Abstract
Plasmids represent a powerful tool to rapidly introduce genes into bacteria and help them reach high expression levels. In vaccine development, with live vaccine vectors, this allows greater flexibility and the ability to induce larger antigen amounts through multiple gene copies. However, plasmid retention often requires antibiotic resistance markers, the presence of which has been discouraged in clinical applications by the Food and Drug Administration. Therefore, we developed a Listeria monocytogenes-Escherichia coli shuttle plasmid that is retained by complementation of d-alanine racemase-deficient mutant strains both in vitro and in vivo. Our technology potentially allows the production of antibiotic resistance marker-free DNA vaccines as well as bacterial vaccine vectors devoid of engineered antibiotic resistances. As a proof of concept, we applied the d-alanine racemase complementation system to our Listeria cancer vaccine platform. With a transplantable tumor model, we compared the efficacy of the new Listeria vector to that of an established vector containing a conventional plasmid carrying a tumor-specific antigen. Both vaccine vector systems resulted in long-term regression of established tumors, with no significant difference between them. Thus, the Listeria vaccine vector presented here potentially complies with Food and Drug Administration regulations and could be developed further for clinical use.
The revolution in genetic engineering over the last two decades has allowed the development of recombinant DNA vaccines and bacterial antigen delivery systems, opening up new horizons in immunization against infectious diseases and even cancer. Recombinant antigens can be presented to the immune system after immunization with DNA directly or with bacterial vectors expressing vaccine antigens. Both approaches have the advantages of potentially low per-dose costs, ease of distribution and storage, and rational vaccine design. However, the required molecular engineering techniques often rely on the use of antibiotic resistance markers for retention of the recombinant gene. For example, the production of plasmid-based DNA vaccines in Escherichia coli usually relies on kanamycin resistance, which ensures plasmid stability during culture in the presence of the antibiotic. Bacterial delivery systems carrying plasmids also contained antibiotic resistance markers for in vitro selection in addition to in vivo retention systems (7, 11-13, 26). Although the use of plasmids could sometimes be circumvented by integrating the recombinant gene into the bacterial genome by homologous recombination, this process is tedious and time-consuming, resulting in reduced vaccine flexibility.
A recent accelerated chromosomal integration technology developed for Listeria spp. relied again on the use of antibiotic markers (20). The Food and Drug Administration, however, discouraged the in vivo use of genes transferring antibiotic resistance in several communiqués because of the associated potential risk of environmental spread (9, 10). An exception has been made in the past for DNA vaccines, as kanamycin was considered essential for production. Particularly when using live bacterial vectors in a clinical setting, antibiotic resistance markers may represent an environmental problem, as the spread of multiantibiotic resistances in clinical sites is a well-documented issue (21, 23).
Therefore, we have developed an alternative bacterial vector system devoid of any plasmid-borne antibiotic resistance genes with potential applications for DNA vaccines as well. While using Listeria spp. as a proof of principle, the technology presented may be universally applicable to other bacteria. We employed a d-alanine racemase-deficient mutant strain complemented by the gene located on a plasmid carrying the vaccine antigen. d-Alanine is essential for bacterial growth, and its availability is controlled by d-alanine racemases, which are often conserved across different bacterial species (27). d-Alanine metabolism in Listeria spp. is controlled by two genes, dal and dat (29). We hypothesized that the Listeria dal gene could potentially also complement deficient E. coli. Listeria monocytogenes strain Lmdd contained deletions in both dal and dat, rendering it dependent on the addition of exogenous d-alanine or complementation by a plasmid-expressed dal gene (29). A similar d-alanine racemase-deficient E. coli strain, MB2159, has also been published (28). We introduced the Listeria dal gene under the control of Listeria promoter p60 into shuttle plasmid pGG55 (13), replacing both chloramphenicol acetyltransferase (CAT) genes. We used the new shuttle plasmid to complement d-alanine racemase-deficient E. coli MB2159 as well as Listeria monocytogenes Lmdd.
Since prior studies from our group had demonstrated the successful application of L. monocytogenes cells carrying pGG55 as a cancer vaccine in established tumors in mice (13), we also tested the new dal-based plasmid in a cervical cancer model.
From 80 to 95% of cervical tumors have been associated with human papillomavirus (HPV) infection, with HPV-16 present in >50% of the cases (5). HPV proteins E6 and E7 are constitutively expressed in infected cells and are sufficient for immortalization (14, 16). Therefore, we previously developed a potential cervical cancer vaccine based on E7 antigen delivered by an L. monocytogenes-based bacterial vector, L. monocytogenes LLO-E7 (13). Upon infection of antigen-presenting cells in vivo with the L. monocytogenes vaccine vector, the cancer antigen was secreted from the bacterial cell first into the phagolysosome and, upon escape of L. monocytogenes, also into the host cell cytosol, so that E7 was presented on major histocompatibility complex class I and class II complexes on the antigen-presenting cell surface. The antigen-specific CD8+ T-cell response then resulted in tumor regression. L. monocytogenes LLO-E7, however, contains shuttle plasmid pGG55, whose two CAT genes for in vitro selection in gram-negative (E. coli) and gram-positive (L. monocytogenes) bacteria may reduce the application of this vector in a clinical setting. Thus, we improved L. monocytogenes LLO-E7 with the dal complementation system in order to remove both CAT genes. As a measure of biological efficacy, we compared antitumor efficacy between the original and the new delivery system.
