Abstract
Staphylococcus capitis (S. capitis) has been implicated in a large proportion of coagulase-negative staphylococcal infections in very-low-birth-weight infants. To identify potential therapeutic targets, the S. capitis genome was probed for the presence of genes encoding microbial surface components recognizing adhesive matrix molecules (MSCRAMM). By using Southern blot analysis, an S. capitis gene, designated sdrX, that contained sequence motifs consistent with the Sdr family of MSCRAMM proteins was identified. By using monospecific antisera in Western blot and flow cytometry, SdrX was demonstrated to be expressed on the surface of S. capitis. Human collagen type VI was found to bind both the recombinant A domain of SdrX and viable S. capitis expressing SdrX. SdrX is the first collagen-binding Sdr protein described and is the first MSCRAMM protein identified in S. capitis.
Nosocomial infections result in considerable morbidity and mortality, increased hospitalization, and an increase in healthcare utilization. These infections are especially problematic in premature infants. Late-onset sepsis, an invasive infection occurring in neonates after 72 h of life, occurs in 21% of very-low-birth-weight infants (30). Coagulase-negative staphylococci (CoNS) are considered the leading cause of late-onset infections in this population, accounting for 48% of all infections (30). Staphylococcus epidermidis is often reported as the most frequent isolate among the CoNS causing infections in very-low-birth-weight infants. Other species of CoNS have also been shown to cause sepsis in this very susceptible patient population. A recent report (31) described a bloodstream infection in a premature infant that was caused by Staphylococcus capitis. Subsequent analysis of blood cultures from neonates between 1997 and 2000 revealed that approximately 20% of the isolates were S. capitis (31). In addition, the S. capitis isolates were shown to be heteroresistant to vancomycin, making antibiotic therapy more problematic. These findings prompted us to determine if S. capitis expresses surface proteins that could serve as potential antigenic targets for the development of novel antibody-based therapies.
MSCRAMM proteins are cell surface adhesins (24) that recognize and specifically bind to distinct extracellular components of host tissues or to serum-conditioned implanted biomaterials such as catheters, artificial joints, and vascular grafts (25). Clumping factor (ClfA) is an MSCRAMM protein expressed by Staphylococcus aureus that promotes binding of fibrinogen and fibrin to the bacterial cell surface (19, 20). ClfA is the prototype of a multigene family of cell surface proteins characterized by a common domain composed of a serine-aspartate dipeptide repeat (Sdr) (18). Other members of this family that are expressed by S. aureus include ClfB (21), Pls (27), SdrC, SdrD, and SdrE (12). S. epidermidis also expresses a series of Sdr proteins including SdrF, SdrG, and SdrH (8, 18). Three additional Sdr family genes have been cloned and sequenced and include SdrY and SdrZ from Staphylococcus caprae and SdrI from Staphylococcus saprophyticus. GenBank accession numbers for these gene sequences are AY048593, AY048595, and AF402316, respectively. Because of their critical role in bacterial adhesion and the expression of these proteins on the bacterial surface, Sdr family proteins represent attractive targets for immunotherapy. In fact, it has been shown in animal models that ClfA is an excellent target for both active and passive antibody therapies against S. aureus-induced sepsis, septic arthritis, and endocarditis (7, 13, 32).
To determine if additional members of the Sdr protein family are present in S. capitis, a gene fragment incorporating the Sdr repeat region of the S. epidermidis sdrG gene was used to probe the S. capitis genome. In the present study, we report the identification and characterization of a novel Sdr family protein from S. capitis. The data demonstrate that this new gene, sdrX, encodes a surface-expressed protein with sequence motifs in common with other Sdr proteins from staphylococci. Additionally, SdrX was found to be the first Sdr protein to bind collagen, and antibodies against SdrX were shown to inhibit collagen type VI binding activity associated with S. capitis in vitro.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strain XL10-Gold ultracompetent cells (Stratagene, La Jolla, Calif.) and Topo10F′ competent cells (Invitrogen, Carlsbad, Calif.) were used as hosts for DNA transformation. Plasmid pUC18 was used for cloning genomic DNA fragments. Plasmid pQE-30 (QIAGEN, Valencia, Calif.) was used for cloning the A domain of SdrX. The bacterial strain M15(pREP4) (QIAGEN) was used for the expression of the recombinant SdrX A domain. S. capitis strains ATCC 27840, ATCC 27841, ATCC 27842, ATCC 27843, ATCC 35661, ATCC 49324, ATCC 49325, ATCC 49326, and ATCC 49327 were obtained from the American Type Culture Collection (Manassas, Va.). S. capitis strains 004102 and 012106 were clinical isolates from neonatal intensive care unit patients. S. epidermidis strain K28 was a gift from M. Hook.
Southern hybridization.
Genomic DNA from S. epidermidis K28 and S. capitis ATCC 49326 was prepared by using a G/Nome DNA kit (Bio-101, Carlsbad, Calif.) with the addition of 2 mg of lysozyme and 0.1 mg of lysostaphin (Sigma, St. Louis, Mo.)/ml to the cell suspension solution. The hybridization probe was made from the genomic DNA of S. epidermidis K28 by PCR and labeled with digoxigenin (Roche Applied Science, Indianapolis, Ind.). The PCR primers spanned the B and R regions of sdrG (forward primer, 5′-CCGCTTAGTAATGTATTG-3′; reverse primer, 5′-TCTTATCTGAGCTATTG-3′). For Southern blotting, 1 μg of genomic DNA was digested with 20 U of HindIII at 37°C overnight and separated in a 0.8% agarose gel. The Southern transfer, hybridization, and washing were performed according to the instruction manual for the Zeta-probe GT blotting membrane (Bio-Rad, Hercules, Calif.), except that hybridization and washing were performed at 45°C. After washing, the membrane was incubated with an anti-digoxigenin-POD antibody (Roche Applied Science), and the signal was detected with Supersignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.).
Genomic DNA library preparation and screening.
