Abstract
Information from comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis bacillus Calmette-Guérin (BCG) principally allows prediction of potential vaccine candidates. Thirty-six M. tuberculosis DNA vaccine candidates identified by comparative proteome analysis were evaluated in the mouse model for protection against low-dose aerosol M. tuberculosis infection. We identified the DNA vaccine candidate Rv3407 as a protective antigen and analyzed putative major histocompatibility complex class I epitopes by computational predictions and gamma interferon Elispot assays. Importantly, we discovered that the DNA vaccine Rv3407 improved the efficacy of BCG vaccination in a heterologous prime-boost vaccination protocol. Our data demonstrate the rationale of a combination of proteomics, epitope prediction, and broad screening of putative antigens for identification of novel DNA vaccine candidates. Furthermore, our experiments show that heterologous prime-boost vaccination with a defined antigen boost “on top” of a BCG primer provides superior protection against tuberculosis over vaccination with BCG alone.
Together with AIDS and malaria, tuberculosis remains a major health threat, particularly in developing countries (50). Frequently, chemotherapy with at least three drugs over a period of several months is hampered by insufficient compliance, resulting in dramatically increasing incidences of multidrug-resistant strains of Mycobacterium tuberculosis (9). The currently available vaccine, Mycobacterium bovis bacillus Calmette-Guérin (BCG), is the most widely used viable vaccine worldwide (2, 11, 12, 43). BCG apparently fails to protect against the most common form of the disease, pulmonary tuberculosis in adults, but it prevents miliary tuberculosis in newborns and toddlers (11, 25). This raises two issues: first, novel tuberculosis vaccine candidates, which protect against adulthood tuberculosis, are urgently needed (25, 38); second, BCG vaccination of newborns should be continued and not be given up prematurely. A solution which fulfills both requirements can be offered by heterologous prime-boost vaccination strategies comprised of priming with BCG and boosting with a novel vaccine candidate (4, 14).
In preclinical trials, genetic immunization with naked DNA has proven to be a powerful antigen discovery tool and vaccination protocol (8, 16). DNA vaccines use episomal vectors expressing antigens under eukaryotic promoters in the vaccinated host and therefore can elicit potent humoral and cellular immune responses comprising both CD4 and CD8 T cells (8, 16). Several mycobacterial DNA vaccines have been shown to elicit protective immune responses in mice (7, 10, 21, 23, 31, 35, 45-48). However, considerable amounts of DNA are required, particularly in nonhuman primates and humans. In clinical trials, multiple high doses of DNA were required for the induction of immune responses (5, 27, 49).
Elucidation of the complete genome of M. tuberculosis has provided a blueprint for the search for novel vaccine candidates (6). However, the genome sequence alone cannot provide information about the protective value of candidate antigens. Hence, we have devised a two-step strategy in which genes encoding candidate antigens are predicted and then verified experimentally in preclinical vaccine trials in animal models. This strategy could be complemented by definition of correlates of protection.
We have developed such a strategy beginning with comparative proteomics for prediction of antigens which are uniquely present in the proteome of M. tuberculosis but absent in that of BCG (22), followed by large-scale screening of predicted antigen candidates with naked DNA vaccine constructs. Assessment of vaccine efficacy in experimental animals by reduction of bacterial load in the lungs was complemented by in vitro determination of gamma interferon (IFN-γ) production. Our strategy identified one vaccine candidate out of 36 which provided protection similar to that afforded by BCG. This antigen also proved a promising candidate for heterologous prime-boost vaccination strategies that included priming with BCG and boosting with the DNA vaccine. With this strategy, we achieved a protection superior to that conferred by BCG vaccination alone against tuberculosis.
MATERIALS AND METHODS
Mice.
BALB/c mice were bred at the central animal facilities of the Max Planck Institute for Infection Biology at the Bundesinstitut für gesundheitlichen Verbraucherschutz und Veterinärmedizin (Berlin, Germany). Animals were kept under specific-pathogen-free conditions and fed autoclaved food and water ad libitum. In these experiments, female mice were used at 8 weeks of age. Groups of at least five mice were used in all experiments.
Bacteria and cells.
M. bovis BCG strain Danish 1331 (Statens Serum Institute, Copenhagen, Denmark) was cultured in Dubos broth base (Difco, Detroit, Mich.) supplemented with Dubos medium albumin (Difco) at 37°C with shaking until bacterial growth reached an optical density at 600 nm of approximately 0.7 (equivalent to a cell density of approximately 108 cells per ml). M. tuberculosis H37Rv was grown at 37°C on Middlebrook 7H9 agar (Difco) supplemented with oleic acid, albumin, dextrose, and catalase enrichment 1339 (Difco) after passage through mice and subsequently in Middlebrook 7H9 liquid medium containing albumin, dextrose, and catalase enrichment (Difco) under shaking until bacterial growth reached an optical density at 600 nm of approximately 0.7. Mycobacteria were harvested by centrifugation, washed with phosphate-buffered saline without Ca2+ and maintained in 10% glycerol at −70°C until use. Cellular proteins were prepared from whole-cell lysates as described previously (33). The sonicate was stored at −70°C. P815 mastocytoma cells were obtained from the American Type Culture Collection (Manassas, Va.) and cultured in RPMI 1640 (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 U/ml), and 2-mercaptoethanol (RP10).
DNA vaccine construction.
M. tuberculosis genes were cloned into the DNA vaccine vector pCMVtPA (Chiron-Behring, Emeryville). It encodes DNA vaccine antigens as tissue plasminogen activator leader peptide fusion proteins under the control of the cytomegalovirus promoter. The DNA vaccine antigens were amplified by PCR with M. tuberculosis H37Rv chromosomal DNA as the template with gene-specific oligonucleotide primers containing additional cloning sites at their 5′ ends.
PCR amplification was carried out in an Eppendorf Mastercylcer (Eppendorf, Hamburg, Germany) for 30 cycles with the following conditions: 94°C for 1 min, 56°C for 45 s, and 72°C for 1 min. The amplification products were purified by agarose gel electrophoresis and subcloned into the SmaI restriction site of the cloning vector pUC18. The resulting plasmids carrying the subcloned PCR products were identified by restriction endonuclease digestion, and the inserted sequences were confirmed by DNA sequencing. The verified sequences were cleaved from the pUC18 backbone by restriction endonuclease digestion specific for the incorporated 5′ sites of the PCR products and subsequently ligated to the DNA vaccine vector pCMVtpa, resulting in the conclusive DNA vaccine constructs. The inserted sequences were verified by DNA sequencing. For DNA vaccination experiments, the DNA vaccines encoding unique M. tuberculosis antigens were purified with the EndoFree plasmid purification kit (Qiagen, Hilden, Germany).