MATERIALS AND METHODS
All standard molecular methods were performed following published protocols (1). Electrocompetent E. coli MB2159 (a kind gift from Michael Benedik and Ulrich Strych, University of Houston) and L. monocytogenes Lmdd were prepared as described (1, 24) and stored in single-use aliquots at −70°C until electroporation with pTV3. Heat shock-competent E. coli TOP10 was obtained from Invitrogen for transformation with pCR2.1- and pETblue1-based constructs. Plasmid DNA was prepared with the Qiafilter Midi Kit (Qiagen, Valencia, Calif.) following the manufacturer's recommendations. Chemicals were purchased from Sigma (St. Louis, Mo.), restriction enzymes and ligase from New England Biolabs (Beverly, Mass.), and disposables from Fisher (Pittsburgh, Pa.) unless noted otherwise.
Construction of pTV3.
Plasmid pGG55 (13) was used as a basis to construct antibiotic resistance marker-free shuttle vector pTV3. The replication region for gram-positive bacteria in pGG55 was amplified by PCR (primer 1, 5′-GTC GAC GGT CAC CGG CGC CAC TAA CTC AAC GCT AGT AG-3′; primer 2, 5′-TTA ATT AAG CTA GCC AGC AAA GAA AAA CAA ACA CG-3′) to introduce additional restriction sites for EheI and NheI (bold type). The PCR product was ligated into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) following the manufacturer's protocols and the sequence was verified (Nucleic Acid Facility, University of Pennsylvania). The L. monocytogenes dal gene (primer 1, 5′-CCA TGG TGA CAG GCT GGC ATC-3′; primer 2, 5′-GCT AGC CTA ATG GAT GTA TTT TCT AGG-3′) and a minimal p60 promoter sequence (18) (primer 1. 5′-TTA ATT AAC AAA TAG TTG GTA TAG TCC-3′; primer 2, 5′-GAC GAT GCC AGC CTG TCA CCA TGG AAA ACT CCT CTC-3′) were isolated by PCR amplification from bacteria. The primers introduced a PacI site upstream of the p60 sequence, an NheI site downstream of the dal sequence (restriction sites in bold type), and an overlapping dal sequence downstream of the p60 promoter for subsequent fusion of p60 and dal by splice overlap extension-PCR.
The resulting p60-dal expression cassette was ligated into pCR2.1-TOPO and transformed into d-alanine racemase-negative strain MB2159. Upon complementation of MB2159, the expression cassette was also sequence verified. The p60-dal gene was removed from pCR2.1 by PacI and NheI digestion, and the gram-positive replication region was removed from pCR2.1 by PacI and EheI digestion. In order to remove both CAT genes from pGG55, the plasmid was digested with EheI and NheI. All digests were purified by agarose gel to isolate the sequences of interest (replication, p60-dal, LLO-E7-prfA) with the gel extraction kit (Qiagen) following the manufacturer's recommendations. The p60-dal cassette, the gram-positive replication region, and the remainder of pGG55 containing the E. coli origin of replication and the listeriolysin (LLO)-E7-prfA cassette were ligated in a three-way ligation at 4°C for 20 h with T4 DNA ligase (Roche). The resulting plasmid, pTV3, was electroporated into E. coli MB2159. After selection on Luria-Bertani broth (LB) plates, plasmid DNA was prepared from colonies, and complete ligation was verified by restriction digestion. L. monocytogenes Lmdd was electroporated with pTV3 DNA prepared from E. coli, and transformed cells were also selected on LB plates.
Preparation of DNA for real-time PCR.
Total L. monocytogenes DNA was prepared with the Masterpure total DNA kit (Epicentre). Briefly, L. monocytogenes was cultured for 24 h at 37°C and 250 rpm in 25 ml of LB. Bacterial cells were pelleted by centrifugation, resuspended in phosphate-buffered saline supplemented with 5 mg of lysozyme per ml and incubated for 20 min at 37°C. Subsequently DNA was isolated following the manufacturer's recommendations.
In order to obtain standard target DNA for real-time PCR, the LLO-E7 gene was PCR amplified from pGG55 (primer 1, 5′-ATG AAA AAA ATA ATG CTA GTT TTT ATT AC-3′; primer 2, 5′-GCG GCC GCT TAA TGA TGA TGA TGA TGA TGT GGT TTC TGA GAA CAG ATG-3′) and cloned into vector pETblue1 (Novagen). Similarly, the plcA amplicon (primers as used for real-time PCR, sequence listed below) was cloned into pCR2.1. E. coli was transformed with pET-LLO-E7 and pCR-plcA, and purified plasmid DNA was prepared for use in real-time PCR.
Bacterial culture and in vivo passaging of L. monocytogenes.
E. coli was cultured following standard methods as described in the literature (1). Listeriae were grown at 37°C and 250 rpm in LB supplemented with 50 μg of streptomycin per ml and harvested during their exponential phase of growth. L. monocytogenes LLO-E7 was cultured in the presence of an additional 37 μg/ml of chloramphenicol. In order to determine growth kinetics, bacteria were grown for 16 h in 10 ml of LB plus antibiotics. The OD600 was measured and adjusted to be similar for the different strains. The culture was then diluted 1:50 into fresh LB medium supplemented with suitable antibiotics and d-alanine if applicable, and the OD600 was measured every 30 min. L. monocytogenes was passaged in C57BL/6 mice as described previously (25). Briefly, 108 CFU were injected intraperitoneally. On day 3, spleens were isolated and homogenized in phosphate-buffered saline. An aliquot of the spleen suspension was plated on LB plates supplemented with antibiotics as applicable. Several colonies were expanded and mixed to establish an injection stock of Lmdd(pTV3).
Plasmid stability testing.
Plasmid maintenance in vitro was determined by serial passage under selective and nonselective conditions. Bacteria were cultured in 50 ml of LB and transferred daily at a 1:1,000 dilution into fresh medium. L. monocytogenes LLO-E7 was cultured in the presence and the absence of 34 μg of chloramphenicol per ml, and Lmdd(pTV3) was cultured with or without supplemental d-alanine at a concentration of 100 μg/ml. The CFU for each culture were determined daily on BHI plates and on BHI plus 34 μg of chloramphenicol per ml for L. monocytogenes LLO-E7 and BHI plus 100 μg of d-alanine per ml for Lmdd(pTV3). All liquid medium and plates contained an additional 50 μg of streptomycin per ml to select for listeriae and reduce potential growth of contaminants.
