Abstract
Glycosylphosphatidylinositol (GPI)-anchored proteins are abundantly expressed in the infective and intracellular stages of Trypanosoma cruzi and are recognized as antigenic targets by both the humoral and cellular arms of the immune system. Previously, we demonstrated the efficacy of genes encoding GPI-anchored proteins in eliciting partially protective immunity to T. cruzi infection and disease, suggesting their utility as vaccine candidates. For the identification of additional vaccine targets, in this study we screened the T. cruzi expressed sequence tag (EST) and genomic sequence survey (GSS) databases. By applying a variety of web-based genome-mining tools to the analysis of ∼2,500 sequences, we identified 348 (37.6%) EST and 260 (17.4%) GSS sequences encoding novel parasite-specific proteins. Of these, 19 sequences exhibited the characteristics of secreted and/or membrane-associated GPI proteins. Eight of the selected sequences were amplified to obtain genes TcG1, TcG2, TcG3, TcG4, TcG5, TcG6, TcG7, and TcG8 (TcG1-TcG8) which are expressed in different developmental stages of the parasite and conserved in the genome of a variety of T. cruzi strains. Flow cytometry confirmed the expression of the antigens encoded by the cloned genes as surface proteins in trypomastigote and/or amastigote stages of T. cruzi. When delivered as a DNA vaccine, genes TcG1-TcG6 elicited a parasite-specific antibody response in mice. Except for TcG5, antisera to genes TcG1-TcG6 exhibited trypanolytic activity against the trypomastigote forms of T. cruzi, a property known to correlate with the immune control of T. cruzi. Taken together, our results validate the applicability of bioinformatics in genome mining, resulting in the identification of T. cruzi membrane-associated proteins that are potential vaccine candidates.
American trypanosomiasis is a disease of humans that is caused by the protozoan Trypanosoma cruzi of the family Trypanosomatidae. T. cruzi is estimated to affect ∼20 million people, mainly in the areas of South and Central America, Mexico, and the southern United States (47, 48). Upon infection, human patients exhibit acute flu-like symptoms and then enter an indeterminate phase wherein they exhibit no clinical disease symptoms (19, 35). Approximately 40% of the infected patients, however, progress to the chronic phase, which is characterized by myocardial inflammation, fibrosis and necrosis, and cardiac dysfunction, eventually leading to heart failure (19, 33). Chemotherapies for the treatment of both acute and chronic patients are of limited use due to high toxicity and poor efficacy of the available drugs (40). No vaccines are available against T. cruzi.
An understanding of the protective immune responses that can effectively arrest pathogen survival in the mammalian host is critical for the development of vaccines against any infectious disease. Fortunately, a number of studies conducted in the last decade have defined the effector mechanisms required for the control of T. cruzi infection (6, 24, 41). These studies have led to the conclusion that defined antigens capable of inducing strong neutralizing lytic antibody and cytotoxic T-cell responses would likely constitute an effective vaccine that can provide protection from T. cruzi infection.
In recent studies, we and others have shown the potential utility of T. cruzi surface antigens as vaccine candidates. Several reports demonstrated the capacity of complement regulatory protein, paraflagellar rod protein, and trans-sialidases (TS), all expressed as surface antigens in T. cruzi, to elicit antiparasite immune responses capable of enhancing the survival of infected mice (9, 29, 31, 37, 38). We have shown the protective efficacy of the TS family members, namely, amastigote surface proteins ASP-1 and ASP-2 and trypomastigote surface antigen TSA-1, as DNA vaccines. DNA immunization plasmids incorporating these genes, singly or together, provided substantial resistance to T. cruzi infection in mice that was further enhanced by codelivery of cytokine adjuvants (16, 46). Altogether, these results have established the immunogenic potential of parasite surface antigens and suggested their utility as vaccine candidates in controlling T. cruzi infection and disease.
It is, however, important that the protective immunity afforded by all of the vaccination regimens that have been tested thus far is partial and fails to prevent infection or death in 100% of the immunized animals. Researchers failed to induce sterile immunity with TSA-1, ASP-1, and ASP-2 in C57BL/6 and BALB/c mice (16, 46) or any immunity in C3H/HeSnJ mice (15). This may be because these genes comprise a very restricted subcomponent of the TS gene family and represent only a minor proportion of all of the possible target molecules in T. cruzi. The immune responses elicited by these antigens might not, therefore, be of sufficient magnitude to efficiently control T. cruzi. Identification of additional vaccine candidates would enhance efforts toward developing a multicomponent DNA vaccine capable of increasing the protective immunity induced by TS family members and other antigenic targets.
In this study, we aimed to identify by computational analysis of T. cruzi sequence databases novel, parasite-specific surface antigens that are not members of large gene families and might have utility as vaccine candidates. We employed a combination of web-based bioinformatic tools for global similarity searching and software predictive of targeting, localization, and expression in different cellular compartments. The mining of ∼2,500 sequences in expressed sequence tag (EST) and genomic sequence survey (GSS) databases identified 19 sequences that exhibited characteristics of secreted and/or membrane-associated glycosylphosphatidylinositol (GPI) proteins. We report the antigenic characteristics of eight of the selected candidates (TcG1, TcG2, TcG3, TcG4, TcG5, TcG6, TcG7, and TcG8 [TcG1-TcG8]). TcG1-TcG8 genes are highly conserved in diverse, clinically relevant strains of T. cruzi and encode surface antigens that are expressed in multiple stages of the parasite. Antibodies to these antigens were induced in immunized mice, of which a majority exhibited trypanolytic activity. Together, our results validate the applicability of bioinformatics in rapid genome mining and the identification of trypanosome membrane-associated proteins that are potential vaccine candidates.
MATERIALS AND METHODS
Parasites and mice.