Vaccination and challenge infection.
BALB/c mice were immunized three times intramuscularly with 100 μg of DNA per mouse (50 μg per quadricep) at 21-day intervals. DNA vaccines encoding 37 M. tuberculosis antigens were tested. As a positive control, mice were vaccinated with BCG by intravenous injection into the tail vain (39). Naïve mice were used as a negative control. At 120 days post-BCG vaccination and 21 days after the last booster immunization with DNA vaccine candidates, mice were challenged with M. tuberculosis H37Rv by an aerosol exposure system (Glas-Col) with an estimated infection dose of 200 bacilli per lung. For prime-boost experiments, animals were vaccinated intravenously with 106 BCG (prime). At 120 days after the BCG prime, the animals were twice boost-vaccinated with 100 μg of DNA per mouse at an interval of 21 days. Mice were then aerosol challenged 21 days after the last booster vaccination. Protection was monitored on day 30 and day 60 postinfection by enumeration of bacterial CFU in lungs by plating serially diluted organ homogenates on Middlebrook 7H9 agar.
Identification of antigenic peptide epitopes encoded by the DNA vaccines.
Possible major histocompatibility complex (MHC) I immunogenic peptide epitopes for the antigens Rv1511, Rv2520c, and Rv3407 were computed by the program MHC-I Antigenic Peptide Processing Prediction (MAPPP), which is available on the internet (http://mpiib-berlin.mpg.de/MAPPP). A combination of the proteasomal processing software FRAGPREDICT and PAProC with the MHC binding ligand predictions SYFPEITHI and BIMAS in the expert mode of MAPPP were used (17). Parameters for FRAGPREDICT were set at 0.5 (minimal residue cleavage probability and minimal fragment cleavage probability) and at 0.15 in PAProC. We used the murine haplotypes H2Kd, H2Dd and H2Ld and a fragment length of 8 to 10 amino acids. The threshold was set at an overall score of 0.9 except for the gene product Rv3407, for which we chose an overall score of 0.84. All peptides predicted were either 9- or 10-mers. Purified peptides (Jerini, Berlin, Germany) were used in the Elispot assay with splenocytes of vaccinated and control animals.
Cytokine Elispot assay.
Frequencies of IFN-γ-secreting T lymphocytes with specificity for the Rv1511, Rv2520c, and Rv3407 MHC class I epitopes were determined by a modified enzyme-linked immunospot (Elispot) technique (15). Briefly, 96-well Millititer HA nitrocellulose plates (Millipore) were coated with 5 mg of the anti-mouse IFN-γ monoclonal antibody R4 (B&D, Heidelberg, Germany)/ml in 100 μl of carbonate buffer, pH 9.6. After overnight incubation at 4°C, plates were washed twice with phosphate-buffered saline and blocked at 37°C for 2 h with 100 μl of 1% bovine serum albumin in phosphate-buffered saline. Splenocytes (105) from vaccinated mice were pulsed with 10 μg of specific peptides and then cultured in 100 μl of RP10 medium per well. P815 cells were coated with 10 μg of peptides per ml in phosphate-buffered saline at 37°C for 1 h and then washed twice with RP10.
Concanavalin A-stimulated splenocytes served as a positive control. Coated or uncoated P815 cells (105) were added to splenocytes in 100 μl of RP10, and after 20 h of incubation at 37°C and 5% CO2 in the presence of 30 U of interleukin-2 per ml, the plates were washed 10 times with 0.05% Tween 20 in phosphate-buffered saline (washing buffer). To detect IFN-γ-positive spots, 0.25 μg of biotinylated anti-mouse IFN-γ monoclonal antibody XMG1.2 (B&D) per ml in 100 μl of washing buffer was added and incubated at 37°C for 2 h. Plates were washed 10 times and incubated for 1 h at 37°C in 100 μl of a 1:20,000 dilution of alkaline phosphatase-coupled streptavidin (B&D). After five washes, spots of IFN-γ-secreting cells were visualized by adding 50 μl of the ready-to-use substrate 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT; Sigma) dissolved in water. The reaction was stopped after 15 min at 37°C by several washes with distilled water. After drying, spots were counted under a dissecting microscope at threefold magnification or counted automatically with a Bioreader 2000 (Biosys, Karpen, Germany). Frequencies of peptide-specific T cells are expressed as spot-forming units of IFN-γ-secreting cells per 105 splenocytes.
For detection of Rv15101-, Rv2520c-, and Rv3407-specific MHC class II-restricted T cells, a slightly modified protocol was employed (15). To induce specific cytokine secretion by antigen-specific T cells, 105 splenocytes per well were stimulated with 10 μg of heat-denatured protein lysate of H37Rv per ml in 100 μl of RP10 for 3 days. The J774A0.1 macrophage-like cells were pulsed with 10 μg of heat-denatured protein lysate of H37Rv per ml in RP10 at 37°C for 1 h and subsequently washed twice with RP10. Coated or uncoated J774A.1 cells (105) were added to splenocytes in 100 μl of RP10 and after 20 h of incubation at 37°C, 5% CO2 in the presence of 30 U of interleukin-2 per ml, plates were washed 10 times with washing buffer. Plates for IFN-γ detection were prepared as described above. Incubation times and detection followed the Elispot protocol for MHC class I-restricted T cells.
ELISA.
Antibody titers were measured by enzyme-linked immunosorbent assay (ELISA) of pooled sera. Briefly, 96-well microtiter plates were coated overnight with 1 μg of M. tuberculosis protein lysate in 0.1 M carbonate buffer. Blood was collected from vaccinated mice. The sera were diluted, and the endpoint titer was determined as described previously (15). We used the endpoint dilution of 1:50. For determination of isotypes, peroxidase-conjugated rabbit anti-mouse immunoglobulin G1 and immunoglobulin G2a (1:2,500) were used. These antibodies were diluted and incubated sequentially on the plates at 37°C for 2 h. Sera from nonvaccinated mice and BCG-vaccinated animals served as controls.
Statistical analysis.
