahaemolyticus, we examined the enterotoxic activity of the parent and mutant strains in rabbit ileal loops. The bacterial cells (108 CFU) were injected into the ligated ileal loops of rabbits, and then the fluid accumulation in each loop was measured 15 h after the challenge. After the removal of small pieces of the intestine, the fluid accumulation in the ligated ileal loops was measured (Fig. 4). POR-1 demonstrated apparent fluid accumulation (Fig. 4), as previously reported (19). Although the amount of fluid accumulation of the vcrD1 mutant strain was indistinguishable from that of the POR-1 strain, the vcrD2 deletion mutant showed little fluid accumulation and was similar to the negative control, PBS (Fig. 4). We dissected small intestinal pieces from each ileal loop of the rabbit to observe the histology. Sections from tissue infected with the parent and vcrD1 deletion mutant showed a complete loss of villous architecture, with denudation of the surface epithelium in places. Massive hemorrhage with neutrophilic infiltration in the lamina propria extending up to the submucosa with congested and dilated crypts was seen (Fig. 5). In contrast, sections from tissues treated with the vcrD2 deletion mutant did not manifest significant histological changes and maintained the normal villous contour. Mutant vscC1 and vscN1 strains were phenotypically similar to vcrD1, whereas the vscC2 and vscN2 mutant strains were phenotypically similar to the vcrD2 mutant strain. These results strongly suggest that TTSS2 is involved in the enterotoxicity of V. parahaemolyticus, whereas TTSS1 is not.
FIG. 4.
Fluid accumulation in the rabbit ileal loop test. Fluid accumulation (FA) is amount of accumulated fluid (in milliliters) per length (in centimeters) of ligated rabbit small intestine. Bars: 1, parent strain; 2, vcrD1 deletion mutant; 3, vcrD2 deletion mutant; 4, vcrD2 complementation into vcrD2 deletion mutant; 5, PBS. Data represented are means ± standard deviations. The fluid accumulation of 0.2 μg of cholera toxin (Sigma), used as a positive control, was 0.71 ± 0.12 ml/cm. Asterisks indicate significant differences from the results obtained with the parent strain (P < 0.01).
FIG. 5.
Hematoxylin and eosin staining of tissue from rabbit intestinal loops infected with 108 CFU of the parent strain (B), vcrD1 mutant (C), vcrD2 mutant (D), and PBS as a negative control (A) for 15 h. The parent and vcrD1 mutant strains showed blunting of villi, submucosal edema, hemorrhage, and severe inflammation.
Secretion of proteins by mutant strains.
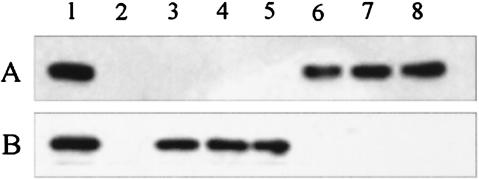
The secretion of VopP (VPA1346), a virulence-related protein encoded in the proximity of TTSS2 with homology to Yersinia YopP, and VopD (VP1656), a protein encoded in TTSS1 region with homology to Yersinia YopD, by the mutant strains was analyzed. The presence or absence of VopD and VopP in the culture supernatant was examined by Western blotting with the antibodies against each protein (Fig. 6). VopD was detected in supernatants of the parent and TTSS2-related mutant strains, such as vcrD2, vscC2, and vscN2. However, in the supernatant of the TTSS1-related mutant strains, such as vcrD1, vscC1, and vscN1, VopD was not detected. The VopP protein was detected in the supernatants of TTSS1-related mutant strains, but they were not detected in TTSS2-related mutants (Fig. 6). These data strongly suggest that VopD was secreted in a TTSS1-dependent manner while VopP was secreted via TTSS2. In other words, the two TTSSs in V. parahaemolyticus, TTSS1 and TTSS2, recognize distinct proteins for secretion.
FIG. 6.
Western blot analysis of proteins in culture supernatants of the parent and TTSS-related mutant strains grown in heart infusion broth medium for 5 h (for VopD) or in LB medium for 16 h (for VopP) at 37°C. Western blot analysis was performed by using the polyclonal antibodies against VopD (A) and VopP (B). Lane 1, parent strain (POR-1); lane 3, vcrD1 mutant; lane 4, vscC1 mutant; lane 5, vscN1 mutant; lane 6, vcrD2 mutant; lane 7, vscC2 mutant; lane 8, vscN2 mutant. Lane 2 in panel A is the vopD mutant, and lane 2 in panel B is the vopP mutant.