Genomic DNA from S. capitis ATCC 49326 was digested with HindIII and separated in a 0.8% agarose gel. DNA fragments ranging from 4 to 6 kb were purified from the gel, ligated into HindIII-digested pUC18, and transformed into XL10-Gold ultracompetent E. coli (Stratagene). The bacterial colonies were blotted onto an 85-mm C/P Lift membrane (Bio-Rad) and lysed with 0.5 N NaOH-1% sodium dodecyl sulfate (SDS) for 10 min. The membrane was then washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and baked at 80°C for 30 min. Colony hybridization was performed with the digoxigenin-labeled hybridization probe under the same conditions as those for the Southern hybridization.
DNA sequencing and analysis.
The cloned DNA fragments or PCR products were sequenced by primer extension sequencing (Seqwright, Houston, Tex.). DNA and amino acid sequences were analyzed by using Lasergene software (DNASTAR, Inc., Madison, Wis.). The BLAST network service (http://www.ncbi.nlm.nih.gov/) was used for sequence homology searches.
Genomic DNA PCR.
Genomic DNA was prepared from mid-log-phase cultures by using a MicroLysis kit (Microzone Ltd., West Sussex, United Kingdom). Lysis of bacterial cells was achieved through three thermal cycles (65°C for 5 min, 96°C for 2 min, 65°C for 4 min, 96°C for 1 min, 65°C for 1 min, and 96°C for 30 s) with a GeneAmp PCR System 2400 (Perkin-Elmer, Wellesley, Mass.). The clarified supernatant was collected and amplified by PCR for 30 cycles at 94°C for 30 s, 47°C for 30 s, and 72°C for 1 min with primers specific for sdrX (sdrX-AF [5′-CGGGATCCGAGACTTCAACTGAATTAAC-3′] and sdrX-AR [5′-AACTGCAGCGCGTATAAATCGCAATCTG-3′]).
Reverse transcription-PCR (RT-PCR) analysis of sdrX expression.
Total RNA was isolated from early log, mid-log, and stationary phases of bacteria cultures as follows. Two volumes of the RNAprotect bacteria reagent (QIAGEN) were added to 1 volume of cell culture and treated at room temperature for 5 min. Total RNA was isolated by using an RNeasy mini kit (QIAGEN) according to the manufacturer's instructions, except that cells were lysed with 2 mg of lysozyme and 0.1 mg of lysostaphin (Sigma)/ml for 1 h at 37°C. Reverse transcription was carried out with MLV reverse transcriptase (Promega, Madison, Wis.) in the presence of deoxynucleoside triphosphates, 2 μg of total RNA, and the primer for sdrX (5′-AACTGCAGCGCGTATAAATCGCAATCTG-3′) or 16S rRNA (5′-AACTTTATGGGATTTGCT-3′). The resulting cDNA was amplified by PCR with primers specific for 16S rRNA and sdrX (16S rRNA primers, 5′-TTGAAACTCAAAGGAATTG-3′ and 5′-AACTTTATGGGATTTGCT-3′; sdrX primers, 5′-GGTATGCCATTAGAAGATTTAC-3′ and 5′-AACTGCAGCGCGTATAAATCGCAATCTG-3′).
Cloning the SdrX A domain.
The A domain of SdrX was amplified by PCR from the genomic DNA of S. capitis ATCC 49326 (forward primer, 5′-CGGGATCCGAGACTTCAACTGAATTAAC-3′; reverse primer, 5′-AACTGCAGCGCGTATAAATCGCAATCTG-3′). PCR was carried out with Pfu Turbo DNA polymerase (Stratagene) for 30 cycles at 94°C for 30 s, 45°C for 30 s, and 72°C for 1 min. The PCR product was gel purified by using a Qiaquick gel extraction kit (QIAGEN), digested with BamHI and PstI, and ligated into pQE-30 (QIAGEN) using T4 DNA ligase (New England Biolabs, Beverly, Mass.). The resulting plasmid (pQE-30/sdrX-A) was transformed into bacterial strain M15(pREP4), and the transformants were selected on Luria broth plates supplemented with ampicillin (100 μg/ml) and kanamycin (25 μg/ml) (Sigma).
Expression and purification of the SdrX A domain.
E. coli M15(pREP4) carrying pQE-30/sdrX-A was cultured in Luria broth supplemented with ampicillin (100 μg/ml) and kanamycin (25 μg/ml) at 37°C to an optical density at 600 nm of 0.9. Gene expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Cells were harvested and resuspended in the lysis buffer containing 25 mM Tris (pH 8.0), 0.5 M NaCl, and 5 mM imidazole. The recombinant SdrX A domain (rSdrX-A) protein was purified as previously described (7).
Preparation of polyclonal antiserum and purified hyperimmune antibodies.
The purified rSdrX-A protein was used as an immunogen to generate polyclonal antiserum in both mice and rabbits. Serum was separated from blood collections, and for some applications, the immunoglobulin G (IgG) fraction from the serum was purified by using protein G affinity chromatography.
Flow cytometry.
The expression of SdrX by S. capitis cells in early-log-phase and mid-log-phase cultures and cultures grown overnight was determined by flow cytometry with a FACSCalibur flow cytometer (B-D Biosciences, San Jose, Calif.) equipped with an argon-ion laser (488 nm) as previously described (7).
Detection of SdrX by Western blot analysis.