The significance of differences was calculated according to Student's t test and the nonparametric two-tailed Mann-Whitney test with a confidence interval at 95% and a P value of < 0.05 considered significant. The individual groups in each experiments contained either five to six mice for the initial screening procedure or at least seven and, occasionally, 10 mice per time point for verification. Mean values of in vitro studies are based on three replicates.
RESULTS
Selection and characterization of protective antigens.
A proteome comparison was performed between M. tuberculosis and M. bovis BCG to identify genes which are expressed in M. tuberculosis but absent in M. bovis on the protein level (22, 28). More than 60 proteins were identified which were differentially expressed in M. tuberculosis and BCG (22, 28). Of these, 36 were tested as naked DNA vaccine candidates in an initial screening for their protective efficacy in a mouse model. Table 1 shows the protein expression profile. Note that only seven of the 36 differentially expressed proteins were absent from the genome of BCG, while 29 were encoded in the genome of BCG but differentially expressed as proteins. Most of these candidates induced reduction of CFU in lungs after low-dose M. tuberculosis aerosol challenge at day 14 postinfection, while only a few protected at later time points.
TABLE 1.
Protective efficacy of 36 DNA vaccine candidates preselected by comparative proteomics
| Antigen | BCG
|
Log CFU reduction in lungs 30 days post- challenge | |
|---|---|---|---|
| Genome | Relative expressiona | ||
| BCG | ≥0.8 | ||
| Rv3804cb | Encoded | ± | Efficient (≥0.5) |
| Rv1511 | Deleted (RD6) | Absence28 | |
| Rv2520c | Encoded | − | |
| Rv3407 | Encoded | ↓28 | |
| Rv0068 | Encoded | ↔22 | Intermediate (≥0.3) |
| Rv0685 | Encoded | ± | |
| Rv1926c | Encoded | ± | |
| Rv2802c | Deleted | Absence | |
| Rv3710 | Encoded | ↓22, 28 | |
| Rv0036c | Encoded | ↓22, 28 | Low (<0.3) |
| Rv0120c | Encoded | ↓28 | |
| Rv0222 | Deleted (RD4) | Absence28 | |
| Rv0566c | Encoded | ↓ | |
| Rv0952 | Encoded | ↔22, 28 | |
| Rv1130 | Encoded | ↓ | |
| Rv1392 | Encoded | ↔22, 28 | |
| Rv1477 | Encoded | ↓ | |
| Rv1478 | Encoded | ↓ | |
| Rv1558 | Encoded | ↓ | |
| Rv1596 | Encoded | ↓22, 28 | |
| Rv1643 | Encoded | ↓ | |
| Rv1856c | Encoded | ↓28 | |
| Rv1978 | Deleted (RD2) | Absence28 | |
| Rv2005c | Encoded | ↓ | |
| Rv2031c | Encoded | ↓ | |
| Rv2068c | Encoded | ↓ | |
| Rv2557 | Encoded | ↓28 | |
| Rv2558 | Encoded | ↓ | |
| Rv2770c | Encoded | ↓ | |
| Rv2780 | Encoded | ↓22, 28 | |
| Rv2883c | Encoded | ↓28 | |
| Rv2971 | Encoded | ↔28 | |
| Rv3275c | Encoded | ↓ | |
| Rv3673c | Encoded | ↓ | |
| Rv3871 | Deleted (RD1) | Absence | |
| Rv3873 | Deleted (RD1) | Absence | |
| Rv3874 | Deleted (RD1) | Absence | |
In comparison to M. tuberculosis by two-dimensional gel electrophoresis analysis (±, small or no difference; −, not detected; ↓, lower; ↔, difference in electrophoretic protein mobility).
Used as a control (Ag 85A).
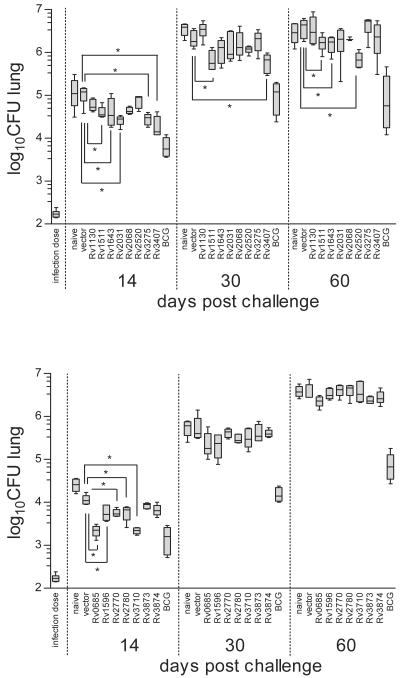
Figure 1 shows representative data for 15 vaccine candidates in lungs. CFU counts in spleens revealed similar results, indicating no difference in dissemination of the different DNA vaccine candidates (data not shown). We repeated the experiment with the DNA vaccine candidates which showed protective effects and subsequently grouped all candidates according to their vaccine efficacy (Table 1). Verification revealed that three DNA vaccine candidates provided appreciable levels of protection comparable to the control antigen 85 (Ag85; Rv3804) at day 30 and day 60 postchallenge (Fig. 2). Preliminary histology after challenge infection from the lungs of DNA-vaccinated animals did not show any difference from control animals (data not shown).
FIG. 1.
Screening for protective antigens with DNA vaccination. BALB/c mice were immunized three times at days −63, −42, and −21 with 100 μg of the DNA vaccine candidates. Mice were aerosol challenged at day 0 with M. tuberculosis (approximately 200 bacteria per lung). Protection was measured as CFU in the lungs at days 14, 30, and 60 postchallenge. Two experiments from the initial screening comprising 15 out of 37 DNA vaccine candidates are shown. All efficient, some intermediate, and several DNA vaccine candidates with low efficacy (see Table 1) are shown in a box and whiskers graph. The box extends from the 25th percentile to the 75th percentile, with a line at the median. The whiskers extend above and below the box to show the highest and lowest values. Statistical analysis and P values were determined by the two-tailed Mann-Whitney test, and significant differences (P < 0.05) are indicated by an asterisk. At least five mice per group were used.
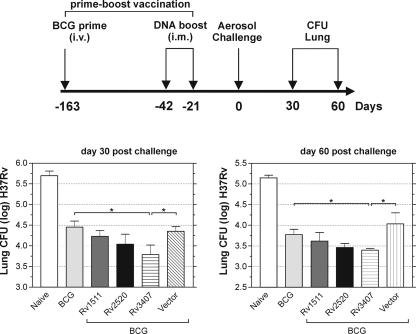
FIG. 2.