Plasmid maintenance in vivo was determined by intraperitoneal injection of L. monocytogenes LLO-E7, Lmdd(pTV3), or wild-type 10403S in C57BL/6 mice. Viable bacteria were isolated from spleens homogenized in phosphate-buffered saline at 16, 24, 48, 72, and 120 h. CFU for each sample were determined at each time point on BHI plates and on BHI plus 34 μg of chloramphenicol per ml for L. monocytogenes LLO-E7 and BHI plus 100 μg of d-alanine per ml for Lmdd(pTV3).
Real-time PCR.
Taqman primer-probe sets (Applied Biosystems) were designed with the ABI PrimerExpress software (Applied Biosystems) with E7 as a plasmid target (primer 1, 5′-GCA AGT GTG ACT CTA CGC TTC G-3′; primer 2, 5′-TGC CCA TTA ACA GGT CTT CCA-3′; probe, 5′-FAM [6-carboxyfluorescin]-TGC GTA CAA AGC ACA CAC GTA GAC ATT CGT AC-TAMRA [6-carboxytetramethylrhodamine]-3′) and the one-copy gene plcA (primer 1, TGA CAT CGT TTG TGT TTG AGC TAG-3′; primer 2, 5′-GCA GCG CTC TCT ATA CCA GGT AC-3′; probe, 5′-TET [tetrachlorofluorescin]-TTA ATG TCC ATG TTA TGT CTC CGT TAT AGC TCA TCG TA-TAMRA-3′) as a listerial genome target. Although the probes were differently labeled (E7 probe with FAM, plcA probe with TET), real-time PCR was not performed in a multiplex format.
For PCR, 0.4 μM primer and 0.05 mM probe were mixed with Pure Taq RTG PCR beads (Amersham) as recommended by the manufacturer. Standard curves were prepared for each target with purified plasmid DNA, pET-LLO-E7, and pCR-plcA and used to calculate gene copy numbers in unknown samples. Mean ratios of E7 copies to plcA copies were calculated based on the standard curves and calibrated by dividing the results for Lmdd(pTV3) and L. monocytogenes LLO-E7 with the results from L. monocytogenes E7, a strain with a single copy of the E7 gene integrated into the genome. All samples were run in triplicate in each quantitative PCR assay, which was repeated three times. Variation between samples was analyzed by two-way analysis of variance with the KyPlot software. Results were deemed statistically significant at P < 0.05.
Tumor regression.
Tumor regression experiments were performed as described (13) with the TC-1 cervical cancer model. TC1 is a C57BL/6 epithelial cell line that was immortalized with HPV E6 and E7 and transformed with activated Ras, forming tumors upon subcutaneous implantation (22). Briefly, 105 TC-1 cells were implanted subcutaneously into syngeneic C57BL/6 mice on day 0 and allowed to reach a measurable size of 4 to 5 mm in diameter. On days 7 and 14, mice were injected intraperitoneally with 1/10th the 50% lethal dose (LD50) of the Lmdd(pTV3) or L. monocytogenes LLO-E7 vector. The naïve group did not receive any treatment, and each group consisted of eight mice. Tumor diameters were measured twice weekly with an electronic caliper.
T-cell analysis.
T cells from spleen and tumor-infiltrating T cells were analyzed for CD8 and CD4 surface markers and E7 specificity as described (13). Briefly, C57BL/6 mice were immunized intraperitoneally as described above with Lmdd(pTV3) or L. monocytogenes LLO-E7. Splenocytes and tumors were harvested 5 days after the boost and stained at room temperature with H-2Db tetramers loaded with the E7 peptide (RAHYNIVTF) or a control (human immunodeficiency virus Gag) peptide. Tetramers were provided by the National Institute of Allergy and Infectious Diseases Tetramer Core Facility and the National Institutes of Health AIDS Research and Reference Reagent Program.
Three-color flow cytometry for CD8+ (53-6.7, phycoerythrin conjugated), CD62− E7 H-2Db tetramer+ was performed with a FACSCalibur flow cytometer with CellQuest software (Becton Dickinson, Mountain View, Calif.). Intracellular gamma interferon (IFN-γ) staining was performed as described previously (13, 19) on a second subset of cells. Before staining for the cell surface antigens and IFN-γ production, lymphocytes were stimulated in vitro by culturing in the presence of monensin (BD Golgi Stop reagent; BD Biosciences) as recommended by the manufacturer to accumulate intracellular IFN-γ in the Golgi apparatus. After culture for 5 h in RP-10 supplemented with interleukin-2 (50 U/ml) and 1 μl of brefeldin A (monensin) per ml, the cells were surface stained for effector markers at 4°C for 20 min with phycoerythrin-conjugated anti-CD8 (PharMingen) and antigen-presenting cell-conjugated MEL-14 (anti-CD62 ligand). Cells were gated on CD62 ligand low to select activated cells before being analyzed for CD8+ IFN-γ+ populations.
RESULTS
Construction of antibiotic resistance factor-free plasmid pTV3.
The recombinant shuttle plasmid had to be maintained in three different host environments: in E. coli during cloning manipulations in vitro, in L. monocytogenes in vitro during cloning and during the production of immunization material, and in L. monocytogenes in vivo after immunization. We chose an auxotroph complementation system based on d-alanine racemase to ensure plasmid retention in all three scenarios. The E. coli strain is an alr dadX mutant that is not able to synthesize d-alanine racemase (27). The L. monocytogenes dal dat mutant strain Lmdd (29) similarly is not able to synthesize d-alanine racemase due to partial deletions of the dal and dat genes. Both MB2159 and Lmdd required exogenous d-alanine for growth, but d-alanine racemase functions could be restored by expressing the alr gene (E. coli) (23) or the dal gene (listeriae) from a plasmid (27, 29).