S. capitis cells from early-log-phase and mid-log-phase cultures and cultures grown overnight were washed in water and resuspended in the cell suspension solution (G/Nome DNA kit; Bio-101) containing 1× proteinase inhibitor cocktail (Sigma). Bacteria were lysed by sonication five times for 10 s each with a Sonic Dismembrator 550 (Fisher Scientific, Hampton, N.H.). The lysate was cleared by centrifugation at 20,800 × g for 10 min. The supernatant was collected as the cytoplasm fraction. The pellet was resuspended in the cell suspension solution supplemented with 1× proteinase inhibitor cocktail and 2 mg of lysozyme and 0.2 mg of lysostaphin (Sigma)/ml and incubated at 37°C for 3 h. The tube was centrifuged for 5 min at 20,800 × g, and the supernatant was collected. Proteins were separated by electrophoresis in a 10% Novex Bis-Tris gel (Invitrogen) and transferred onto a polyvinylidene difluoride membrane (Invitrogen). The membrane was incubated with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBS-T) and 5% milk for 1 h at room temperature. Mouse anti-SdrX hyperimmune serum was added at a 1:200 dilution. For competition, 250 μg of rSdrX-A was added to the SdrX antibody. The membrane was incubated overnight at 4°C, washed three times with PBS-T, and incubated with the horseradish peroxidase-conjugated goat anti-mouse antibody at a 1:5,000 dilution for 1 h at room temperature. After being washed three times in PBS-T, the membrane was incubated with SuperSignal West Pico chemiluminescent substrate (Pierce) for 5 min and exposed to X-ray film.
rSdrX-A domain ligand binding.
The rSdrX-A protein was covalently coupled to Fluoresbrite YG carboxylate microspheres (1.0 μm; Polysciences, Warrington, Pa.). A total of 96 μg of rSdrX-A protein/ml was absorbed on the microspheres as determined by using the bicinchoninic acid protein assay method. Human collagen type I, II, III, IV, V, or VI (Rockland Immunochemicals, Gilbertsville, Pa.) and fibrinogen (Enzyme Research Laboratories, South Bend, Ind.), fibronectin, vitronectin, laminin (Sigma), or elastin (CalBiochem, San Diego, Calif.) were diluted in 1× PBS, pH 7.4 (GIBCO BRL, Rockville, Md.), to 5 μg/ml. The extracellular matrix (ECM) proteins were coated onto 96-well Costar EIA plates (Corning Incorporated, Corning, N.Y.) at 4°C overnight in duplicate. Plates were washed four times with 1× PBS, pH 7.4, containing 0.05% Tween 20 (1× PBS-T) and blocked with a 1% (wt/vol) solution of bovine serum albumin (BSA) in distilled water for 1 h at room temperature. The enzyme-linked immunosorbent assay (ELISA) plates were washed after blocking. Fluorescent microspheres covalently coupled with rSdrX-A were diluted in 1× PBS-T containing 0.1% BSA and added to the ECM protein-coated ELISA plates for 2 h at room temperature. Nonadherent microspheres were removed by washing three times with 1× PBS-T. Following the last wash, 1× PBS-T was added to all of the wells. The fluorescence generated by adherent microspheres was read with an excitation wavelength at 440 nm, an emission wavelength at 486 nm, and a cutoff wavelength at 475 nm by using a Spectra-MAX M2 plate reader (Molecular Devices Corporation, Sunnyvale, Calif.).
Bacterial adherence assay.
Adherence assays were performed as previously described (8), with modifications. A 1:100 dilution of a culture of S. capitis ATCC 35661 grown overnight was incubated for 4 h at 37°C in tryptic soy broth with rotation. The early-log-phase cultures were centrifuged at 2,620 × g for 10 min at 2 to 8°C. The bacterial pellet was washed twice with 20 ml of 1× PBS (Life Technologies, Inc., Rockville, Md.) by centrifuging as described above. After washing, the bacterial suspension was adjusted to an optical density at 600 nm of 2.7 in 1× PBS. Costar ELISA plates (Corning Incorporated) were coated overnight at 2 to 8°C in duplicate with 20-μg/ml solutions of human collagen types I, II, III, V, and VI (Rockland Immunochemicals). The coated plates were washed three times with 1× PBS buffer, blocked with 1% BSA for 1 h at room temperature, and washed again. S. capitis ATCC 35661 cells were serially diluted and added to the ECM-coated plates. The bacteria were incubated on the plates for 2 h at 37°C. Following the incubation, the plates were washed to remove nonadherent bacteria. The adherent bacteria were fixed with 25% formaldehyde at room temperature for 30 min. The formaldehyde was removed by washing, and the adherent bacteria were stained with crystal violet (prepared in 5% acetic acid solution) for 1 min at room temperature. Excess crystal violet was removed by washing four times with 1× PBS. The absorbance was read at 570 nm by using a Spectra-MAX M2 plate reader (Molecular Devices Corporation).
Nucleotide sequence accession number.
The nucleotide sequence of the sdrX gene has been deposited in the GenBank database under accession number AY10088.
RESULTS
Cloning and sequencing sdrX.
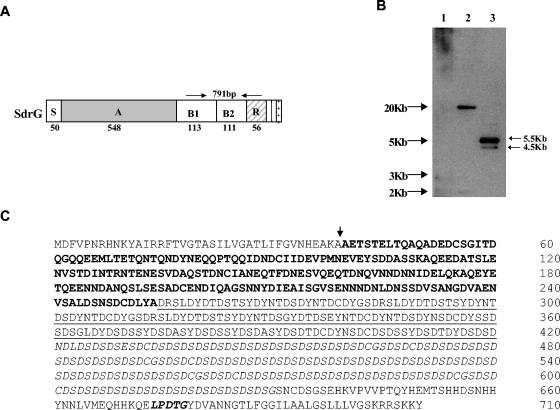
Genomic DNA was isolated from S. epidermidis strain K28. This DNA was used as a template for PCR to generate a 791-bp gene fragment spanning the B1 through R regions of the sdrG gene (Fig. 1A). The resulting PCR product was used as a hybridization probe (R region probe) to determine whether the S. capitis genome contained genes which shared sequence similarities with Sdr family proteins. A genomic Southern blot of DNA prepared from S. capitis strain ATCC 49326 was probed with the sdrG R region fragment by using low-stringency hybridization. Two HindIII fragments of approximately 5.5 and 4.5 kb hybridized with the probe (Fig. 1B). In order to isolate and identify these potential Sdr family gene sequences, a mini-genomic library was constructed by digesting genomic DNA from S. capitis strain ATCC 49326 with HindIII and cloning size-fractionated fragments (3 to 6 kb) into the pUC18 vector. Inserts of 5.5 and 4.5 kb were subsequently cloned by colony hybridization and were sequenced.