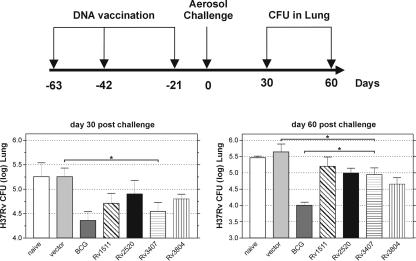
Protection against tuberculosis by DNA vaccination. BALB/c mice were immunized three times with 100 μg of the DNA vaccine candidates at the indicated time points. Mice were aerosol challenged with M. tuberculosis (approximately 200 bacteria per lung) 21 days after the last boost. Top, experimental schedule. Bottom, protection achieved by different DNA constructs. Bars represent the mean ± standard deviation of the CFU counts of 7 mice per group. Shown is one representative out of two similar experiments. P values were determined by Student's t test, and significant differences (P < 0.05) are indicated by an asterisk.
General agreement exists that Ag 85 is one of the most efficacious vaccine candidates known (22, 28). The three antigens which provided appreciable levels of protection were Rv2520c, which encodes a transmembrane protein of unknown function with a mass of 8.6 kDa; Rv1511, which is a putative guanine diphosphate-d-mannose dehydratase of 40 kDa; and Rv3407, which encodes a protein of unknown function with a mass of 10 kDa. The candidate Rv3407 was identified previously as being slightly expressed in virulent M. tuberculosis but absent in BCG (22, 28). In the experiment shown in Fig. 2, BCG vaccination, which was used as a positive control, provided significant protection of 0.8 log at both time points, whereas the empty-vector control resulted in no protection. The Rv3407-encoding DNA vaccine induced comparable protection (0.7 log) at day 30 postchallenge, and then slightly decreased to 0.5 log by day 60. The vaccine candidate Rv2520c achieved intermediate protection which was, however, not significantly different from the vector control in most experiments.
Frequencies of IFN-γ-secreting T lymphocytes after DNA vaccination.
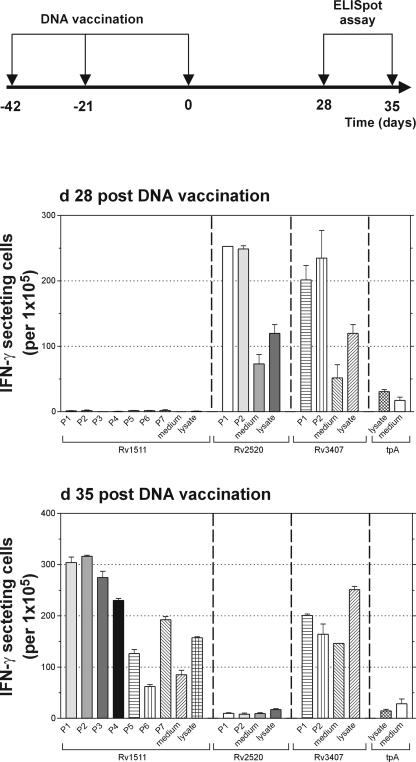
Antigen-specific IFN-γ secretion by spleen cells from vaccinated and control mice was analyzed by Elispot assay (Fig. 3). Splenocyte responses were stimulated either by soluble protein lysate of M. tuberculosis or by peptides representing predicted dominant MHC class I epitopes (Table 2). The prediction was done by MAPPP (17), which combines existing prediction tools for proteasomal processing and MHC class I anchoring. This combination enhances the accuracy and significance of the results. IFN-γ responses were measured 28 and 35 day after the second booster vaccination. At day 28 postvaccination, the vaccine candidate Rv1511 failed to induce any specific IFN-γ secretion, whereas vaccine candidates Rv2520c and Rv3407 induced IFN-γ production in the form of peptides and protein lysate. At day 35 postvaccination, Rv1511 induced IFN-γ secretion in response to most predicted peptides. Similarly, vaccine candidate Rv1511 induced an IFN-γ response with M. tuberculosis lysate at this time point. The vaccine candidate Rv3407 induced specific IFN-γ responses with both, peptide and lysate, although at this time point, only 1 of the 2 predicted peptides induced IFN-γ secretion after restimulation. In contrast, the vaccine candidate Rv2520c failed to stimulate IFN-γ secretion at this later time point. We assume that the failure of Rv2520c and Rv1511 to consistently induce protection was related to the missing IFN-γ responses at early (Rv1511) or late (Rv2520c) time points. In contrast, consistent protection induced by Rv3407 correlated well with the capacity to induce IFN-γ responses at early as well as late time points.
FIG. 3.
Numbers of IFN-γ-positive cells in mice immunized with Rv1511, Rv2520c, or Rv3407 as determined by the Elispot assay. Splenocytes were prepared at day 28 and day 35 after the last DNA vaccine boost. They were incubated in the presence of P815 or J774A.1 cells either unpulsed or pulsed with peptide (see Table 2) or M. tuberculosis lysate. IFN-γ-secreting cells are depicted as mean numbers per 105 immune splenocytes with standard deviation error bars for triplicate measurements. Shown is one representative out of two similar experiments.
TABLE 2.
Predicted MHC epitopes for vaccine candidates Rv1511, Rv2520, and Rv3407
| Candidate and peptide no. | Epitope | Position | MHC type | n-mer | MHC binding score | Overall score |
|---|---|---|---|---|---|---|
| Rv1511 | ||||||
| P1 | TFNTSRIDHL | 36 | H2_Kd | 10 | 0.642857142857143 | 0.9404761904 |
| P2 | TRLVTLLSTI | 67 | H2_Kd | 10 | 0.571428571428571 | 0.9285714285 |
| P3 | ASPPPQNEL | 135 | H2_Kd | 9 | 0.67741935483871 | 0.9454424787 |
| P4 | FYPRSPYGAA | 146 | H2_Kd | 10 | 0.571428571428571 | 0.9285714285 |
| P5 | TFVTRKITRA | 190 | H2_Kd | 10 | 0.714285714285714 | 0.9523804724 |
| P6 | VYMGNLDAV | 211 | H2_Kd | 9 | 0.526315789473684 | 0.9210526315 |
| P7 | QYVKFDQRYL | 271 | H2_Kd | 10 | 0.821428571428571 | 0.9702380952 |
| Rv2520 | ||||||
| P1 | DPNTIKQEI | 4 | H2_Ld | 9 | 0.516129032258065 | 0.9193548387 |
| P2 | RVIAFLRKPI | 42 | H2_Kd | 10 | 0.5 | 0.9166666666 |
| Rv3407 | ||||||
| P1 | IPARRPQNL | 64 | H2_Ld | 9 | 0.774193548387097 | 0.9623655913 |
| P2 | RPQNLLDVT | 68 | H2_Ld | 9 | 0.0641025641025641 | 0.8440170940 |
Specific antibody responses.