T. cruzi epimastigotes were cultivated axenically at 28°C in liver infusion-tryptose medium (46) supplemented with 5% heat-inactivated fetal bovine serum (HyClone). The C2C12 cells were purchased from the American Type Culture Collection and maintained in complete RPMI 1640 medium (HyClone). Trypomastigotes of T. cruzi were maintained and propagated by continuous in vitro passage of parasites in monolayers of C2C12 cells (32). Amastigotes were obtained by incubation of the freshly harvested trypomastigotes in RPMI-10% fetal bovine serum medium, pH 5.0, at 37°C and 5% CO2 for 2 h (43). C3H/HeN male mice (6 to 8 weeks old) were obtained from Harlan Laboratories. Animal experiments were performed according to the National Institutes of Health Guide for Care and Use of Experimental Animals and approved by the University of Texas Medical Branch Animal Care and Use Committee.
In silico analysis.
T. cruzi EST and GSS databases from the European Bioinformatics Institute consisted of ∼22,000 and ∼10,000 sequences, respectively, and were analyzed from July 2000 to October 2001. We performed TBLASTX analysis (1, 2) of the T. cruzi sequences against the nonredundant (NR) sequence database provided by the National Center for Biotechnology Information (NCBI), National Institutes of Health, Bethesda, Md. A cutoff “expect” value of 10−5 was used for all database searches. T. cruzi sequences that exhibited no homology to sequences in public databases were translated into amino acid sequence by a TRANSLATE tool at the Expert Protein Analysis System (ExPASy) website (http://www.expasy.com) (3). The open reading frames (ORFs) were analyzed for the identification of protein-sorting signals and localization sites by PSORT (27). The N-terminal signal sequence and the signal peptide cleavage site were identified by SignalP (28). The GPI signal sequence and the cleavage-attachment (C/A) site were determined by DGPI from D. Buloz and J. Kronegg at the Swiss Institute of Bioinformatics, University of Geneva, Geneva, Switzerland (http://www.expasy.com). Prediction of domains or motifs suggestive of putative functions of proteins was performed by using SMART (Simple Modular Architecture Research Tool) and CDD (Conserved Domain Database) tools available at the NCBI website. The major histocompatibility complex (MHC) class I and class II epitopes were identified by Propred (39) and the HLA Bind tool available at the ExPASy website (3). Sequence alignment was performed by ClustalW (http://www.expasy.com).
RNA isolation and cDNA synthesis.
For total RNA isolation, T. cruzi epimastigotes were harvested by centrifugation at 3,000 × g for 10 min, washed twice with cold phosphate-buffered saline (PBS), and suspended in guanidine-phenol solution (109 parasites/ml). Parasites were homogenized, and the total RNA was extracted as described previously (8), with slight modifications (14). First-strand cDNA was synthesized by incubation of total RNA (5 μg) with 2.5 U of Moloney murine leukemia virus reverse transcriptase (New England Biolabs) and oligo(dT)16 at 42°C for 1 h in a 20-μl reaction volume and stored at −20°C until further use.
Cloning and sequencing of TcG1-TcG8.
Genes TcG1-TcG8 were amplified from epimastigote cDNA in a PCR with universal splice leader, present upstream to the start codon (ATG) in mRNA-cDNA of trypanosomes (25), as forward primer and gene-specific reverse primer (listed in Table 1). Individual amplicons (10 μl) were resolved on 1% agarose gels, stained with ethidium bromide, and visualized and photographed with a FluorChem imaging system (Alpha Innotech). Amplicons were purified by using a DNA extraction kit (QIAGEN) and cloned into a pCR2.1 T/A cloning vector (Invitrogen). All cloned sequences were confirmed by restriction digestion and sequencing at the Recombinant DNA Core Facility at the University of Texas Medical Branch and analyzed with various web-based software.
TABLE 1.
Oligonucleotides used in this studya
| Gene and use | Forward primer sequence (5′ → 3′) | Reverse primer sequence (5′ → 3′) | Size (bp) |
|---|---|---|---|
| Gene amplificationb | |||
| TcG1 | CACCAGTTTCTGTACTATATTG | CGTTCGAGATGCGCTTCT | 538 |
| TcG2 | CACCAGTTTCTGTACTATATTG | CCCAACAGCGGTGGAA | 839 |
| TcG3 | CACCAGTTTCTGTACTATATTG | AATCCCCTGATACGTCG | 1,518 |
| TcG4 | CACCAGTTTCTGTACTATATTG | TGACAGAACGTGAATGGG | 600 |
| TcG5 | CACCAGTTTCTGTACTATATTG | TGAAGAAGAGCGTCGAGTGC | 1,427 |
| TcG6 | CACCAGTTTCTGTACTATATTG | CACAGCAAGGGAGCAAC | 799 |
| TcG7 | CACCAGTTTCTGTACTATATTG | CTTTTGCAATGGTCTTTGCG | 636 |
| TcG8 | CACCAGTTTCTGTACTATATTG | TCACTGTGGTACAACGCTGACC | 1,256 |
| TcGP18 | AAGCTTCGAGCATTGTCTATGTGCCTTGAA | CTCGAGCTACAGCAGGTCATATTGTACATC | 450 |
| Genomic conservation, expression analysis, and cloningc | |||
| TcG1 | GGATCCATGGTGAAGGCGAACTATATT | GGGTCTAGATCACGTTCGAGATGCGCTTC | 499 |
| TcG2 | GGATCCATGTCGCTTTCATTTATCGAGTCAGGG | GGGTCTAGATCACCCAACAGCGGTGGAA | 662 |
| TcG3 | GGATCCATGCTTCAGCGTACCTGCAGC | GGGTCTAGATCAGCTTGACACTTCGC | 1,011 |
| TcG4 | GGATCCATGTCAGCCAAGGCTCCC | GGGTCTAGATCACTTTTCAAGCGCC | 276 |
| TcG5 | GGATCCATGGGGAAGGAAAAGGTGC | GGGTCTAGATCACTTCTTAGCGGC | 1,350 |
| TcG6 | AAGGCTATGCTGGCGACAC | GGGTCTAGATCACACAGCAAGGG | 756 |
| TcG7 | GGATCCATGCTGGCGACACACGG | GGGTCTAGACTACATCCATCCTCGCC | 351 |
| TcG8 | GGATCCATGTCCGATAACCATCAACTGG | GGGTCTAGATCACTGTGGTACAACGCTG | 1,188 |
| TcGP18 | AAGCTTCGAGCATTGTCTATGTGCCTTGAA | CTCGAGCTACAGCAGGTCATATTGTACATC | 450 |
Expression at the mRNA level.