Distribution of TTSS genes among Vibrio species.
In a previous study, Makino et al. analyzed the distribution of the TTSS1 and TTSS2 genes among V. parahaemolyticus strains. The TTSS1 genes were present in all of the V. parahaemolyticus strains while the TTSS2 genes were present only in KP-positive strains (14). To analyze the distribution of TTSS genes in other Vibrio species, dot hybridization studies were performed with a number of Vibrio species under stringent conditions. Six probes specific for the vcrD, vscC, or vscN genes of TTSS1 and TTSS2 in V. parahaemolyticus strain RIMD2210633 were prepared. Among a total of 16 Vibrio species tested, TTSS1-related probes (vcrD1, vscC1, and vscN1) exhibited a positive signal in the Vibrio alginolyticus, Vibrio harveyi, and Vibrio tubiashii strains similar to that of the positive control RIMD2210633. Other Vibrio species gave no positive signal (Table 1). The probes for the TTSS2 genes (vcrD2, vscC2, and vscN2) did not hybridize with any of the Vibrio species tested except for the KP-positive V. parahaemolyticus strains. These data, along with a previous report (14), suggest that the TTSS2 genes are unique for KP-positive V. parahaemolyticus. In contrast, TTSS1 genes were present in all the V. parahaemolyticus strains tested, and the homologues are also present in some other Vibrio species.
TABLE 1.
Distribution of TTSS1 and TTSS2 genes in Vibrio spp.a
| Strain (n) | Result forb:
|
|
|---|---|---|
| TTSS1 | TTSS2 | |
| V. parahaemolyticus | ||
| KP positive (25/25)c | + | + |
| KP negative (32/32) | + | |
| V. aerogenes (1) | ||
| V. alginolyticus (4/4) | + | |
| V. anguillarum (1) | ||
| V. carchariae (1) | ||
| V. cholerae (5) | ||
| V. costicola (1) | ||
| V. fluvialis (1) | ||
| V. harveyi (2/2) | + | |
| V. mimicus (2) | ||
| V. natriegens (1) | ||
| V. hollisae (1) | ||
| V. pectenicida (1) | ||
| V. proteolyticus (4) | ||
| V. vulnificus (5) | ||
| V. tubiashii (1/1) | + | |
Dot hybridization was carried out at 42°C under high-stringency conditions.
A + indicates that all three probes (vcrD, vscC, and vscN) produced the position signal. The absence of a + indicates a negative signal against all three probes.
Number of strains positive (or negative)/number of strains tested.
DISCUSSION
The bacterial TTSS, which is found in animal and plant pathogens, is composed of more than 20 proteins that form a macromolecular assembly. The TTSS delivers bacterial virulence effectors not only across the bacterial inner and outer membranes but also directly into the host cell (3, 11). The apparatus proteins of TTSS are highly conserved among the bacteria that possess the system. In contrast, the effector proteins secreted via the TTSS are not conserved among those bacteria, and hence, the biological effects vary widely in target host cells (11). Genes encoding the TTSS apparatus are generally found on PAIs or virulence plasmids (11, 27). The TTSS has been identified in numerous gram-negative pathogenic bacteria including Yersinia spp., Salmonella enterica serovar Typhimurium, Shigella flexneri, and enteropathogenic E. coli (11).
Genome sequencing of the clinical V. parahaemolyticus strain RIMD2210633 revealed that the strain possesses two sets of genes for the TTSS (14). This was the first report of the TTSS in bacteria belonging to the genus Vibrio. In RIMD2210633, one set of the TTSS genes (TTSS1) was located on chromosome 1 while TTSS2 was located on chromosome 2 (14). The gene organization of TTSS1 was very similar to that of the TTSSs reported for Yersinia species. In contrast, TTSS2 is not similar to any particular TTSS of other bacteria reported to date. TTSS2 seems to contain at least essential genes for protein secretion. The distribution of those genes among various V. parahaemolyticus strains and the G+C content of the DNA regions containing TTSS1 or TTSS2 suggested that TTSS1 is intrinsic in the species while TTSS2 has a feature of laterally transferred DNA (14).