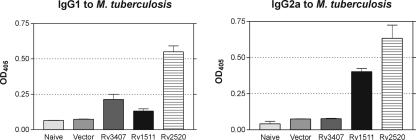
Sera from three mice per group were collected at day 35 after the last booster vaccination. Serum dilutions were prepared and an endpoint titer of 1:50 was determined. The three vaccine candidates induced specific antibody responses (Fig. 4). Although vaccine candidate Rv2520c induced high IgG1 and IgG2 antibody titers, vaccine candidate Rv3407 produced only weak antibody responses of the IgG1 isotype and no IgG2 response. In contrast, vaccine candidate Rv1511 also induced IgG2a antibody secretion. Thus, the capacity to induce profound antibody responses did not correlate with the protective capacity of the three vaccine candidates.
FIG. 4.
Serum antibodies in mice immunized with Rv1511, Rv2520c, or Rv3407 as determined by ELISA. Plates were coated with lysates from M. tuberculosis. Sera from mice were diluted 1:50, and the OD405 values were determined with alkaline phosphatase-coupled anti-mouse IgG1 or IgG2a as the secondary antibody. Bars represent the mean, with standard deviation error bars for triplicate measurements.
BCG prime-naked DNA vaccine boost protocol against tuberculosis.
Mice were prime vaccinated with BCG and after 120 days, two additional booster vaccinations with the naked DNA vaccine candidates were performed (Fig. 5). At day 21 after the last booster vaccination, mice were aerosol challenged with a low M. tuberculosis inoculum. At day 30 and day 60 after challenge infection, bacterial counts in lungs were determined. Vaccination with BCG alone achieved protection of approximately 1 log and additional boost with Rv1511 or Rv2520c did not increase protection significantly. Importantly, boost vaccination with Rv3407 significantly (P < 0.05) increased protection by BCG prime by 0.23 log or 0.27 log at day 30 or day 60, respectively. Thus, the heterologous BCG prime-Rv3407 boost vaccination induced protection superior to BCG alone. These results exemplify the potential of this heterologous prime-boost vaccination schedule and suggest that it should be further exploited for control of tuberculosis.
FIG. 5.
Protection against tuberculosis by BCG prime-DNA boost vaccination. BALB/c mice were immunized with 106 CFU of BCG Danish 1331 and boost immunized 120 days later twice with 100 μg of the DNA vaccine candidates at an interval of 21 days. Mice were aerosol challenged with M. tuberculosis (approximately 200 bacteria per lung) 21 days after the last boost. Bars represent the mean ± standard deviation of the CFU counts of seven mice per group. Shown is one representative out of two similar experiments. P values were determined by Student's t test, and significant differences (P < 0.05) are indicated by an asterisk.
DISCUSSION
This paper demonstrates that heterologous prime-boost vaccination significantly improves protection afforded by BCG alone. Previously, numerous prime-boost vaccination protocols have been employed in various infectious disease models with varying success. Prime-boost vaccination protocols in experimental tuberculosis include DNA/modified vaccinia virus Ankara (MVA) primer-booster (30), DNA/protein primer-booster (45, 48), BCG/protein prime-boost (4), DNA/BCG prime-boost (41, 42) and BCG/MVA prime-boost (14) vaccination schedules. These prime-boost protocols used ESAT6 (30, 41), MPT63 (30), Ag85A (4, 14, 42, 45), Ag85B (45), and HSP60 (48) as antigens. The prime-boost vaccination regime comprising naked DNA priming followed by a boost with recombinant MVA has been successful in AIDS (1, 18, 20, 44) and malaria (29, 34). However, only a few protocols considered the fact that BCG priming needs to be included in clinical tuberculosis vaccine trials, as BCG immunization is done in early childhood. Our approach, therefore, began with a BCG prime.
Currently, two major avenues of vaccine design against tuberculosis are being followed (24): First, viable vaccines either as gene deletion mutants of M. tuberculosis or as recombinant BCG expressing additional antigens or immunomodulators. Second, subunit vaccines either as protein adjuvant formulations or naked DNA vaccines. Some viable vaccines induce better protection than BCG. However, application of recombinant viable vaccines to humans is hampered by regulatory and safety issues. In contrast, subunit vaccines generally induce lower and rarely equal protection to BCG.
In order to achieve superior protection to BCG by a subunit vaccine and to fulfill requirements for subsequent clinical field trials, we decided to use a BCG primer followed by boost vaccination with a single-subunit vaccine candidate. We combined this strategy with a rational broad-scale search for protective antigens. Although we reported recently that equal protection is induced between postprimary M. tuberculosis infection and BCG vaccination in mice (32), enhancement of the immune response against tuberculosis by various vaccination schemes and combination of different vaccine candidates is conceivable. Our data are consistent with the previously published observation that Ag85 can reestablish protection at prolonged time periods after BCG vaccination (4). Reciprocally, Feng et al. found that protection by DNA priming is augmented by BCG boosting (10). Although this finding is coherent with our notion that prime-boost protocols induce superior protection to BCG vaccination alone, this strategy is less attractive for vaccination trials which are based on childhood BCG vaccination.
Protection against tuberculosis critically depends on T lymphocytes, rendering identification of protective antigens a difficult task (24). Thus far, the criteria, which define a protective T cell antigen, remain elusive. It has been claimed that an antigen's unique presence in the pathogen, abundance, secretion and/or distinct physicochemical properties are of critical importance. Indeed, in experimental listeriosis, immunodominant antigens have been described which induce protective immunity (40). In contrast, identification of immunodominant antigens has not been successful in tuberculosis. The antigens currently used in protection studies include HSP 60 (46), mtb8.4 (7), Ag85 (12, 14), ESAT6 (3, 36), and a fusion protein comprising both ESAT6 and Ag85 (37). These antigens have different features and were used in different vaccination protocols making it difficult to envisage general criteria which define protective antigens. Yet it is also clear that differences exist and that the vast majority of M. tuberculosis proteins fail to induce any protective response.