FIG. 1.
Identification and cloning of S. capitis ATCC 49326 genomic sequences with homology to the repeat region of sdrG. (A) Schematic representation of the SdrG protein and the corresponding region of the gene sequence that was used as a hybridization probe (arrows). (B) Southern hybridization. Genomic DNA was digested with HindIII, separated on a 1% agarose gel, and transferred onto a Zeta-probe membrane. The blot was hybridized with a digoxigenin-labeled probe from the B and R regions of sdrG. Lane 1: 1-kb DNA molecular weight marker; lane2: genomic DNA from S. epidermidis K28; lane 3: genomic DNA from S. capitis ATCC 49326. (C) Deduced amino acid sequence of SdrX from S. capitis ATCC 49326. The vertical arrow indicates the signal peptide cleavage site (amino acid 39). The A region (amino acids 40 to 254) is shown in boldface type. The B repeat region (BX) (amino acids 255 to 420) is underlined. The R region (amino acids 421 to 630) containing the SD repetitive sequence is in italics. The cell wall-anchoring motif LPDTG (amino acids 674 to 678) is in italic boldface type.
The 5.5-kb insert contained an open reading frame of 2,133 bp encoding a protein of 710 amino acids with a calculated molecular weight of 76.71 kDa. The deduced protein sequence (Fig. 1C) included a long serine-aspartate repeat (Sdr) region characteristic of the Sdr protein family. In keeping with current naming conventions, the newly identified protein was named SdrX. SdrX contained other protein sequence features typical of other Sdr proteins (18, 27). The N-terminal portion of the encoded protein contained a putative signal peptide of 39 amino acids as predicted by SignalP software (http://www.cbs.dtu.dk/services/SignalP). Following the signal sequence, there was a 215-amino-acid region which we have designated as the putative A domain. The A domain (amino acids 40 to 254) of SdrX was used to perform a protein-protein BLAST search against the nonredundant GenBank database of the National Center for Biotechnology Information. The search returned an alignment with the fibronectin binding autolysin (AtlC) from S. caprae (1). The two sequences shared an identity of 24% and a similarity of 42% in the region spanning amino acids 25 to 168 in the A domain of SdrX and amino acids 56 to 191 of AtlC. A second BLAST search for short, nearly exact matches against the nonredundant GenBank database resulted in a significant alignment with the Aas surface protein from S. saprophyticus (10). A sequence identity of 35% and a similarity of 47% between a portion of the A domain of SdrX (in the region from amino acids 67 to 201) and the Aas protein (amino acids 81 to 198) were observed. However, the A domain of SdrX has little sequence similarity to previously described Sdr proteins.
The A region was followed by a B region containing a series of short tandem repeats (Fig. 1C). Further downstream, a highly repetitive region of 210 amino acid residues followed, composed of tandemly repeated serine-aspartate residues with other residues such as cysteine, glutamic acid, and glycine found sporadically through the region. The C-terminal portion of the protein contained the sequence LPDTG, which corresponds to the cell wall-anchoring motif LPXTG found in all Sdr family proteins (18, 27). A membrane-spanning region and a positively charged tail, features also common to most Sdr family proteins, were identified in the C terminus of the protein sequence (Fig. 1C).
Prevalence of the sdrX gene in S. capitis strains.
Genomic DNAs from nine different American Type Culture Collection strains of S. capitis and two clinical isolates of S. capitis were amplified by PCR using primers specific for the sdrX gene. All of the strains tested were found to contain the sdrX gene (data not shown).
Transcription of sdrX at different stages of bacterial growth.
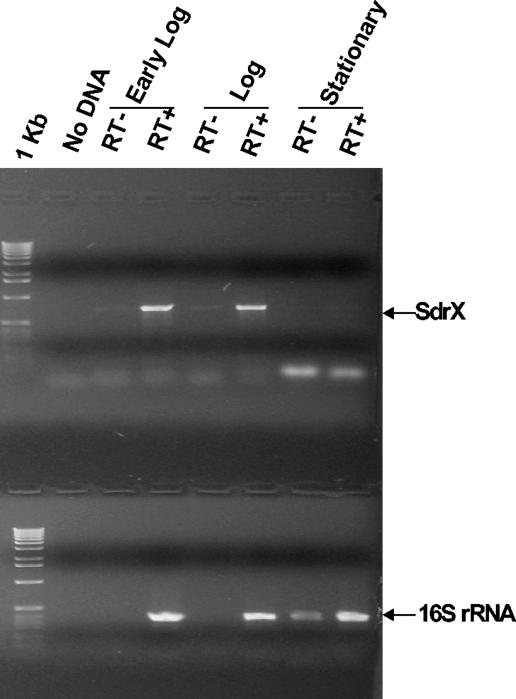
To determine if sdrX gene transcription is regulated during the growth cycle, total RNA was isolated from S. capitis ATCC 49326 cultures at early log, mid-log, and stationary phases. RT-PCR was performed by using primers specific for sdrX. RT-PCR with primers specific for 16S rRNA was also performed as a control. As shown in Fig. 2, the sdrX gene was transcribed at both early log and mid-log phases, but sdrX-specific RNA could not be detected at stationary phase, indicating that the transcription of sdrX is regulated during the different stages of bacterial growth.
FIG. 2.
Detection of sdrX mRNA by RT-PCR. Total RNA was isolated from S. capitis ATCC 49326 culture at early log, mid-log, and stationary phases. 16S rRNA and sdrX RNA were converted into cDNA by using sequence-specific primers and amplified by RT-PCR. RT+ and RT− indicate with and without reverse transcriptase, respectively. The positions of the sdrX and 16S rRNA products are indicated.
Expression and purification of the A domain of SdrX.