There have been some reports of bacteria that possess two sets of the TTSS in the same bacterium (5, 8, 9, 21, 24). The most-studied bacterium is S. enterica, which possesses two sets of the TTSS, SPI-1 and SPI-2 (9). SPI-1 mediates enterocyte invasion while SPI-2 influences survival within macrophages. Enterohemorrhagic E. coli O157:H7 has been known to possess a TTSS named LEE on the chromosome which is involved in the formation of attaching and effacing lesions in epithelial cells (12). Genome sequencing of enterohemorrhagic E. coli O157:H7 (8, 20) revealed that the organism possesses another TTSS named ETT2. The function of ETT2 is unknown. The presence of multiple TTSSs in the same bacterium was also reported in the genera Yersinia and Burkholderia (5, 21, 24). However, except for the case of the TTSSs in Salmonella spp., the roles of multiple TTSSs in a single bacterium have not yet been elucidated.
The present study demonstrated that both TTSS1 and TTSS2 are functional in V. parahaemolyticus strain RIMD2210633. It was revealed that TTSS1 is involved in the cytotoxicity of the organism in HeLa cells while TTSS2 has a role in enterotoxicity in a rabbit model. These data raise the possibility that these TTSSs may be involved in pathogenicity in humans. It is noteworthy that TTSS2 was discovered exclusively in human-pathogenic (KP-positive) V. parahaemolyticus strains but not in environmental strains (14), suggesting the relevance of TTSS2 to pathogenicity in humans. On the other hand, TTSS1 was present in all of the V. parahaemolyticus strains, irrespective of clinical or environmental isolates. In other words, even V. parahaemolyticus strains which are nonpathogenic to humans possess TTSS1. This raises a question as to whether TTSS1 is really involved in pathogenicity in humans. We do not have an answer to this question at the moment, but it is possible that TTSS1 alone is not sufficient to cause disease in humans and it synergistically acts in pathogenesis when some other bacterial factors are present. Since we have identified a PAI on chromosome 2 of a clinical V. parahaemolyticus that is present only in human-pathogenic strains (14), the putative additional factors may be encoded in that region. This issue is to be explored in the future.
In this study, we determined that TTSS1 and TTSS2 recognize and secrete distinct proteins (Fig. 6). Several mechanisms have been proposed in which TTSS recognizes proteins selectively (1). How TTSS-secreted proteins are targeted to one of the two TTSSs in a single bacterial cell of V. parahaemolyticus is an interesting issue, and it remains to be elucidated.
We found that the homologues of the TTSS genes are present not only in V. parahaemolyticus but also in some other Vibrio species, such as V. harveyi, V. alginolyticus, and V. tubiashii (Table 1). In all cases, the genes are homologues of the TTSS1 genes but not of the TTSS2 genes. These vibrios are known to live in marine or estuarine environments, often associating with phytoplanktons, zooplanktons, or animals, including fish and shellfish. The TTSS is an apparatus which can inject bacterial proteins into eukaryotic cells; therefore, it would be interesting to know the natural targets for protein injection by the TTSS of vibrios. This would be a clue to an understanding of the life cycle of Vibrio species in natural environments, which is not yet well understood. In the present study, we carried out dot hybridization for the TTSS genes in Vibrio species under stringent conditions (Table 1) to make sure we detected genes with a high homology to the V. parahaemolyticus TTSS genes. Similar screening of the genes under less stringent conditions may identify the homologues of the TTSS genes in more Vibrio species, indicating the possession of the TTSS by a wider range of vibrios.
The present study revealed that both sets of TTSSs in V. parahaemolyticus are functional and involved in distinct phenotypes. Identifying the effector proteins responsible for those phenotypes and analyzing the conditions for expression of and secretion by the two TTSSs will lead to a better understanding of the role of the TTSSs in V. parahaemolyticus. Studies on the pathogenic mechanism of V. parahaemolyticus have thus far mainly focused the research on a hemolysin, TDH. The present study demonstrated that the TTSSs possessed by V. parahaemolyticus may be involved in the pathogenicity of V. parahaemolyticus in humans. This finding could open a door to new concept on the pathogenic mechanism of the organism. It is possible that detailed analysis of the newly identified TTSSs of V. parahaemolyticus could lead to the development of novel preventive and therapeutic means to fight infection by the pathogen.