We based our strategy for identifying protective antigens on the assumption that proteins which are differentially expressed by M. tuberculosis compared with BCG can possess protective potential. To this end, we performed proteome analysis of M. tuberculosis and BCG and subsequently identified at least 60 proteins which were more abundantly expressed by M. tuberculosis than by BCG. The genes encoding for 36 proteins were used for construction of naked DNA vaccine candidates which were subsequently tested in a high throughput vaccination protocol. Of the antigens tested, three appeared promising in that they induced protection of approximately 0.5 log. Subsequent more detailed analysis revealed that antigen Rv3407 was able to induce significant protection at about the level afforded by BCG and Ag85 (Rv3804). In contrast, the two other antigens, Rv1511 and Rv2520c, failed to induce reliable levels of protection.
Antigen Rv3407 is interesting in that the coding gene is present in the genome of BCG. However, it is not detectable in the proteome of BCG cultured in vitro. We assume that Rv3407 has a protective capacity which is, however, impaired because it is only expressed in low abundance by BCG. We suppose that overexpression of Rv3407 under the control of a strong promoter could increase the vaccine efficacy of BCG. More importantly, we succeeded in improving the vaccine efficacy of BCG by overcoming the weak antigenicity of Rv3407 with low abundance in BCG by prime-boost vaccination.
We also determined IFN-γ and antibody responses induced by these vaccine candidates. Consistent with the robust protection induced by Rv3407, prominent IFN-γ production and low antibody responses were observed. It is generally accepted that both CD4 and CD8 T lymphocytes are required for protective immunity against tuberculosis (13, 24). In vitro stimulation with protein antigens almost exclusively stimulates CD4 T cells due to the fact that soluble protein antigens are primarily processed via the MHC class II pathway. In contrast, MHC class I-restricted CD8 T cells are virtually unresponsive in this protocol. CD8 T lymphocytes, however, can be stimulated by antigen-presenting cells pulsed with peptides representing MHC class I epitopes.
MHC class I epitopes of the DNA vaccines used were calculated by the prediction tool MAPPP (17). We determined the protein and peptide responses induced by the vaccine candidates. In accordance with the profound protection induced by Rv3407, IFN-γ was induced by proteins and peptides at both time points measured. In contrast, Rv2520c and Rv1511 produced appreciable IFN-γ only at day 28 or day 35, respectively. Further analyses will be needed to address the differential kinetics of IFN-γ induction by these DNA vaccine candidates. Nevertheless, these antigens were potent inducers of antibody responses, supporting the notion that IFN-γ responses represent the best possible correlates of vaccine-induced protection to date and arguing against a correlation of protection with antibody responses.
Our finding that boosting with Rv3407 improved the vaccine efficacy of BCG suggests that future clinical trials can be performed in BCG vaccinated individuals and, moreover, that subunit vaccination can be used to improve preexisting protection evoked by BCG. Although initial studies in animal models demonstrated potent activity of DNA vaccines in various infection models, studies in humans were disappointing (19, 26). More recent protocols comprised of priming with naked DNA and booster vaccination with recombinant MVA show that naked DNA performs better in macaques and humans if administered as part of a prime-boost regime (29). Therefore, the capacity of BCG prime and naked DNA boost vaccination in humans should be exploited for three reasons: (i) BCG needs to be included in future vaccine trials against tuberculosis; (ii) naked DNA vaccination has been shown to be effective in humans with prime-boost vaccination protocols; and (iii) this vaccination regime was capable of improving BCG-induced protection. In conclusion, our results provide evidence that a BCG-naked DNA prime-boost vaccination protocol represents a valuable candidate for future vaccine trials targeted at one of the major health problems worldwide.
Acknowledgments
Financial support from the European Union (EC-FP6, TB-VAC) and the German Research Society (priority program on novel vaccination strategies) is gratefully acknowledged.
Editor: D. L. Burns
REFERENCES
- 1.Amara, R. R., J. M. Smith, S. I. Staprans, D. C. Montefiori, F. Villinger, J. D. Altman, S. P. O'Neil, N. L. Kozyr, Y. Xu, L. S. Wyatt, P. L. Earl, J. G. Herndon, J. M. McNicholl, H. M. McClure, B. Moss, and H. L. Robinson. 2002. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J. Virol. 76:6138-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom, B. R., and P. E. Fine. 1994. The BCG experience: implications for future vaccines against tuberculosis, p. 531-557. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection and control. American Society for Microbiology, Washington, D.C.