We determined the expression of the selected genes by traditional semiquantitative reverse transcription-PCR (RT-PCR). Total RNA and cDNA from the epimastigote, trypomastigote, and amastigote forms of T. cruzi were obtained as described above. The cDNA (2 μl) from each stage was subsequently amplified by PCR in a 50-μl reaction volume using 2.5 U of Taq polymerase and 1 μl of 20 μM gene-specific forward and reverse primers (Table 1). Individual amplicons (10 μl) were electrophoresed and imaged as described above. GAPDH (22) and GPI8 (N. Garg, unpublished data) genes that are constitutively expressed in all three stages of T. cruzi were used as positive controls.
Phylogenetic conservation.
Genomic DNA was isolated from different T. cruzi strains as described previously (26). PCR amplification of the selected genes was carried out for 35 cycles in a 50-μl reaction volume with 100 ng of genomic DNA and 1 μl of 20 μM gene-specific forward and reverse primers (Table 1). PCR-amplified products were resolved and imaged as described above. Genomic DNA from Leishmania major and Trypanosoma brucei were also used as templates in parallel reactions.
DNA immunization and serum collection.
The cDNAs encoding TcG1 (1 to 166 amino acids), TcG2 (1 to 220 amino acids), TcG3 (1 to 337 amino acids), TcG4 (1 to 92 amino acids), TcG5 (1 to 450 amino acids), and TcG6 (1 to 252 amino acids) were amplified by PCR. Forward and reverse oligonucleotides for the amplification of TcG1-TcG6 cDNAs were designed to incorporate BamHI, Hind III, or XbaI restriction sites for directional cloning (Table 1). The amplified products were first cloned into the pCR2.1 T/A vector and subcloned into eukaryotic expression vector pCDNA3.1 (Invitrogen) at the BamHI/XbaI or HindIII/XbaI site. The eukaryotic expression plasmid encoding murine granulocyte-macrophage colony-stimulating factor (pCMVI.GM-CSF) was provided by S. A. Johnston of the University of Texas Southwestern Medical Center, Dallas, Tex.
Male C3H/HeN mice (four mice per group) were injected in the quadriceps muscle with pCDNA3.1 containing TcG1-TcG6 (individually) plus granulocyte-macrophage colony-stimulating factor-encoding plasmid (33 μg of each DNA per mouse). Mice were boosted twice at 3-week intervals and sacrificed 1 week after the last immunization. Polyclonal serum was collected by centrifugation of blood samples at 3,000 × g at 4°C for 10 min. Hyperimmune serum was obtained from mice infected twice with culture-derived trypomastigotes (10,000 parasites per mouse) at a 30-day interval.
ELISA.
Culture-derived T. cruzi lysates (70% amastigotes and 30% trypomastigotes, 1.0 × 109 parasites/ml) (34) were used as a source of T. cruzi antigens for capturing serum antibodies (16). Briefly, 96-well polyvinyl chloride plates (BD Biosciences) were coated overnight at 4°C with 100 μl of T. cruzi antigen/well (5 × 105 parasite equivalents/well). Plates were blocked with 200 μl of 1% nonfat dry milk/well in PBS, washed with PBS-0.05% Tween 20 and PBS, and then incubated for 2 h with test serum (1:20 dilution, 100 μl/well) added in twofold dilutions in triplicate. After washing, plates were incubated for 30 min with 100 μl of horseradish peroxidase-labeled goat anti-mouse immunoglobulin G (1:5,000 dilution in PBS-0.05% Tween 20-1% nonfat dry milk) (Cappel)/well. Color was developed with 100 μl of Sure Blue TMB substrate (Kirkegaard & Perry Laboratories)/well, and the optical density was measured at 650 nm by using an automated enzyme-linked immunosorbent assay (ELISA) microplate reader (Bio-Rad).
Flow cytometry.
Surface expression of the selected antigens was determined by flow cytometric analysis as described previously (17). Briefly, parasites were harvested and washed in PBS containing 0.1% bovine serum albumin and 0.1% sodium azide (PAB). For each assay, 106 parasites were suspended in 50 μl of PAB and sequentially incubated on ice for 30 min with the serum sample (1:2 dilution) and fluorescein isothiocyanate-labeled goat anti-mouse immunoglobulin antibody (1:50 dilution in PAB). Following incubation, parasites were fixed with 2% paraformaldehyde and analyzed by flow cytometry on a FACScan apparatus (BD Biosciences). Parasites stained with anti-MHC antibody (Y3), anti-GPI8 antibody, or normal mouse serum (NMS) were used as negative controls. Positive control antibody was C10 (kindly provided by M. Fresno, Centro de Biologia Molecular, Universidad Autonoma de Madrid, Madrid, Spain) against GP50/55 protein, known to be constitutively expressed as GPI-anchored protein in all stages of parasite development (18). In some experiments, we used parasites stained with serum from chronically infected mice (chronic mouse serum [CMS]) (180 days postinfection) as a positive control. Flow data were analyzed by Cell Quest software (BD Biosciences).
Agglutination-trypanocidal assay.