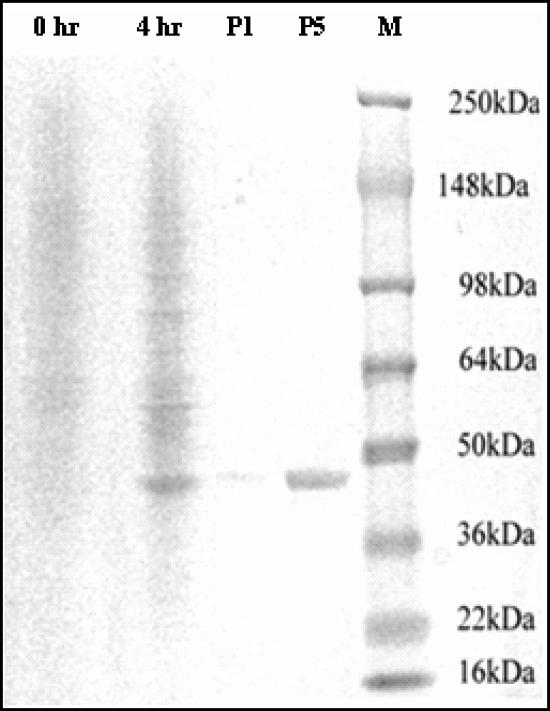
The A domain of SdrX was cloned into the pQE-30 vector and expressed as a His6-tagged fusion protein in E. coli M15(pREP4). One prominent 45-kDa band was detected in the cell extract after induction for 4 h with 1 mM IPTG (Fig. 3). This band was not present in the absence of induction. Although the apparent molecular weight of the protein by SDS-polyacrylamide gel electrophoresis (PAGE) was significantly different from the predicted molecular weight for the recombinant protein, mass spectroscopy analysis confirmed a molecular weight of 25.7 kDa. The migration pattern of the rSdrX-A in SDS-PAGE is consistent with other recombinant A domains of previously identified MSCRAMM proteins (our unpublished data).
FIG. 3.
Expression and purification of the recombinant A domain of SdrX. The A domain of SdrX was cloned into pQE-30 and expressed as a His-tagged fusion protein in M15(pREP4). Cell extracts were purified on a chelating HiTrap column. SDS-PAGE analysis was performed with the crude cell extracts before (0 h) and after (4 h) induction. Data from an analysis of purified recombinant protein are shown at loading levels of 1 μg (P1) and 5 μg (P5). SeeBluePlus2 was used as the molecular weight marker (M).
Localization of SdrX expression.
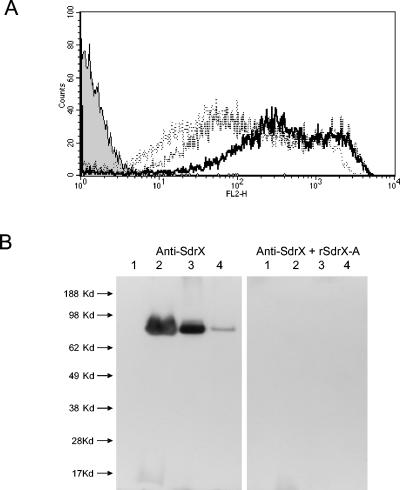
The presence of a cell wall-anchoring motif, LPDTG, in the cell wall-spanning domain of SdrX suggested that the protein was likely to be associated with the cell wall (28). To determine if SdrX is indeed expressed on the surface of S. capitis, a panel of strains was analyzed by flow cytometry with an rSdrX-A-specific antiserum. The anti-rSdrX-A serum recognized the surfaces of all of the strains tested. Strain ATCC 35661 was found to have the highest level of antibody recognition (data not shown). This strain was selected for a time course experiment to determine if SdrX protein expression varied with time in culture. S. capitis cultures were analyzed at early log, mid-log, and stationary growth phases by flow cytometry (Fig. 4A). The highest mean fluorescence was observed with early-log-phase cultures. Analysis of later stages of growth resulted in lower levels of immunofluorescence, suggesting reduced levels of antigen expression. This finding was corroborated by Western immunoblotting analysis. Cytoplasmic and cell wall protein fractions were prepared from early-log-, mid-log-, and stationary-phase S. capitis cultures. A band of approximately 80 kDa was detected in the cell wall fractions at all time points. However, signal intensity was highest in early-log- and mid-log-phase culture samples (Fig. 4B). The signal from the 80-kDa band was completely eliminated when the Western blot was carried out in the presence of soluble rSdrX-A, indicating that the antibody signal was specific for SdrX. SdrX protein was not observed in the cytoplasmic fraction, suggesting that the vast majority of SdrX protein was cell wall associated.
FIG. 4.
Surface expression of SdrX. (A) Detection of surface localization of SdrX by flow cytometry. S. capitis strain ATCC 35661 was grown to early log (bold line), mid-log (broken line), or stationary (dotted line) phase. Bacteria were stained with rabbit anti-rSdrX-A followed by phycoerythrin-conjugated donkey anti-rabbit antibody (heavy and light chain). Background staining with a normal rabbit serum control is indicated by the shaded grey histogram. (B) Western immunoblotting analysis of SdrX. Proteins from cell extracts were separated in 10% Bis-Tris gel in MES (morpholineethanesulfonic acid)-SDS NuPage running buffer and transferred to a polyvinylidene difluoride membrane. The Western blots were probed with hyperimmune serum generated against the recombinant A domain of SdrX (rSdrX-A). Western blotting was carried out in the presence (right panel) or absence (left panel) of soluble rSdrX-A as a competitor to demonstrate specificity. SeeBluePlus2 was used as the molecular weight marker. Lane 1: cytoplasm fraction from early-log-phase culture of S. capitis ATCC 35661. Cell wall fractions from S. capitis ATCC 35661 cultures at early log (lane 2), mid-log (lane 3), and stationary (lane 4) phases are shown.
Recognition of ECM proteins by rSdrX-A and S. capitis.