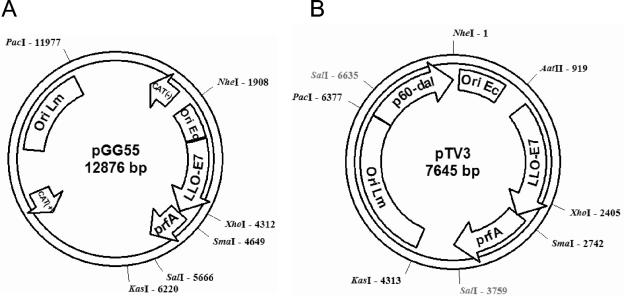
We used the Listeria dal gene under the control of the Listeria p60 promoter to construct a shuttle plasmid that was retained in both bacteria. Plasmid pGG55 (13), which is based on E. coli-Listeria shuttle vector pAM401 (32), was modified by removing both CAT genes and initially also the Listeria replication region by restriction with NheI and EheI. A PCR-amplified Listeria replication gene and the p60-dal expression cassette were then ligated with the remaining E. coli p15 origin of replication and the cancer antigen expression cassette consisting of the hly promoter, LLO-E7 fusion gene, and Listeria prfA gene (Fig. 1). The resulting plasmid, pTV3, was stably maintained in E. coli strain MB2159 as well as in L. monocytogenes strain Lmdd. Bacterial growth on LB medium that was not supplemented with additional d-alanine indicated that the dal expression cassette derived from the Listeria dal gene under control of the Listeria p60 promoter was also active in gram-negative E. coli. Both E. coli and strain Lmdd transformed with pTV3 remained sensitive to chloramphenicol, indicating successful removal of both CAT genes from the plasmid (data not shown).
FIG. 1.
Schematic map of E. coli-Listeria monocytogenes shuttle plasmids pGG55 (A) and pTV3 (B). CAT(−), chloramphenicol acetyltransferase for E. coli; CAT(+), chloramphenicol acetyltransferase for Listeria monocytogenes; Ori Lm, replication region for Listeria monocytogenes; Ori Ec, p15 origin of replication for Escherichia coli; prfA, Listeria monocytogenes pathogenicity-regulating factor A; LLO-E7, fusion between the gene encoding a C-terminally truncated listeriolysin O, including its promoter, and the gene encoding HPV E7; p60-dal, expression cassette of the p60 promoter and Listeria monocytogenes dal gene. Selected restriction sites of interest are depicted.
As the p15 origin of replication of the original shuttle plasmid pAM401 was not manipulated during cloning, we did not expect the plasmid copy number to change in E. coli. Similarly, we did not expect the copy number of pTV3 to be different in L. monocytogenes compared to that of parent plasmid pGG55, since the initially deleted replication region was added back in and the permanently deleted copy control region had already been inactive in pGG55. However, because of the extent of the changes to the plasmid, the copy number per listerial cell was compared between L. monocytogenes LLO-E7 and Lmdd(pTV3) by real-time PCR with E7 on the plasmid as a target and plcA in the genome. Real-time PCR results were calibrated by using L. monocytogenes E7 (13), which contains a single-copy genomic integration of the E7 gene. We found no statistically significant difference (P < 0.05) in plasmid copy numbers between Lmdd(pTV3) and L. monocytogenes LLO-E7 grown in vitro in the absence [Lmdd(pTV3)] or the presence (L. monocytogenes LLO-E7) of chloramphenicol, indicating stable plasmid retention in both strains (data not shown).
We also tested antigen expression in vitro by Western blot. When analyzing equal amounts of total protein from bacterial culture supernatants, Lmdd(pTV3) cultures contained approximately twice as much total antigen as L. monocytogenes LLO-E7 cultures (data not shown). This difference may be a result of a higher overall metabolic load in Lmdd-LLO-E7 impaired by the larger size of the plasmid (12.8 kb) compared to Lmdd(pTV3) (7.6 kb).
Shuttle vector pTV3 complements deficient strain Lmdd in vitro and in vivo.
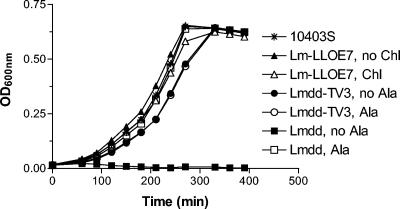
In order to verify the complementation of bacterial functions, we compared in vitro growth kinetics between Lmdd(pTV3) and L. monocytogenes LLO-E7. Lmdd(pTV3) was able to grow in alanine-free medium, whereas noncomplemented Lmdd required the addition of d-alanine (Fig. 2). Additional exogenous d-alanine did not enhance in vitro growth of Lmdd(pTV3) further. Thus, the d-alanine gene located on the plasmid fully complemented in vitro growth of strain Lmdd. Some minor growth delay could be observed between strain Lmdd complemented with d-alanine and plasmid-complemented Lmdd(pTV3). This may be due to the metabolic load imposed by the need to synthesize the plasmid.
FIG. 2.
Growth kinetics of Listeria monocytogenes strains LLO-E7, Lmdd(pTV3), and Lmdd. All strains were cultured for 16 h, and subsequently the culture OD600 was adjusted to be similar for all strains. The cultures were diluted 1:50 in fresh medium and incubated at 37°C and 250 rpm. The OD600 was measured every 30 min. +Ala, additional d-alanine was added to the culture at 100 μg/ml final concentration; +Chl, additional chloramphenicol was added to the culture at 34 μg/ml. The experiment was repeated with similar results (data not shown).