T. cruzi epimastigotes or trypomastigotes were washed and suspended in RPMI 1640 medium (5 × 104 parasites/25 μl). Epimastigotes were incubated at 37°C with a 25-μl twofold dilution of serum samples. After 4 h of incubation, the live, freely moving parasites were counted by light microscopy. All samples were analyzed in triplicate, and the percent agglutination of epimastigotes was calculated by the following equation: (total parasites − freely moving, unagglutinated parasites after incubation/total parasites) × 100.
To determine the trypanocidal activity of the antigen-specific antibodies, trypomastigotes were incubated with a twofold dilution of serum samples (as described above) ± human complement (25 μl/well). After incubation, live parasites in the presence of 0.03% trypan blue-PBS were counted. Controls included parasites incubated with preimmune serum ± heat-inactivated complement or complement alone. The percent trypanocidal efficiency was calculated by the following equation: (total trypomastigotes − live trypomastigotes after incubation/total parasites) × 100.
Nucleotide sequence accession numbers.
The nucleotide sequences of the genes encoding the selected antigens described here have been submitted to GenBank under accession numbers AY727914, AY27915, AY727916, AY727917, AY727921, AY727918, AY727919, and AY727920 for TcG1 to TcG8, respectively.
RESULTS
Bioinformatic analysis of the EST and GSS databases.
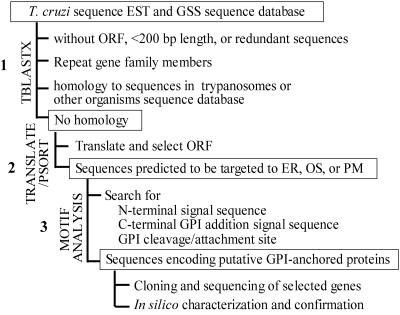
We conducted bioinformatic analysis of the T. cruzi sequence databases to identify novel parasite-specific genes encoding potential vaccine candidates. Our first criterion was the identification of sequences that lack homology to the genes already known in trypanosomes, to the members of the repeat gene families like trans-sialidases and mucins, and to the housekeeping genes or other genes present in higher eukaryotes. The primary criterion for selection as a putative vaccine candidate was the presence of signal sequences or motifs predictive of expression as secreted or membrane-associated proteins. The strategy for this analysis is shown in Fig. 1 and discussed below.
FIG. 1.
Overall strategy for the identification of putative vaccine candidates in T. cruzi. Abbreviations: ER, endoplasmic reticulum; PM, plasma membrane; OS, outside.
We first analyzed the T. cruzi EST and GSS sequence databases for homology to sequences in the NR sequence database at NCBI by using BLASTN and TBLASTX (1, 2). As shown in Table 2, 57.2% of GSS sequences were identified as members of the repeat gene families, including TS, mucins, short interspersed repetitive elements, histones, retrotransposons, heat shock proteins, and ribosomal DNA (4, 13, 45). These large gene families consist of hundreds to thousands of members that share significant sequence homology and interfere with the identification of novel genes. In addition, ∼16% of GSS sequences either were too short (<200 bp), lacked an ORF due to the presence of several stop codons, or consisted of repeats that were not family members but represented genes sequenced and reported multiple times. A majority of the GSS database (>73%), therefore, represented redundant sequences that were not useful for identification of novel genes. In comparison, only 15% of the EST sequences were identified as the repeat gene family members. After filtering out the ESTs that were less than 200 bp or that lacked the ORF, ∼66% of the EST database was NR and considered useful in identifying novel, parasite-specific genes (Table 2).
TABLE 2.
Database match categories of T. cruzi EST and GSS sequences
| Sequence category and subcategory | GSS sequences
|
EST
|
||
|---|---|---|---|---|
| No. | % | No. | % | |
| Total | 1,493 | 100 | 924 | 100 |
| Redundant | 1,094 | 73.2 | 305 | 33.0 |
| Repeat gene family members | 855 | 57.2 | 145 | 15.6 |
| Less than 200 bp or repeat sequences | 225 | 15.0 | 109 | 11.7 |
| No ORF | 14 | 0.93 | 51 | 5.5 |
| Nonredundant | 399 | 26.8 | 619 | 66.0 |
| Database homology to trypanosomes only | 58 | 3.8 | 48 | 5.1 |
| Database homology to trypanosomes and other organisms | 81 | 5.4 | 223 | 24.1 |
| No database homologya | 260 | 17.4 | 348 | 37.6 |
Cutoff for homology was set at an expect value of ≤10−5.
The BLAST search of the NR sequences, i.e., 399 GSS (27%) and 619 EST (66%) sequences, identified homologues in public databases for 29.2% of the EST and 9.2% of the GSS sequences (Table 2). Approximately 5% of the EST and 4% of the GSS sequences included in this group exhibited homology to trypanosomatid sequences in a parasite-specific manner and might be good candidates for drug design. Next, we screened the T. cruzi ORFs exhibiting no homology in public databases, i.e., 348 EST and 260 GSS sequences (Table 2), for the motifs predictive of targeting and cellular localization by PSORT (27), SignalP (28), and DGPI algorithms. This in silico analysis identified 17 EST and 2 GSS sequences exhibiting motifs typical of membrane-associated GPI proteins and/or secreted proteins, which are listed in Table 3.
TABLE 3.