- 3.Brandt, L., M. Elhay, I. Rosenkrands, E. B. Lindblad, and P. Andersen. 2000. ESAT-6 subunit vaccination against Mycobacterium tuberculosis. Infect. Immun. 68:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, J. V., A. A. Frank, M. A. Keen, J. T. Bellisle, and I. M. Orme. 2001. Boosting vaccine for tuberculosis. Infect. Immun. 69:2714-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calarota, S., G. Bratt, S. Nordlund, J. Hinkula, A. C. Leandersson, E. Sandstrom, and B. Wahren. 1998. Cellular cytotoxic response induced by DNA vaccination in HIV-1-infected patients. Lancet 351:1320-1325. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Coler, R. N., A. Campos-Neto, P. Ovendale, F. H. Day, S. P. Fling, L. Q. Zhu, N. Serbina, J. L. Flynn, S. G. Reed, and M. R. Alderson. 2001. Vaccination with the T cell antigen Mtb 8.4 protects against challenge with Mycobacterium tuberculosis. J. Immunol. 166:6227-6235. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly, J. J., J. B. Ulmer, J. W. Shiver, and M. A. Liu. 1997. DNA vaccines. Annu. Rev. Immunol. 15:617-648. [DOI] [PubMed] [Google Scholar]
- 9.Espinal, M. A. 2003. The global situation of MDR-TB. Tuberculosis 83:44-51. [DOI] [PubMed] [Google Scholar]
- 10.Feng, C. G., U. Palendira, C. Demangel, J. M. Spratt, A. S. Malin, and W. J. Britton. 2001. Priming by DNA immunization augments protective efficacy of Mycobacterium bovis bacille Calmette-Guérin against tuberculosis. Infect. Immun. 69:4174-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fine, P. E. 1995. Bacille Calmette-Guérin vaccines: a rough guide. Clin. Infect.. Dis. 20:11-14. [DOI] [PubMed] [Google Scholar]
- 12.Fine, P. E. M. 1995. Variation in protection by Bcg-implications of and for heterologous immunity. Lancet 346:1339-1345. [DOI] [PubMed] [Google Scholar]
- 13.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 14.Goonetilleke, N. P., H. McShane, C. M. Hannan, R. J. Anderson, R. H. Brookes, and A. V. S. Hill. 2003. Enhanced immunogenicity and protective efficacy against Mycobacterium tuberculosis of bacille Calmette-Guérin vaccine using mucosal administration and boosting with a recombinant modified vaccinia virus Ankara. J. Immunol. 171:1602-1609. [DOI] [PubMed] [Google Scholar]
- 15.Grode, L., M. Kursar, J. Fensterle, S. H. Kaufmann, and J. Hess. 2002. Cell-mediated immunity induced by recombinant Mycobacterium bovis Bacille Calmette-Guérin strains against an intracellular bacterial pathogen: importance of antigen secretion or membrane-targeted antigen display as lipoprotein for vaccine efficacy. J. Immunol. 168:1869-1876. [DOI] [PubMed] [Google Scholar]
- 16.Gurunathan, S., D. M. Klinman, and R. A. Seder. 2000. DNA vaccines: Immunology, application, and optimization. Annu. Rev. Immunol. 18:927-974. [DOI] [PubMed] [Google Scholar]
- 17.Hakenberg, J., A. K. Nussbaum, H. Schild, H. G. Rammensee, C. Kuttler, H. G. Holzhütter, P. M. Kloetzel, S. H. E. Kaufmann, and H. J. Mollenkopf. 2003. MAPPP: MHC class I antigenic peptide processing prediction. Appl. Bioinform. 2:155-158. [PubMed] [Google Scholar]
- 18.Hanke, T., C. Barnfield, E. G. T. Wee, L. Agren, R. V. Samuel, N. Larke, and P. Liljestrom. 2003. Construction and immunogenicity in a prime-boost regimen of a Semliki Forest virus-vectored experimental HIV clade A vaccine. J. Gen Virol. 84:361-368. [DOI] [PubMed] [Google Scholar]
- 19.Hanke, T., R. V. Samuel, T. J. Blanchard, V. C. Neumann, T. M. Allen, J. E. Boyson, S. A. Sharpe, N. Cook, G. L. Smith, D. I. Watkins, M. P. Cranage, and A. J. McMichael. 1999. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by using a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J. Virol. 73:7524-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. X. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huygen, K., J. Content, O. Denis, D. L. Montgomery, A. M. Yawman, R. R. Deck, C. M. DeWitt, I. M. Orme, S. Baldwin, C. Dsouza, A. Drowart, E. Lozes, P. Vandenbussche, J. P. VanVooren, M. A. Liu, and J. B. Ulmer. 1996. Immunogenicity and protective efficacy of a tuberculosis DNA vaccine. Nat. Med. 2:893-898. [DOI] [PubMed] [Google Scholar]
- 22.Jungblut, P. R., U. E. Schaible, H. J. Mollenkopf, U. Zimny-Arndt, B. Raupach, J. Mattow, P. Halada, S. Lamer, K. Hagens, and S. H. E. Kaufmann. 1999. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 33:1103-1117. [DOI] [PubMed] [Google Scholar]
- 23.Kamath, A. T., C. G. Feng, M. Macdonald, H. Briscoe, and W. J. Britton. 1999. Differential protective efficacy of DNA vaccines expressing secreted proteins of Mycobacterium tuberculosis. Infect. Immun. 67:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann, S. H. E. 2001. How can immunology contribute to the control of tuberculosis? Nat. Rev. Immunol. 1:20-30. [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann, S. H. E. 2000. Is the development of a new tuberculosis vaccine possible? Nat. Med. 6:955-960. [DOI] [PubMed] [Google Scholar]
- 26.MacGregor, R. B., R. Ginsberg, K. E. Ugen, Y. Baine, C. U. Kang, X. M. Tu, T. Higgins, D. B. Weiner, and J. D. Boyer. 2002. T-cell responses induced in normal volunteers immunized with a DNA-based vaccine containing HIV-1 env and rev. AIDS 16:2137-2143. [DOI] [PubMed] [Google Scholar]
- 27.MacGregor, R. R., J. D. Boyer, K. E. Ugen, K. E. Lacy, S. J. Gluckman, M. L. Bagarazzi, M. A. Chattergoon, Y. Baine, T. J. Higgins, R. B. Ciccarelli, L. R. Coney, R. S. Ginsberg, and D. B. Weiner. 1998. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J. Infect.. Dis. 178:92-100. [DOI] [PubMed] [Google Scholar]
- 28.Mattow, J., P. R. Jungblut, U. E. Schaible, H. J. Mollenkopf, S. Lamer, U. Zimny-Arndt, K. Hagens, E. C. Muller, and S. H. E. Kaufmann. 2001. Identification of proteins from Mycobacterium tuberculosis missing in attenuated Mycobacterium bovis BCG strains. Electrophoresis 22:2936-2946. [DOI] [PubMed] [Google Scholar]