Although in vitro assays help to determine individual bacterial functions, we found that these assays do not always predict in vivo virulence as determined by LD50 values when comparing a number of bacterial strains from our group. Therefore, we based our comparison of strain virulence on LD50 values, which were found to be similar between the two vectors Lmdd(pTV3 (0.75 × 109) and L. monocytogenes LLO-E7 (109).
Recombinant plasmid is maintained in vitro and in vivo.
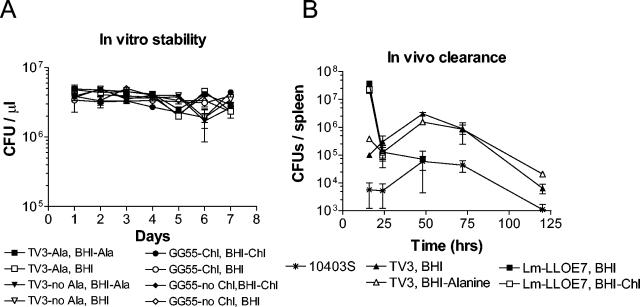
To determine plasmid stability, L. monocytogenes LLO-E7 and Lmdd(pTV3) were cultured for 70 generations in vitro in the presence and absence of selective pressure. CFU were determined daily on selective and nonselective plates for each culture. Upon plasmid loss, a greater number of colonies were expected on nonselective plates, BHI plus d-alanine for Lmdd(pTV3), BHI only for L. monocytogenes LLO-E7, versus selective plates, BHI only for Lmdd(pTV3), BHI plus chloramphenicol for L. monocytogenes LLO-E7. No difference in CFU was detected between nonselective and selective plates, indicating stable maintenance of the plasmid throughout the culture for at least 70 generations, when the experiment was terminated.
In addition, we tested plasmid stability in vivo in C57BL/6 mice by isolating viable bacteria at different time points after injection. Again, CFU counts on selective and nonselective plates were used to determine plasmid maintenance among the isolated bacteria (Fig. 3). No differences in CFU were detected on selective and nonselective plates for each construct, indicating the stable presence of the recombinant plasmid in all bacteria isolated. Since viable Lmdd(pTV3) bacteria were isolated at least until day 5, plasmid loss in vivo followed by early clearance of injected bacteria can be excluded as a possible reason for the low virulence as observed by LD50 values. L. monocytogenes LLO-E7 was cleared very rapidly in vivo within 48 to 72 h. Despite having an LD50 value similar to that of Lmdd(pTV3), L. monocytogenes LLO-E7 may be more attenuated in other virulence factors.
FIG. 3.
Plasmid maintenance in vitro (A) and in vivo (B). To determine in vitro stability, strains were cultured with (GG55-Chl) and without (GG55-no Chl) chloramphenicol (L. monocytogenes LLO-E7) or with and without d-alanine [Lmdd(pTV3)]. The cultures were diluted 1:1,000 daily into fresh LB. The CFU of the cultures were determined daily on BHI (BHI) and on BHI with chloramphenicol (BHI-Chl) for L. monocytogenes LLO-E7 or on BHI with d-alanine (BHI-Ala) for Lmdd(pTV3). All liquid medium and plates contained an additional 50 μg of streptomycin per ml, to which Listeria monocytogenes strain 10403S is naturally resistant. To determine in vivo plasmid maintenance, L. monocytogenes was injected intraperitoneally at a dose of 1/10 the LD50 in C57BL/6 mice. Spleens were harvested at different time points postinjection and homogenized in phosphate-buffered saline. CFU counts were prepared on BHI plates with and without d-alanine for Lmdd(pTV3), on BHI plates with and without chloramphenicol for L. monocytogenes LLO-E7, and on BHI plates only for wild-type 10403S.
Lmdd(pTV3) regresses established tumors.
The efficacy of both vectors to serve as potential cervical cancer vaccines was compared in a tumor regression model. Since we were using the HPV E7 antigen and direct comparison between the two vectors required similar tumor sizes between the different groups, the cotton-tail rabbit papillomavirus model with virally induced tumors and a cotton-tail rabbit papillomavirus target was not suitable. We instead chose the TC-1 cell line model, which is well characterized for HPV vaccine development and which allowed controlled comparison of the regression of established tumors of similar size after immunization with Lmdd(pTV3) or L. monocytogenes LLO-E7.
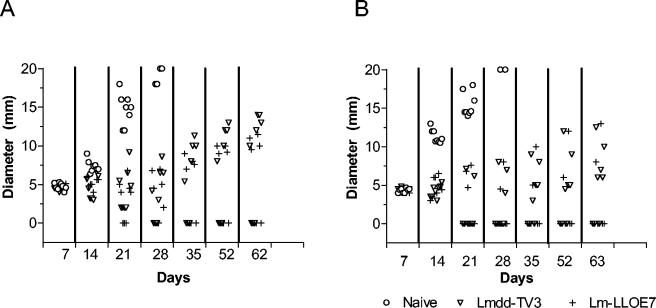
In two separate experiments, immunization of mice with Lmdd(pTV3) and L. monocytogenes LLO-E7 resulted in similar tumor regression (Fig. 4), with no statistically significant difference (P < 0.05) between the vaccinated groups. All immunized mice were still alive after 63 days, whereas mice in the naïve group had to be sacrificed when their tumors reached 20 mm in diameter. Cured mice remained tumor free until the termination of the experiment. Previously observed in vitro differences between Lmdd(pTV3) and L. monocytogenes LLO-E7 (see above) were not reflected in vaccine efficacy in vivo.
FIG. 4.