T. cruzi sequences exhibiting characteristics of membrane-associated or secreted proteinsa
| NCBI accession no. | Selected ORF | PSORT localization (P value) | N-terminal signal sequence P value | GPI C/A site | GPI addition signal sequence |
|---|---|---|---|---|---|
| AI0810133 | 5′-3′ frame 2 | PM (0.6) | 0.75 | + | + |
| AQ910890 | 5′-3′ frame 1 | PM (0.6) | 1.0 | + | − |
| AW325178 | 3′-5′ frame 2 | PM (0.4) | 1.0 | − | + |
| AW325227 | 5′-3′ frame 2 | PM (0.6) | 0.75 | + | − |
| AW325291 | 3′-5′ frame 3 | PM (0.6) | 0.25 | + | − |
| AW329906 | 5′-3′ frame 3 | OS (0.3) | 1.0 | + | − |
| AW330105 | 5′-3′ frame 3 | PM (0.4) | 1.0 | + | − |
| AW330110 | 5′-3′ frame 3 | PM (0.6) | 1.0 | + | − |
| AW330132 | 3′-5′ frame 2 | PM (0.6) | 0.75 | + | − |
| AW330166 | 3′-5′ frame 1 | PM (0.9) | 1.0 | + | + |
| AW330173 | 5′-3′ frame 2 | PM (0.3) | 0.5 | + | − |
| AW330187 | 5′-3′ frame 1 | OS (0.8) | 0.75 | + | − |
| AW330209 | 5′-3′ frame 3 | PM (0.4) | 0.5 | + | + |
| AW330228 | 5′-3′ frame 2 | PM (0.7) | 0.75 | + | + |
| AW330247 | 3′-5′ frame 3 | PM (0.4) | 0.75 | + | + |
| AW330324 | 5′-3′ frame 1 | OS (0.8) | 1.0 | − | + |
| AW330353 | 3′-5′ frame 1 | PM (0.6) | 1.0 | + | + |
| AW330404 | 3′-5′ frame 2 | PM (0.6) | 0.0 | + | + |
| AZ050159 | 3′-5′ frame 1 | OS (0.8) | 1.0 | + | − |
As predicted by computational and bioinformatic analysis of EST and GSS databases. Localization of the encoded products to plasma membrane (PM) and outside (OS) was determined by PSORT analysis (probability score, 0 to 1). The presence of N-terminal signal sequence was assessed by SignalP (probability score, 0 to 1), the score of 1 being considered maximum for both PSORT and SignalP. The GPI cleavage-attachment (C/A) site of three amino acids (ω, ω + 1, ω + 2) and a GPI addition signal sequence in the C-terminal region of the ORF were predicted by DGPI. “+” and “−” signs represent the probability of the presence or absence, respectively, of a GPI C/A site or GPI addition signal.
We validated the accuracy of the bioinformatics approach for the identification of ESTs encoding putative membrane-associated proteins by conducting a similar analysis of the TS and mucin gene family members. Trans-sialidases and mucins are well-characterized antigens known to be expressed as GPI-anchored proteins in T. cruzi (10, 11, 36). Of the 30 TS family members analyzed, 17 (56.6%) were predicted by PSORT to be targeted to the plasma membrane. SignalP and DGPI identified an N-terminal signal sequence in 19 (63.3%), a C/A site in 28 (93.3%), and a GPI addition signal in 13 (43.3%) TS family members. Of the 21 mucin gene family members analyzed, all (100%) were predicted to contain an N-terminal signal sequence and a C/A site, 11 (52.3%) were identified to contain motifs for a GPI addition signal, and 15 (71.4%) were identified to be localized on plasma membrane. Collectively, >90% of the TS and mucin family members were accurately predicted by at least two algorithms as membrane-associated GPI proteins.
Molecular characterization of the selected ORFs.
By using universal splice leader as forward primer and the gene-specific reverse primers (designed on the basis of the sequence of the selected 19 EST or GSS sequences), we successfully amplified 13 amplicons by traditional RT-PCR. Sequencing revealed that 5 of the 13 amplicons contain multiple stop codons with dispersed short ORFs and may encode pseudogenes. The remaining amplicons encoded ORFs (TcG1-TcG8) of varying size, with TcG4 and TcG7 being <500 bp; TcG1, TcG2, and TcG6 being 500 to 1,000 bp; and TcG3, TcG5, and TcG8 being >1,000 bp in size (Table 4). Bioinformatic analysis of the cloned sequences (TcG1-TcG8) identified at least two motifs in each encoded protein suggestive of targeting to the secretory pathway and expression as a membrane-associated protein. Interestingly, though no homologues were identified by BLAST screening of the selected EST and GSS sequences, TBLASTX analysis of the amplified products (TcG1-TcG8) resulted in identification of motifs or domains suggestive of functional homologues to four of the cloned genes (Table 4). It is likely that BLAST search efficiency is limited with short EST and GSS sequences of 200 to 400 bp. TcG2, TcG4, TcG6, and TcG7 exhibited no clear paralogue in the NR database. These ORFs, however, showed a significant similarity to chromosome sequence clones from Leishmania, T. brucei, and T. cruzi (Table 4), suggesting that the encoded hypothetical proteins are not an artifact of cloning and in fact exist in T. cruzi as well as in other parasitic protozoans.
TABLE 4.