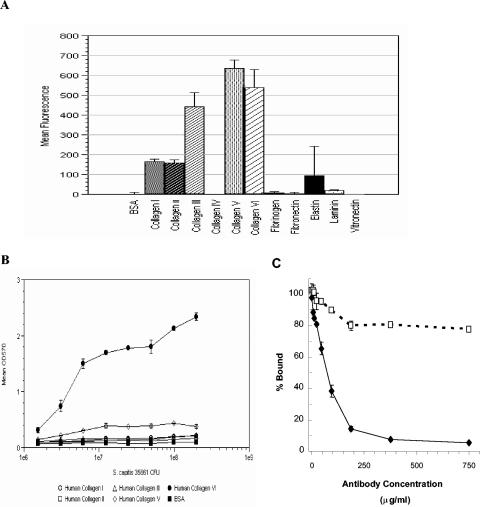
To identify potential ligands for SdrX, a fluorescent bead-based assay was developed. The rSdrX-A protein was covalently coupled to fluorescent beads and incubated with a panel of immobilized ECM proteins including human collagen type I, II, III, IV, V, or VI, fibrinogen, fibronectin, vitronectin, laminin, and elastin. The rSdrX-A-coated beads were captured by most of the human collagen including types I, II, III, V, and VI (Fig. 5A). To determine if any of these potential ligands for SdrX could also act as ligands for S. capitis adhesion, mid-log-phase cultures of S. capitis strain ATCC 35661 were incubated on microtiter plates coated with human collagen types I, II, III, V, and VI or BSA (Fig. 5B). S. capitis bound specifically to plates coated with collagen type VI, which increased as a function of bacterial concentration. However, the bacteria did not adhere to any of the other human collagen types that were tested. To determine if SdrX contributed to the collagen type VI binding activity observed in S. capitis, the bacteria were preincubated with increasing concentrations of rabbit anti-rSdrX-A polyclonal antibody prior to performance of the adherence assay. The anti-rSdrX-A antibody was able to inhibit S. capitis binding to collagen type VI by approximately 95%, whereas equal concentrations of normal rabbit IgG marginally reduced bacterial adhesion to the collagen type VI-coated substrate (Fig. 5C). This result suggests that most if not all of the collagen type VI binding activity observed in S. capitis was mediated by SdrX.
FIG. 5.
Identification of the SdrX ligand. (A) Screening of ECM proteins by a fluorescent bead-based assay. The rSdrX-A protein was covalently coupled to fluorescent beads and incubated with the immobilized human ECM proteins listed on the horizontal axis. Immobilized BSA served as a negative control. The vertical bars indicate the mean fluorescence and standard deviations measured in duplicate wells. (B) Bacterial adherence assay. S. capitis strain ATCC 35661 was bound to immobilized human collagen I, II, III, V, and VI and BSA. Symbols indicate the mean absorbance and standard deviations of duplicate wells. (C). Inhibition of binding of S. capitis strain ATCC 35661 to immobilized collagen VI. Bacteria were incubated with rabbit anti-rSdrX-A (diamonds) or normal rabbit IgG (squares). Symbols indicate the mean percent bound and standard deviations of duplicate wells.
DISCUSSION
A molecular approach was used to determine if Sdr family proteins exist in S. capitis. A DNA fragment corresponding to the B and R regions of the sdrG gene was used to probe the S. capitis genome. A novel member of the Sdr family of MSCRAMMs was identified, cloned, and sequenced. The deduced protein sequence was compared to the published protein sequences of other Sdr family molecules. The overall structure of the coding region was found to follow the general pattern observed in other Sdr family proteins (18) and included a signal sequence, an A domain, a repetitive domain termed BX, an SD repeat region, a cell wall anchor region with an LPXTG motif sequence (LPDTG, amino acids 674 to 678), a hydrophobic membrane-spanning region, and a series of positively charged residues at the C terminus. Individual domains of SdrX were compared to other members of the Sdr family by using Clustal W analysis.
A comparison of the SdrX signal sequence showed the greatest homology (∼52%) with SdrC, SdrD, and SdrF. The A domain of SdrX was compared to other Sdr protein sequences and showed little or no homology (less than or equal to 11%). The A domain sequence was also used to perform a BLAST search of the public database of the National Center for Biotechnology Information. Two protein sequences, AtlC from S. caprae (1) and Aas from S. saprophyticus (10), were found to have homologies to the A domain of SdrX. The nature and extent of the relationship of SdrX to either of these proteins are currently not known. Although a conserved sequence (TYTFTDYVD) has been reported to be present in the A domains of all S. aureus Sdr proteins (12, 22) and in the S. epidermidis proteins SdrF and SdrG (18), this sequence was not present in the SdrX A domain. The absence of this sequence, taken together with the sequence homology data, suggests that the A domain of SdrX represents a unique structure unrelated to previously described Sdr protein A domains.
The repetitive region of 166 amino acids found between the A domain and the SD dipeptide repeat region in SdrX is made up of short repeated sequences that vary in length. The repeats are high in serine (S) and aspartic acid (D) (56% SD overall) but are sufficiently divergent from the dipeptide repeat R region to be categorized as a separate domain. This sequence is considerably divergent from the B regions described for other Sdr proteins. This region in SdrX was therefore named BX to distinguish it from previously described B regions.
The R region of SdrX is typical in size (210 amino acids) for R regions found throughout the Sdr protein family. The presence of this domain places SdrX unequivocally in the Sdr family of staphylococcal proteins.