ADDENDUM IN PROOF
Recently, genes encoding TTSS components in Vibrio harveyi have been reported (J. M. Henke and B. L. Bassler, J. Bacteriol. 186:3794-3805, 2004).
Acknowledgments
This work was supported by the Research for the Future Program (00L01411) of the Japan Society for the Promotion of Science and Grants-in-Aid for Scientific Research on Priority Areas and Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Editor: D. L. Burns
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2001. How and when are substrates selected for type III secretion? Trends Microbiol. 9:209-214. [DOI] [PubMed] [Google Scholar]
- 2.Cantarelli, V. V., A. Takahashi, I. Yanagihara, Y. Akeda, K. Imura, T. Kodama, G. Kono, Y. Sato, T. Iida, and T. Honda. 2002. Cortactin is necessary for F-actin accumulation in pedestal structures induced by enteropathogenic Escherichia coli infection. Infect. Immun. 70:2206-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 4.Daniels, N. A., L. MacKinnon, R. Bishop, S. Altekruse, et al. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 5.Foultier, B., P. Troisfontaines, S. Muller, F. R. Opperdoes, and G. R. Cornelis. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55:37-51. [DOI] [PubMed] [Google Scholar]
- 6.Hamashima, H., T. Nakano, S. Tamura, and T. Arai. 1990. Genetic transformation of Vibrio parahaemolyticus, Vibrio alginolyticus, and Vibrio cholerae non O-1 with plasmid DNA by electroporation. Microbiol. Immunol. 34:703-708. [DOI] [PubMed] [Google Scholar]
- 7.Hardy, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 173:1176-1183. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 9.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 10.Honda, T., and T. Iida. 1993. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct heamolysin and related heamolysins. Rev. Med. Microbiol. 4:106-113. [Google Scholar]
- 11.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodama, T., Y. Akeda, G. Kono, A. Takahashi, K. Imura, T. Iida, and T. Honda. 2002. The EspB protein of enterohaemorrhagic Escherichia coli interacts directly with α-catenin. Cell. Microbiol. 4:213-222. [DOI] [PubMed] [Google Scholar]
- 14.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, et al. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 15.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris, J. G., and R. E. Black. 1985. Cholera and other vibrioses in the United States. N. Engl. J. Med. 312:343-350. [DOI] [PubMed] [Google Scholar]
- 17.Nishibuchi, M., A. Fasano, R. G. Russell, and J. B. Kaper. 1992. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60:3539-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura, T., H. Hamashima, and K. Okamoto. 2000. Carboxy terminal region of haemolysin of Aeromonas sobria triggers dimerization. Microbiol. Pathog. 28:25-36. [DOI] [PubMed] [Google Scholar]
- 19.Park, K.-S., T. Ono, M. Rokuda, M.-H. Jang, T. Iida, and T. Honda. 2004. Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol. Immunol. 48:313-318. [DOI] [PubMed] [Google Scholar]
- 20.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 21.Rainbow, L., C. A. Hart, and C. Winstanley. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51:374-384. [DOI] [PubMed] [Google Scholar]
- 22.Sakazaki, R., K. Tamura, T. Kato, Y. Obara, S. Yamai, and K. Hobo. 1968. Studies of the enteropathogenic, facultatively halophilic bacteria, Vibrio parahaemolyticus. III. Enteropathogenicity. Jpn. J. Med. Sci. Biol. 21:325-331. [DOI] [PubMed] [Google Scholar]
- 23.Shirai, H., H. Ito, T. Hirayama, Y. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 25.Takeda, Y. 1983. Thermostable direct hemolysin of Vibrio parahaemolyticus. Pharmacol. Ther. 19:123-146. [DOI] [PubMed] [Google Scholar]
- 26.Twedt, R. M., J. T. Peeler, and P. L. Spaulding. 1980. Effective ileal loop dose of Kanagawa-positive Vibrio parahaemolyticus. Appl. Environ. Microbiol. 40:1012-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winstanley, C., and C. A. Hart. 2001. Type III secretion systems and pathogenicity islands. J. Med. Microbiol. 50:116-126. [DOI] [PubMed] [Google Scholar]