Selected T. cruzi open reading frames (ORFs) for antigenicity studiesa
| Cloned ORF (bp) | No. of amino acids | Analysis
|
P value | |||
|---|---|---|---|---|---|---|
| TMHMM | SMART | CDD | TBLASTX | |||
| TcG1 (499) | 166 | OS | KOW | rRNA | T. brucei, T. congolense, T. vivax, surface protein | >1e−67 |
| L14 | L. donovani Ldp23 surface protein | 3e−59 | ||||
| L. braziliens L14 ribosomal protein mRNA | 7e−61 | |||||
| TcG2 (662) | 220 | OS | None | None | L. major chromosome 35 clone | 7e−16 |
| TcG3 (1,011) | 337 | TM | DnaJ | DnaJ | T. brucei chaperone DnaJ protein | 5e−09 |
| T. cruzi TcJ1, TcJ2, TcJ3, TcJ4, DnaJ proteins | >1e−07 | |||||
| Mus musculus DnaJ (Hsp40) homolog, Dnajc5 mRNA | 2e−08 | |||||
| Homo sapiens DnaJ (Hsp40) homolog | ||||||
| TcG4 (276) | 92 | OS | None | None | L. major LMAJFVI-Im55bo8.X4 clone | 3e−12 |
| TcG5 (1,350) | 450 | OS | GTP-EFTU | GTP-EFTU | T. cruzi elongation factor 1α gene | 2e−111 |
| T. brucei elongation factor 1α mRNA | 1e−107 | |||||
| L. donovani elongation factor 1α mRNA | 9e−103 | |||||
| Homo sapiens elongation factor 1α mRNA | 1e−108 | |||||
| TcG6 (756) | 252 | OS | None | None | T. brucei chromosome IV clone | 9e−50 |
| L. major chromosome 34 clone | 2e−14 | |||||
| TcG7 (351) | 117 | OS | None | None | TENU3810 T. cruzi epimastigote cDNA library clone | 8e−34 |
| GSSTc09759 T. cruzi random genomic library clone | 4e−29 | |||||
| TcG8 (1,188) | 396 | OS | AAA | AAA | L. donovani ATP-dependent zinc metallopeptidase 1 | 1e−118 |
| Mus musculus ATP-dependent zinc metalloprotease | 1e−68 | |||||
| Homo sapiens AFG3 ATPase family gene 3-like 2 (AFG3L2) | 5e−74 | |||||
ORF, open reading frame predicted by ORF and TRANSLATE software. THMMM lists the possibility of targeting of the protein to outside (OS) or membrane (transmembrane [TM]). SMART and CDD predict the presence of motifs or domains in a protein sequence. TBLASTX lists the best protein search homology results against an NR protein database along with its P value. GTP-EFTU, GTP-dependent elongation factor Tu.
Antigenic potential of the selected candidates.
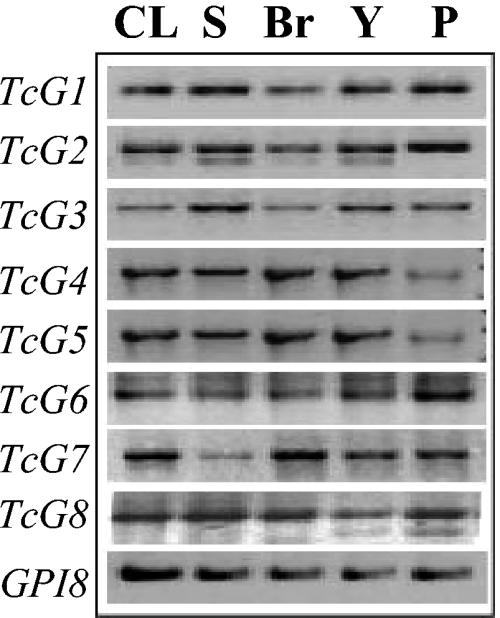
For the selected genes (TcG1-TcG8) to be useful as vaccine candidates, it is essential that they are genomically conserved in a variety of clinically relevant strains and encode antigens that are expressed in different developmental stages of T. cruzi. We performed gene-specific PCR to evaluate the phylogenetic conservation of the selected genes in different T. cruzi strains. We found sequence homologues for all of the eight genes (TcG1-TcG8) in four different strains of T. cruzi by PCR (Fig. 2). No amplification of a product with template DNA incubated with gene-specific forward or reverse primer established the specificity of the PCR (data not shown). Southern blot analysis further confirmed the phylogenetic conservation of TcG1-TcG8 in the clinically relevant parasite strains (data not shown).
FIG. 2.
Phylogenetic conservation of TcG1-TcG8 in various T. cruzi strains. Genes TcG1-TcG8 were amplified in a PCR using gene-specific primers from genomic DNA of CL/Brenner (CL), SylvioX10/4 (S), Brazil (Br), and Y strains of T. cruzi. Plasmid containing the specific gene (P) was used as a template in gene-specific PCR to confirm the accuracy of the reaction. The GPI8 gene was amplified as a positive control. Shown are the amplicons resolved on 1% agarose gel.
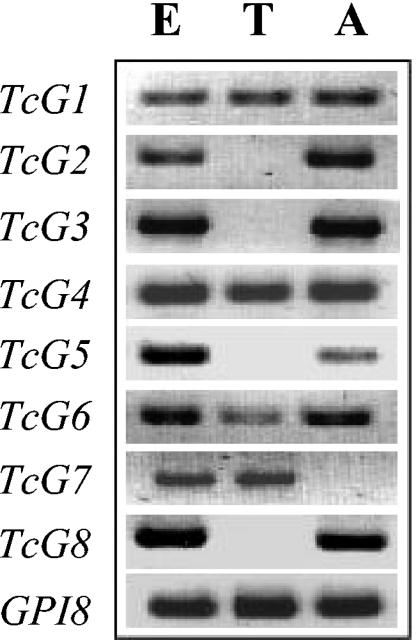
Next, we examined the expression of the selected genes in different developmental stages of T. cruzi. The mRNA level for TcG1-TcG8 was determined by traditional semiquantitative RT-PCR. As shown in Fig. 3, TcG1, TcG4, and TcG6 were amplified from the cDNA of all three stages of T. cruzi. The mRNAs for TcG2, TcG3, TcG5, and TcG8 were detected in epimastigote and amastigote forms, and TcG7 transcripts were amplified from the epimastigote and trypomastigote stages of the parasite. These results suggest that TcG1-TcG8 are expressed in infective (trypomastigote) and/or intracellular (amastigote) stages of the parasite, the clinically relevant mammalian stages. We validated the expression of TcG1-TcG6-encoded antigens in T. cruzi by Western blotting (data not shown).
FIG. 3.