All of the Sdr proteins identified to date also include a conserved sequence motif, LPXTG (18). This sequence is a substrate for sortase, a transpeptidase that cleaves and covalently links the protein to peptidoglycan in the cell wall, allowing for surface expression of the molecule (17, 29). The sequence LPDTG is found in SdrX at position 674. Together, the R region and the BX region provide 419 amino acids between the end of the putative A domain and the LPDTG cell wall-anchoring motif. For ClfA, it has been reported that the R region must be 80 residues in length (112 residues in total from the A domain to LPXTG) to support wild-type clumping function (9). Therefore, the R region of SdrX would appear to be of sufficient length to allow for exposure of the A domain on the surface of S. capitis. Two lines of evidence demonstrate that SdrX is indeed expressed on the cell surface. By Western blot analysis, it was shown that SdrX is present in cell wall fractions prepared from S. capitis but not in cytoplasmic preparations of the same cultures. Also, the A domain of SdrX was found to be accessible to antibodies on the surface of viable S. capitis as measured by flow cytometry. The available data therefore demonstrate that the SdrX protein is a surface-expressed protein, as predicted from the primary sequence.
C terminal to the LPDTG sequence is a 32-amino-acid sequence containing a high percentage of hydrophobic residues (57%) followed by a short, highly polar sequence. Similar regions have been described previously for Sdr proteins (18) and are thought to function as a membrane-spanning region and a cytoplasmic tail. Overall, the analysis of the SdrX protein sequence has led to the conclusion that SdrX is a novel member of the Sdr gene family and the first such protein described for S. capitis.
Beyond the primary sequence data, our investigations of SdrX have focused on the distribution and expression of SdrX. Genomic PCR was carried out to determine if the sdrX gene is widely distributed among S. capitis strains. Panels of 11 different strains were analyzed. All of these strains were found to carry the sdrX gene. Although the panel was relatively small, it included both commonly used laboratory strains as well as two unique clinical isolates collected from a neonatal intensive care unit. The existing data could lead to the prediction that the sdrX gene will be found to be widely distributed among S. capitis strains.
SdrX expression was analyzed at both the RNA and protein levels. Results of RT-PCR demonstrated that the sdrX transcript could be found in early- and mid-log-phase cultures but not in stationary phase. Protein expression data collected by Western blotting and flow cytometry were in agreement with the RT-PCR data inasmuch as the strongest protein signals were obtained in early log phase and the weakest signals were found in bacteria grown to stationary phase. The finding that RNA transcription is no longer occurring in stationary-phase cultures, although protein is still detectable, suggests that the protein remains on the surface of the bacteria in the absence of de novo synthesis and may indicate that the protein is a relatively stable and long-lived molecule.
The recombinant protein rSdrX-A was used to screen ECM proteins for potential ligands. A series of human collagen proteins was identified as having potential candidates for the SdrX ligand. Although the recombinant A domain was able to recognize five different collagen types, assays using S. capitis clearly demonstrated that only one of these proteins, collagen type VI, is a natural target for adherence of the whole cells of S. capitis. These data suggest that some property of the native form of SdrX as expressed on the surface of bacteria is altered when the recombinant A domain is used in the bead binding assay, such that the binding activity is less specific and rSdrX-A is able to interact with several members of the collagen family under the conditions of the assay. However, the critical role of SdrX in the adherence of S. capitis to collagen type VI was confirmed when nearly complete inhibition of this binding activity was achieved with antiserum generated against the A domain of SdrX. Therefore, the data suggest that SdrX is principally responsible for the collagen type VI binding activity observed with S. capitis strain ATCC 35661.
The collagen type VI monomer is made up of three different α chains, each of which consists of a short helical region separating globular domains at the N and C termini (5). The molecule is secreted as a tetramer which then assembles in the ECM to form microfibrils (6). Collagen type VI has been reported to bind to a wide range of molecules including other collagens (types I, II, and IV), decorin, NG2, and integrins (α1β1 and α2β1) (2, 3, 4, 15, 26). Collagen type VI is found in the ECM of virtually all connective tissues including skin, bone, cartilage, nerves, cornea, and skeletal muscle (14, 15). The microfibrillar meshwork formed by collagen VI plays an important role in cell attachment. Indeed, mutations in collagen type VI genes have been linked to an inherited muscular dystrophy, Bethlem myopathy (11, 16, 23). The ubiquitous expression of collagen type VI makes this molecule an ideal ligand for bacterial adherence. Whether collagen type VI binding by S. capitis plays a role in pathogenesis remains to be determined. However, the finding that antibodies can be generated to inhibit the activity of SdrX also suggests that this molecule is a potential candidate for the development of antibody therapies against S. capitis infection.
Acknowledgments
We thank M. Hook for the kind gift of S. epidermidis strain K28. We gratefully acknowledge all the members of the Monoclonal Antibody and Protein Sciences Departments at Inhibitex Inc. for generating antibodies and for purification of recombinant proteins.