Tumor regression. We injected 105 TC-1 tumor cells subcutaneously on day 0 and allowed them to form a tumor for 7 days until approximately 4 to 5 mm in diameter. Listeriae were injected intraperitoneally on days 7 and 14 at a dose of 1/10th the LD50 and tumor diameters were measured twice weekly. Results are depicted as tumor size in individual mice. In experiment A, three of eight mice for Lmdd(pTV3) and four of eight mice for L. monocytogenes LLO-E7 were tumor free at day 62. In experiment B, four of eight mice for L. monocytogenes Lmdd(pTV3) and five of eight mice for L. monocytogenes LLO-E7 were tumor free at day 62. When the tumors reached 20 mm in diameter in the untreated (naïve) group, mice were euthanized in accordance with Institutional Animal Care and Use Committee regulations.
Immunization with the listerial vectors results in antigen-specific tumor-infiltrating T cells.
We have shown previously that antitumor efficacy is closely linked to the ability of the vector to induce antigen-specific tumor-infiltrating lymphocytes (19). To further characterize Lmdd(pTV3) efficacy, we therefore analyzed tumor-infiltrating cytotoxic T cells for E7 antigen specificity (Table 1). The percentage of E7 tetramer-specific T cells infiltrating the tumor was found to be somewhat higher in the Lmdd-LLO-E7 group than in the Lmdd(pTV3) group. Tetramer-positive T cells were functionally characterized by IFN-γ production assays. No significant differences were observed in the percentages of IFN-γ-producing CD8+ T cells in L. monocytogenes LLO-E7-immunized mice versus Lmdd(pTV3)-treated mice. Since both vectors result in similar long-term tumor regression, the number of tumor-infiltrating, antigen-specific T cells in the tumor seems to be sufficient to control tumor growth upon vaccination with either vector.
TABLE 1.
Flow cytometry analysis of tumor-infiltrating T cells
| Mice (n = 3) | Dose (CFU) | % of cells
|
|||
|---|---|---|---|---|---|
| Expt A-
|
Expt B-
|
||||
| CD8+ E7+ | CD8+ IFNγ+ | CD8+ E7+ | CD8+ IFNγ+ | ||
| Naïve | 0 | 8.81 | 1.33 | 4.86 | 0.01 |
| Lmdd (pTV3) | 0.75 × 108 | 20.72 | 7.06 | 14.86 | 5.5 |
| LLO-E7 | 1 × 108 | 27.43 | 5.55 | 20.82 | 7.93 |
DISCUSSION
We have established a potential Listeria-based vaccine delivery system that is free of antibiotic resistance markers to conform to clinical regulatory requirements and improve the environmental safety of live bacterial vectors as well as DNA vaccines. A Listeria dal gene expressed from a plasmid under the control of a constitutive promoter successfully complemented a d-alanine racemase-deficient gram-positive (Listeria monocytogenes) and a gram-negative (Escherichia coli) host. We used this vector to express a cancer antigen fused to Listeria listeriolysin (LLO) as a proof of concept, but our expression cassette could easily be replaced by a multiple cloning site. In order to create a general shuttle vector, pTV3 could be digested with KasI or EheI and AatII, removing the prfA gene, the LLO-E7 fusion gene, and most of the LLO promoter. A multiple cloning site consisting of BamHI, XhoI, XbaI, NotI, SpeI, SmaI, and SacI could then be introduced by ligating the following paired oligonucleotides to the vector backbone: 5′-CGG ATC CCT CGA GCT CAG AGC GGC CGC ACT AGT CCC GGG GAG CTC G 5′-TCG ACG AGC TCC CCG GGA CTA GTG CGG CCG CTC TGA GCT CGA GGG ATC CGA CGT (overhanging ends that are compatible with the vector sites restricted with AatI and SalI are in italics).
Alternatively, the p60-dal expression cassette could be introduced into any commercial plasmid, followed by removal of the antibiotic genes. We have successfully used pCR2.1 (Invitrogen) containing the p60-dal cassette and Listeria replication sequences to complement d-alanine racemase-deficient E. coli strain MB2159 as well as d-alanine racemase-deficient L. monocytogenes strain Lmdd (unpublished data). In order to compare the efficacy of our new vector, Lmdd(pTV3), with the previous version, L. monocytogenes LLO-E7, which served as a gold standard for our evaluation, we characterized both strains in vivo and in vitro. First, plasmid copy numbers between pTV3 in Lmdd(pTV3) and pGG55 in L. monocytogenes LLO-E7 were compared. Although both plasmids contained the same replication region, deletion of more than 50% of the original “unnecessary” sequences could potentially have affected intracellular copy numbers. A specific concern was the copy control region in pGG55 (2, 3, 32), which had already been inactivated previously but was still present in the majority of its sequence. As expected, the evaluation of plasmid E7 copies versus genomic plcA by quantitative PCR showed no significant difference between pTV3 and pGG55 copy numbers in L. monocytogenes.
Next, we compared plasmid maintenance in both vectors in vitro and in vivo. Comparing CFU counts on selective and nonselective plates of samples from serial culture of Lmdd(pTV3) with L. monocytogenes LLO-E7, we found no significant difference. If the plasmid had been lost throughout the culture, we would have expected a higher number of CFU on nonselective plates than under selective conditions. Similar results were obtained in comparing the bacterial populations isolated from the spleens of mice infected with the different vectors. Thus, the lethal-balanced plasmid maintenance system based on d-alanine racemase complementation is stable in vitro as well as in vivo. The new vector displayed less attenuation than L. monocytogenes LLO-E7 in vivo, i.e., Lmdd(pTV3) was cleared more slowly. However, we concluded, based on the LD50 values, that the virulence of Lmdd(pTV3) and L. monocytogenes LLO-E7 is not substantially different.