Expression of TcG1-TcG8 in different developmental stages of T. cruzi. Total RNA isolated from epimastigote (E), trypomastigote (T), and amastigote (A) stages of T. cruzi was reverse transcribed, and the cDNA was amplified by PCR amplification with the gene-specific forward and reverse primers. Amplicons were resolved on 1% agarose gel. The GPI8 gene, constitutively expressed in all three stages of T. cruzi, was amplified as a positive control. No amplification was obtained when template cDNAs were incubated in a PCR with gene-specific forward or reverse primers only.
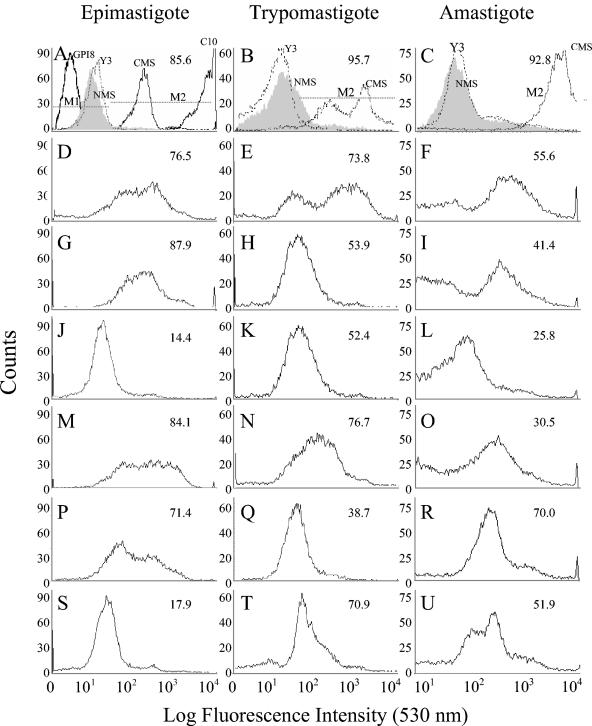
The plasma membrane localization of TcG1-TcG6 in different developmental stages of T. cruzi was determined by flow cytometry. We detected immunofluorescent staining of the epimastigote, trypomastigote, and amastigote forms of T. cruzi with polyclonal serum to the selected antigens (Fig. 4). While polyclonal serum to TcG1 (Fig. 4D, E, and F) stained all parasite stages at a substantial level (>50% positive), maximal staining for TcG2 (Fig. 4G and H) and TcG4 (Fig. 4M and N) was detected at epimastigote and trypomastigote stages. Significant staining with TcG5-specific antibody was detected at epimastigote and amastigote stages (Fig. 4P and R), TcG6-specific antibody recognized trypomastigote and amastigote stages (Fig. 4T and U), and only marginal staining was detected with TcG3-specific antibody (Fig. 4J and L). No fluorescence was detected when parasites were stained with NMS, Y3 antibody to MHC class I, or a polyclonal antibody to intracellular GPI8 protein of T. cruzi (Fig. 4A, B, and C), thus confirming the specificity of the immune staining with antibodies to parasite surface proteins.
FIG. 4.
Flow cytometric analysis to demonstrate surface expression of the selected antigens in T. cruzi. The staining pattern of T. cruzi epimastigote, trypomastigote, and amastigote forms by polyclonal serum to TcG1 (D, E, F), TcG2 (G, H, I), TcG3 (J, K, L), TcG4 (M, N, O), TcG5 (P, Q, R), and TcG6 (S, T, U) was determined by flow cytometry. Background staining with NMS (filled gray areas), antibody to parasite-specific intracellular protein GPI8 (dark solid lines), and nonspecific Y3 antibody to MHC class I (broken lines) are shown in panels A, B, and C. Positive staining with C10 antibody to GP50/55 surface protein (panel A, solid line) and CMS (A, B, C, broken line) is shown. The percentage of fluorescent positive parasites (M2) staining above the background level (M1) are shown in each panel. In panels A, B, and C, the percent positive fluorescence is given for parasites stained with CMS.
Antigenicity of the selected ORFs.
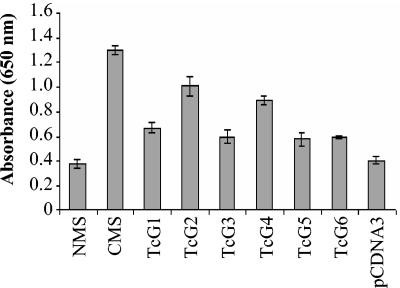
To evaluate the antigenicity of the selected genes, we determined (i) elicitation of the antibody response to TcG1-TcG6-encoded proteins in mice by DNA immunization with antigen-encoding plasmids and (ii) agglutination-trypanocidal activity of the antisera to TcG1-TcG6-encoded proteins. Mice were immunized with the antigen-encoding plasmids, and sera were tested for parasite-specific antibodies by ELISA. As shown in Fig. 5, in all mice immunized with the antigen-encoding plasmid (individually), a parasite-specific antibody response was elicited, with the maximal antibody level elicited in TcG2- and TcG4-immunized mice followed by TcG1-, TcG3-, TcG5-, and TcG6-immunized mice.
FIG. 5.
TcG1-TcG6-encoded antigens elicit parasite-specific antibody response. ELISA was performed with polyclonal sera obtained from mice immunized with TcG1-TcG6-encoding expression plasmids. Sera from normal mice (NMS) and mice immunized with pCDNA 3.1 alone were utilized as negative controls. Serum from chronically infected mice (CMS) was used as a positive control.