Editor: J. T. Barbieri
REFERENCES
- 1.Allignet, J., S. Aubert, K. G. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidanset, D. J., C. Guidry, L. C. Rosenberg, H. U. Choi, R. Timpl, and M. Hook. 1992. Binding of the proteoglycan decorin to collagen type VI. J. Biol. Chem. 267:5250-5256. [PubMed] [Google Scholar]
- 3.Bonaldo, P., V. Russo, F. Bucciotti, R. Doliana, and A. Colombatti. 1990. Structural and functional features of the alpha 3 chain indicate a bridging role for chicken collagen VI in connective tissues. Biochemistry 29:1245-1254. [DOI] [PubMed] [Google Scholar]
- 4.Burg, M. A., E. Tillet, R. Timpl, and W. B. Stallcup. 1996. Binding of the NG2 proteoglycan to type VI collagen and other extracellular matrix molecules. J. Biol. Chem. 271:26110-26116. [DOI] [PubMed] [Google Scholar]
- 5.Engel, J., H. Furthmayr, E. Odermatt, H. von der Mark, M. Aumailley, R. Fleischmajer, and R. Timpl. 1985. Structure and macromolecular organization of type VI collagen. Ann. N. Y. Acad. Sci. 460:25-37. [DOI] [PubMed] [Google Scholar]
- 6.Furthmayr, H., H. Wiedemann, R. Timpl, E. Odermatt, and J. Engel. 1983. Electron-microscopical approach to a structural model of intima collagen. Biochem. J. 211:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall, A. E., P. J. Domanski, P. R. Patel, J. H. Vernachio, P. J. Syribeys, E. L. Gorovits, M. A. Johnson, J. M. Ross, J. T. Hutchins, and J. M. Patti. 2003. Characterization of a protective monoclonal antibody recognizing Staphylococcus aureus MSCRAMM protein clumping factor A. Infect. Immun. 71:6864-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartford, O., L. O'Brien, K. Schofield, J. Wells, and T. J. Foster. 2001. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147:2545-2552. [DOI] [PubMed] [Google Scholar]
- 9.Hartford, O., P. Francois, P. Vaudaux, and T. J. Foster. 1997. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 25:1065-1076. [DOI] [PubMed] [Google Scholar]
- 10.Hell, W., H. G. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 11.Jobsis, G. J., H. Keizers, J. P. Vreijling, M. de Visser, M. C. Speer, R. A. Wolterman, F. Baas, and P. A. Bolhuis. 1996. Type VI collagen mutations in Bethlem myopathy, an autosomal dominant myopathy with contractures. Nat. Genet. 14:113-115. [DOI] [PubMed] [Google Scholar]
- 12.Josefsson, E., K. W. McCrea, D. Ni Eidhin, D. O'Connell, J. Cox, M. Hook, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144(Pt. 12):3387-3395. [DOI] [PubMed] [Google Scholar]
- 13.Josefsson, E., O. Hartford, L. O'Brien, J. M. Patti, and T. Foster. 2001. Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J. Infect. Dis. 184:1572-1580. [DOI] [PubMed] [Google Scholar]
- 14.Keene, D. R., E. Engvall, and R. W. Glanville. 1995. Ultrastructure of type VI collagen in human skin and cartilage suggests an anchoring function for this filamentous network. J. Cell Biol. 107:1995-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo, H.-J. C. L. Maslen, D. R. Keene, and R. W. Glanville. 1997. Type VI collagen anchors endothelial basement membranes by interacting with type IV collagen. J. Biol. Chem. 272:26522-26529. [DOI] [PubMed] [Google Scholar]
- 16.Lamande, S. R., J. F. Bateman, W. Hutchison, R. J. M. Gardner, S. P. Bower, E. Byrne, and H.-H. M. Dahl. 1998. Reduced collagen VI causes Bethlem myopathy: a heterozygous COL6A1 nonsense mutation results in mRNA decay and functional haploinsufficiency. Hum. Mol. Genet. 7:981-989. [DOI] [PubMed] [Google Scholar]
- 17.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 18.McCrea, K. W., O. Hartford, S. Davis, D. N. Eidhin, G. Lina, P. Speziale, T. J. Foster, and M. Hook. 2000. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146:1535-1546. [DOI] [PubMed] [Google Scholar]
- 19.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 20.McDevitt, D., P. Vaudaux, and T. J. Foster. 1992. Genetic evidence that bound coagulase of Staphylococcus aureus is not clumping factor. Infect. Immun. 60:1514-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 22.O'Connell, D. P., T. Nanavaty, D. McDevitt, S. Gurusiddappa, M. Hook, and T. Foster. 1998. The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J. Biol. Chem. 273:6821-6829. [DOI] [PubMed] [Google Scholar]
- 23.Pan, T.-C., R.-Z. Zhang, M. A. Pericak-Vance, R. Tandan, T. Fries, J. Stajich, K. Viles, J. M. Vance, M.-L. Chu, and M. C. Speer. 1998. Missense mutation in a von Willebrand factor type A domain of the α3(VI) collagen gene (COL6A3) in a family with Bethlem myopathy. Hum. Mol. Genet. 7:807-812. [DOI] [PubMed] [Google Scholar]
- 24.Patti, J. M., and M. Hook. 1994. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 6:752-758. [DOI] [PubMed] [Google Scholar]
- 25.Patti, J. M., B. A. Aleen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 45:585-617. [DOI] [PubMed] [Google Scholar]
- 26.Pfaff, M., M. Aumailley, U. Specks, J. Knolle, H. G. Zerwes, and R. Timpl. 1993. Integrin and Arg-Gly-Asp dependence of cell adhesion to the native and unfolded triple helix of collagen type VI. Exp. Cell Res. 206:167-176. [DOI] [PubMed] [Google Scholar]
- 27.Savolainen, K., L. Paulin, B. Westerlund-Wikström, T. J. Foster, T. K. Korhonen, and P. Kuusela. 2001. Expression of pls, a gene closely associated with the mecA gene of methicillin-resistant Staphylococcus aureus, prevents bacterial adhesion in vitro. Infect. Immun. 69:3013-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 30.Stoll, B. J., N. Hansen, A. A. Fanaroff, L. L.Wright, W. A. Carlo, R. A. Ehrenkranz, J. A. Lemons, E. F. Donovan, A. R. Stark, J. E. Tyson, W. Oh, C. R. Bauer, S. B. Korones, S. Shankaran, A. R. Laptook, D. K. Stevenson, L. A. Papile, and W. K. Poole. 2002. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics 110:285-291. [DOI] [PubMed] [Google Scholar]
- 31.Van Der Zwet, W. C., Y. J. Debets-Ossenkopp, E. Reinders, M. Kapi, P. H. M. Savelkoul, R. M. Van Elburg, K. Hiramatsu, and C. M. J. E. Vandenbroucke-Grauls. 2002. Nosocomial spread of a Staphylococcus capitis strain with heteroresistance to vancomycin in a neonatal intensive care unit. J. Clin. Microbiol. 40:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vernachio, J., A. S. Bayer, T. Le, Y.-L. Chai, B. Prater, A. Scnhneider, B. Ames, P. Syribeys, J. Robbins, and J. M. Patti. 2003. Anti-clumping factor A immunoglobulin reduces the duration of methicillin-resistant Staphylococcus aureus bacteremia in an experimental model of infective endocarditis. Antimicrob. Agents Chemother. 47:3400-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]