- 29.McConkey, S. J., W. H. H. Reece, V. S. Moorthy, D. Webster, S. Dunachie, G. Butcher, J. M. Vuola, T. J. Blanchard, P. Gothard, K. Watkins, C. M. Hannan, S. Everaere, K. Brown, K. E. Kester, J. Cummings, J. Williams, D. G. Heppner, A. Pathan, K. Flanagan, N. Arulanantham, M. T. M. Roberts, M. Roy, G. L. Smith, J. Schneider, T. Peto, R. E. Sinden, S. C. Gilbert, and A. V. S. Hill. 2003. Enhanced T-cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat. Med. 9:729-735. [DOI] [PubMed] [Google Scholar]
- 30.McShane, H., R. Brookes, S. C. Gilbert, and A. V. S. Hill. 2001. Enhanced immunogenicity of CD4+ T-cell responses and protective efficacy of a DNA-modified vaccinia virus Ankara prime-boost vaccination regimen for murine tuberculosis. Infect. Immun. 69:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mollenkopf, H. J., D. Groine-Triebkorn, P. Andersen, J. Hess, and S. H. E. Kaufmann. 2001. Protective efficacy against tuberculosis of ESAT-6 secreted by a live Salmonella typhimurium vaccine carrier strain and expressed by naked DNA. Vaccine 19:4028-4035. [DOI] [PubMed] [Google Scholar]
- 32.Mollenkopf, H. J., M. Kursar, and S. H. Kaufmann. 2004. Immune response to postprimary tuberculosis in mice: Mycobacterium tuberculosis and Miycobacterium bovis bacille Calmette-Guérin induce equal protection. J. Infect.. Dis. 190:588-597. [DOI] [PubMed] [Google Scholar]
- 33.Mollenkopf, H. J., J. Mattow, U. E. Schaible, L. Grode, S. H. Kaufmann, and P. R. Jungblut. 2002. Mycobacterial proteomes. Methods Enzymol. 358:242-256. [DOI] [PubMed] [Google Scholar]
- 34.Moorthy, V. S., S. McConkey, M. Roberts, P. Gothard, N. Arulanantham, P. Degano, J. Schneider, C. Hannan, M. Roy, S. C. Gilbert, T. E. A. Peto, and A. V. S. Hill. 2003. Safety of DNA and modified vaccinia virus Ankara vaccines against liver-stage P. falciparum malaria in non-immune volunteers. Vaccine 21:1995-2002. [DOI] [PubMed] [Google Scholar]
- 35.Morris, S., C. Kelley, A. Howard, Z. M. Li, and F. Collins. 2000. The immunogenicity of single and combination DNA vaccines against tuberculosis. Vaccine 18:2155-2163. [DOI] [PubMed] [Google Scholar]
- 36.Olsen, A. W., P. R. Hansen, A. Holm, and P. Andersen. 2000. Efficient protection against Mycobacterium tuberculosis by vaccination with a single subdominant epitope from the ESAT-6 antigen. Eur. J. Immunol. 30:1724-1732. [DOI] [PubMed] [Google Scholar]
- 37.Olsen, A. W., L. A. H. van Pinxteren, L. M. Okkels, P. B. Rasmussen, and P. Andersen. 2001. Protection of mice with a tuberculosis subunit vaccine based on a fusion protein of antigen 85B and ESAT-6. Infect. Immun. 69:2773-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orme, I. M. 1999. Beyond BCG: the potential for a more effective TB vaccine. Mol. Med. Today 5:487-492. [DOI] [PubMed] [Google Scholar]
- 39.Palendira, U., A. G. Bean, C. G. Feng, and W. J. Britton. 2002. Lymphocyte recruitment and protective efficacy against pulmonary mycobacterial infection are independent of the route of prior Mycobacterium bovis BCG immunization. Infect. Immun. 70:1410-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pamer, E. G., A. Sijts, M. S. Villanueva, D. H. Busch, and S. Vijh. 1997. MHC class I antigen processing of Listeria monocytogenes proteins: implications for dominant and subdominant CTL responses. Immunol. Rev. 158:129-136. [DOI] [PubMed] [Google Scholar]
- 41.Skinner, M. A., B. M. Buddle, D. N. Wedlock, D. Keen, G. W. de Lisle, R. E. Tascon, J. C. Ferraz, D. B. Lowrie, P. J. Cockle, H. M. Vordermeier, and R. G. Hewinson. 2003. A DNA prime-Mycobacterium bovis BCG boost vaccination strategy for cattle induces protection against bovine tuberculosis. Infect. Immun. 71:4901-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skinner, M. A., A. J. Ramsay, G. S. Buchan, D. L. Keen, C. Ranasinghe, L. Slobbe, D. M. Collins, G. W. De Lisle, and B. M. Buddle. 2003. A DNA prime-live vaccine boost strategy in mice can augment IFN-gamma responses to mycobacterial antigens but does not increase the protective efficacy of two attenuated strains of Mycobacterium bovis against bovine tuberculosis. Immunology 108:548-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sterne, J. A. C., L. C. Rodrigues, and I. N. Guedes. 1998. Does the efficacy of BCG decline with time since vaccination? Int. J. Tuberc. Lung Dis. 2:200-207. [PubMed] [Google Scholar]
- 44.Tang, Y. Y., F. Villinger, S. I. Staprans, R. R. Amara, J. M. Smith, J. G. Herndon, and H. L. Robinson. 2002. Slowly declining levels of viral RNA and DNA in DNA/recombinant modified vaccinia virus Ankara-vaccinated macaques with controlled simian-human immunodeficiency virus SHIV-89.6P challenges. J. Virol. 76:10147-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanghe, A., S. D'Souza, V. Rosseels, O. Denis, T. H. M. Ottenhoff, W. Dalemans, C. Wheeler, and K. Huygen. 2001. Improved immunogenicity and protective efficacy of a tuberculosis DNA vaccine encoding antigen 85 by protein boosting. Infect. Immun. 69:3041-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tascon, R. E., M. J. Colston, S. Ragno, E. Stavropoulos, D. Gregory, and D. B. Lowrie. 1996. Vaccination against tuberculosis by DNA injection. Nat. Med. 2:888-892. [DOI] [PubMed] [Google Scholar]
- 47.Ulmer, J. B., M. A. Liu, D. L. Montgomery, A. M. Yawman, R. R. Deck, C. M. DeWitt, J. Content, and K. Huygen. 1997. Expression and immunogenicity of Mycobacterium tuberculosis antigen 85 by DNA vaccination. Vaccine 15:792-794. [DOI] [PubMed] [Google Scholar]
- 48.Vordermeier, H. M., D. B. Lowrie, and R. G. Hewinson. 2003. Improved immunogenicity of DNA vaccination with mycobacterial HSP65 against bovine tuberculosis by protein boosting. Vet. Microbiol. 93:349-359. [DOI] [PubMed] [Google Scholar]
- 49.Wang, R., D. L. Doolan, T. P. Le, R. C. Hedstrom, K. M. Coonan, Y. Charoenvit, T. R. Jones, P. Hobart, M. Margalith, J. Ng, W. R. Weiss, M. Sedegah, C. de Taisne, J. A. Norman, and S. L. Hoffman. 1998. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science 282:476-480. [DOI] [PubMed] [Google Scholar]
- 50.World Health Organization. 2002. The World Health Report 2002. Reducing risks, promoting healthy life. Report. World Health Organization, Geneva, Switzerland.