As for any vaccine, in vivo efficacy was the most important aspect of our antibiotic resistance-free vector system. We performed side-by-side tumor regression studies of Lmdd(pTV3) and L. monocytogenes LLO-E7 and found no significant difference in the ability of these vaccines to cause the regression of established tumors in mice. All of the vaccinated mice were alive after more than 60 days, and 40 to 65% of the tumors had disappeared permanently. Analysis of the percentage of E7 tetramer-positive CD8+ T cells in the spleens showed no significant differences between the two vaccines (data not shown). Interestingly, a higher percentage of E7-specific T cells, as measured by tetramer staining, were found in the tumors of L. monocytogenes LLO-E7-immunized mice than in the Lmdd(pTV3) group, although the absolute numbers of E7-specific cells were comparable. In addition, equivalent E7-specific percentages of T cells isolated from tumors from mice treated with either vaccine secreted IFN-γ. Thus, overall, we did not find significant differences in the efficacy of the antibiotic resistance-free system in its ability to serve as a cancer vaccine.
Several systems have been published to develop alternatives to antibiotic resistance markers in gram-positive and gram-negative bacteria, such as active plasmid segregation (12), postsegregational killing (12), repressor titration (6, 31), and the complementation of auxotroph mutants (4, 26). Often, however, these technologies have been developed for only one bacterial species or required the combination of several systems to allow selection in E. coli in vitro and the final host in vitro and in vivo (12), the latter being essential for the efficacy of bacterial vaccine delivery vectors. Such combinations may result in enhanced metabolic load of the bacterial vector. In other examples, antibiotic resistance genes were retained for selection in E. coli (8, 15, 17).
The complementation system presented has a number of advantages and applications. The new plasmid contains only a single expression cassette for selection in E. coli and L. monocytogenes in vitro and in vivo, thereby simplifying the vaccine vector system. We were able to significantly reduce the size of the expression plasmid from 12.8 kb to only 7.6 kb, resulting in less metabolic load for the bacterial vector and easier plasmid manipulation. Although the recombinant plasmid is highly stable in its d-alanine racemase-deficient host, which cannot survive upon plasmid loss, it is not likely to be retained upon potential transfer to other bacteria in the gut. It does not confer an evolutionary advantage on normal cells and does not contain active retention systems such as partition sequences. Thus, outside its deficient host cell, the dal-containing plasmid will most likely be diluted out of the population and ultimately be eliminated over time.
Independent of its use in our Listeria-based vaccine vector, the d-alanine racemase complementation system that we developed could be used to eliminate the requirement of antibiotic resistance markers in the manufacture of DNA vaccines in E. coli. The Listeria-derived dal expression cassette or an E. coli-derived alr gene for that matter could be included in DNA vaccine plasmids instead of kanamycin when using a d-alanine racemase-deficient E. coli strain for vaccine production. This system could be used for plasmids of any copy number without being restricted to high-copy-number plasmids, as is the alternative repressor titration system. There will be no introduction of potentially toxic amounts of d-alanine, which was a concern for the in vivo complementation of strain Lmdd with exogenous d-alanine.
As d-alanine racemases are homologous across many different bacterial species, the complementation system described here could potentially be applied universally in other vector settings with only slight modifications. Bacteria that contain d-alanine racemases similar to those in L. monocytogenes or E. coli include Mycobacterium spp. (28), Bacillus spp. (29), Lactobacillus spp. (4), Shigella spp. (33), and Salmonella spp. (30). We demonstrated the feasibility and efficacy of this approach in animal models of cervical cancer as a proof of concept, but other antigens from other cancer types or from infectious diseases could be targeted. We will also move the prototype cervical cancer vaccine presented here towards its clinical application in the near future. Our antibiotic resistance-marker free Listeria vaccine system with its implications for the antibiotic gene-free production of DNA vaccines represents a powerful tool for the development in clinical application of recombinant vaccine vectors.
Acknowledgments
We thank Micheal Bendik and Ulrich Strych for providing E. coli strain MB2159, Fred Frankel for providing Listeria strain Lmdd, and Toby Hecht for helpful discussions.
This work was supported by grant CA69632 and RAID grant NSC 715814 from the National Institutes of Health.
Yvonne Paterson and Thorsten Verch wish to disclose that they each have a financial interest in Advaxis Inc., a vaccine and therapeutic company that has licensed or has an option to license all patents from the University of Pennsylvania that concern the use of L. monocytogenes as a vaccine vector.
Editor: F. C. Fang
REFERENCES
- 1.Ausubel, K., R. Brent, and R. E. Kingston. 1997. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 2.Behnke, D., M. S. Gilmore, and J. J. Ferretti. 1981. Plasmid pGB301, a new multiple resistance streptococcal cloning vehicle and its use in cloning of a gentamicin/kanamycin resistance determinant. Mol. Gen. Genet. 182:414-421. [DOI] [PubMed] [Google Scholar]
- 3.Brantl, S., D. Behnke, and J. C. Alonso. 1990. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAM beta 1 and pSM19035. Nucleic Acids Res. 18:4783-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bron, P. A., M. G. Benchimol, J. Lambert, E. Palumbo, M. Deghorain, J. Delcour, W. M. De Vos, M. Kleerebezem, and P. Hols. 2002. Use of the alr gene as a food-grade selection marker in lactic acid bacteria. Appl. Environ. Microbiol. 68:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burd, E. M. 2003. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cranenburgh, R. M., J. A. Hanak, S. G. Williams, and D. J. Sherratt. 2001. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 29:E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunstan, S. J., C. P. Simmons, and R. A. Strugnell. 2003. In vitro and in vivo stability of recombinant plasmids in a vaccine strain of Salmonella enterica var. Typhimurium. FEMS Immunol. Med. Microbiol. 37:111-119. [DOI] [PubMed] [Google Scholar]
- 8.Fennelly, G. J., S. A. Khan, M. A. Abadi, T. F. Wild, and B. R. Bloom. 1999. Mucosal DNA vaccine immunization against measles with a highly attenuated Shigella flexneri vector. J. Immunol. 162:1603-1610. [PubMed] [Google Scholar]
- 9. Food and Drug Administration. 1998. Guidance for human somatic cell therapy and gene therapy. Food and Drug Administration, Washington, D.C.
- 10. Food and Drug Administration. 1996. Points to consider on plasmid DNA vaccines for preventive infectious disease indications. Food and Drug Administration, Washington, D.C.
- 11.Galen, J. E., and M. M. Levine. 2001. Can a ′flawless' live vector vaccine strain be engineered? Trends Microbiol. 9:372-376. [DOI] [PubMed] [Google Scholar]
- 12.Galen, J. E., J. Nair, J. Y. Wang, S. S. Wasserman, M. K. Tanner, M. B. Sztein, and M. M. Levine. 1999. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 67:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunn, G. R., A. Zubair, C. Peters, Z. K. Pan, T. C. Wu, and Y. Paterson. 2001. Two Listeria monocytogenes vaccine vectors that express different molecular forms of human papilloma virus-16 (HPV-16) E7 induce qualitatively different T-cell immunity that correlates with their ability to induce regression of established tumors immortalized by HPV-16. J. Immunol. 167:6471-6479. [DOI] [PubMed] [Google Scholar]
- 14.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1992. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 66:2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jabbar, I. A., G. J. Fernando, N. Saunders, A. Aldovini, R. Young, K. Malcolm, and I. H. Frazer. 2000. Immune responses induced by BCG recombinant for human papillomavirus L1 and E7 proteins. Vaccine 18:2444-2453. [DOI] [PubMed] [Google Scholar]
- 16.Kanda, T., S. Watanabe, and K. Yoshiike. 1988. Immortalization of primary rat cells by human papillomavirus type 16 subgenomic DNA fragments controlled by the SV40 promoter. Virology 165:321-325. [DOI] [PubMed] [Google Scholar]
- 17.Kawahara, M., K. Matsuo, T. Nakasone, T. Hiroi, H. Kiyono, S. Matsumoto, T. Yamada, N. Yamamoto, and M. Honda. 2002. Combined intrarectal/intradermal inoculation of recombinant Mycobacterium bovis bacillus Calmette-Guerin (BCG) induces enhanced immune responses against the inserted HIV-1 V3 antigen. Vaccine 21:158-166. [DOI] [PubMed] [Google Scholar]
- 18.Kohler, S., A. Bubert, M. Vogel, and W. Goebel. 1991. Expression of the iap gene coding for protein p60 of Listeria monocytogenes is controlled on the posttranscriptional level. J. Bacteriol. 173:4668-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamikanra, A., Z. K. Pan, S. N. Isaacs, T. C. Wu, and Y. Paterson. 2001. Regression of established human papillomavirus type 16 (HPV-16)-immortalized tumors in vivo by vaccinia viruses expressing different forms of HPV-16 E7 correlates with enhanced CD8+-T-cell responses that home to the tumor site. J. Virol. 75:9654-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaitre, J. P., H. Echchannaoui, G. Michaut, C. Divies, and A. Rousset. 1998. Plasmid-mediated resistance to antimicrobial agents among listeriae. J. Food Prot. 61:1459-1464. [DOI] [PubMed] [Google Scholar]
- 22.Lin, K. Y., F. G. Guarnieri, K. F. Staveley-O'Carroll, H. I. Levitsky, J. T. August, D. M. Pardoll, and T. C. Wu. 1996. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 56:21-26. [PubMed] [Google Scholar]
- 23.National Antimicrobial Resistance Monitoring System. 2001. Annual report. Centers for Disease Control, Atlanta, Ga.
- 24.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 25.Peters, C., and Y. Paterson. 2003. Enhancing the immunogenicity of bioengineered Listeria monocytogenes by passaging through live animal hosts. Vaccine 21:1187-1194. [DOI] [PubMed] [Google Scholar]
- 26.Pilgrim, S., J. Stritzker, C. Schoen, A. Kolb-Maurer, G. Geginat, M. J. Loessner, I. Gentschev, and W. Goebel. 2003. Bactofection of mammalian cells by Listeria monocytogenes: improvement and mechanism of DNA delivery. Gene Ther. 10:2036-2045. [DOI] [PubMed] [Google Scholar]
- 27.Strych, U., and M. J. Benedik. 2002. Mutant analysis shows that alanine racemases from Pseudomonas aeruginosa and Escherichia coli are dimeric. J. Bacteriol. 184:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strych, U., R. L. Penland, M. Jimenez, K. L. Krause, and M. J. Benedik. 2001. Characterization of the alanine racemases from two mycobacteria. FEMS Microbiol. Lett. 196:93-98. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, R. J., H. G. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires d-alanine for growth. Infect. Immun. 66:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasserman, S. A., C. T. Walsh, and D. Botstein. 1983. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 153:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, S. G., R. M. Cranenburgh, A. M. Weiss, C. J. Wrighton, D. J. Sherratt, and J. A. Hanak. 1998. Repressor titration: a novel system for selection and stable maintenance of recombinant plasmids. Nucleic Acids Res. 26:2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoigawa, K., R. Hirasawa, H. Ueno, Y. Okubo, S. Umesako, and K. Soda. 2001. Gene cloning and characterization of alanine racemases from Shigella dysenteriae, Shigella boydii, Shigella flexneri, and Shigella sonnei. Biochem. Biophys. Res. Commun. 288:676-684. [DOI] [PubMed] [Google Scholar]