We monitored the agglutination-trypanolytic activity of the antigen-specific antibodies, since this property correlates with protection from T. cruzi infection (38, 44). Considering that the epimastigote stage is susceptible to complement in an antibody-independent manner (12), we measured the agglutination of the epimastigotes after incubation with the antigen-specific polyclonal serum. Our data show that the agglutination capacity of anti-TcG2 antibody (>80% at a 1:16 dilution) was similar to that induced in chronically infected mice exposed to multiple parasite proteins (Table 5). The polyclonal sera to TcG3 and TcG6 showed a ∼70 to 80% agglutination efficiency, while TcG1-, TcG4-, and TcG5-specific polyclonal sera exhibited a 60 to 70% agglutination efficiency against epimastigotes at a 1:16 serum dilution (Table 5). No agglutination was observed when epimastigotes were incubated with polyclonal antiserum to GPI8 (a parasite-specific intracellular protein), Y3 antibody to nonspecific MHC class I protein, or NMS, suggesting the specificity of the antibodies to the selected parasite surface antigens in initiating epimastigote agglutination.
TABLE 5.
Agglutination and trypanolytic activity of the TcG1-TcG6-specific polyclonal sera
| Antiseruma | % Agglutination activityb at serum dilution of:
|
% Trypanolytic activityc at serum dilution of:
|
||||
|---|---|---|---|---|---|---|
| 1:4 | 1:8 | 1:16 | 1:4 | 1:8 | 1:16 | |
| NMS | 0 | 0 | 0 | 0 | 0 | 0 |
| Y3 | 0 | 0 | 0 | 0 | 0 | 0 |
| GP18 | 0 | 0 | 0 | 0 | 0 | 0 |
| CMS | 96 | 86 | 80 | 100 | 94 | 86 |
| TcG1 | 90 | 82 | 68 | 92 | 68 | 42 |
| TcG2 | 98 | 90 | 82 | 88 | 72 | 38 |
| TcG3 | 92 | 85 | 76 | 42 | 24 | 0 |
| TcG4 | 84 | 80 | 60 | 94 | 84 | 68 |
| TcG5 | 86 | 74 | 68 | 0 | 0 | 0 |
| TcG6 | 92 | 75 | 72 | 38 | 22 | 0 |
Immune sera were obtained from mice immunized with TcG1-TcG6-encoding plasmids. Parasites incubated with NMS, Y3 antibody to nonspecific MHC class 1 molecule, and polyclonal antibody to intracellular GPI8 protein of T. cruzi were used as controls. Parasites incubated with CMS were used as a positive control.
For agglutination assay, epimastigotes (5 × 104 in 25 μl of RPMI medium) were incubated for 4 h at 37°C with a 25-μl serum sample, and free, nonagglutinated epimastigotes were counted.
For trypanolytic assay, trypomastigotes (5 × 104 in 25 μl of RPMI medium) were incubated for 4 h at 37°C with a 25-μl serum sample plus 25 μl of human complement, and free, live trypomastigotes that did not take trypan blue stain were counted. No trypomastigote lysis was observed with heat-inactivated immune serum alone, complement alone, or heat-inactivated immune serum plus heat-inactivated complement.
The complement-dependent trypanocidal efficiency of the antigen-specific antibodies was measured against trypomastigote stage. As shown in Table 5, trypomastigotes were maximally lysed (88 to 94% at a 1:4 dilution of the polyclonal serum plus complement) by TcG1-, TcG2-, and TcG4-specific polyclonal sera. TcG3- and TcG6-specific polyclonal sera exhibited 38 to 42% trypanocidal efficiency (1:4 serum dilution plus complement). No lysis was observed when trypomastigotes were incubated with heat-inactivated immune serum alone, complement alone, or heat-inactivated immune serum plus heat-inactivated complement (data not shown). Furthermore, no lytic activity was detected when trypomsatigotes were incubated with complement plus anti-GPI8 antibody, Y3 antibody to MHC class I molecule, or NMS, thus confirming the specificity of the lytic activity of the antibodies to the selected surface antigens.
DISCUSSION
In this study, we adopted a web-based bioinformatic screening approach to determine the utility of T. cruzi sequence databases to identify potential vaccine candidates. Our approach was based on six main experimental steps: (i) in silico analysis of the T. cruzi EST and GSS databases to determine the utility of each database in identifying both ORFs and genes encoding putative novel proteins destined for the periphery of the parasite, (ii) assessment of genomic conservation of the selected genes in diverse parasite strains and expression in different developmental stages of T. cruzi, (iii) use of DNA immunization to generate antigen-specific mouse immune sera, (iv) analysis of serum specificity by Western blotting and ELISA, (v) validation of antigen localization by flow cytometry, and (vi) assessment of the trypanolytic activity of the antigen-specific mouse immune sera.
By utilizing this screening approach, we have established the utility of the EST database in identification of ORFs encoding putative membrane-associated proteins that are potential vaccine candidates. The survey of ∼2,500 EST and GSS sequences resulted in the identification of 19 sequences encoding novel, parasite-specific, putative membrane-associated proteins, of which 17 were identified from the EST database. An important observation from the literature that validates the predictions of our experimental approach is that TS and mucin family members have been identified as the membrane-associated proteins expressed as GPI proteins in T. cruzi (10, 11, 36). A majority of these family members have also been classified as surface exposed by our analysis.
Eight of the 19 sequences selected by computational analysis were successfully amplified to obtain the TcG1-TcG8 genes and further characterized at the molecular and antigenicity levels. Our data demonstrate amplification of variable-length TcG1-TcG8 genes from the genomic DNA of diverse, clinically relevant T. cruzi strains (Fig. 2). Interestingly, except for TcG4, none of these genes were amplified from the genomic DNA of Leishmania and T. brucei, although sequence homologues to TcG1-TcG8-encoded proteins were found in a kinetoplastid database (Table 4). In an ELISA assay, the antisera to TcG1-TcG6-encoded proteins did not recognize Leishmania and T. brucei antigens (data not shown). Together, these data suggest that the selected TcG1-TcG8 genes are highly conserved in the T. cruzi genome and that the antigens encoded by these genes might be immunologically unique to T. cruzi.
Interestingly, although we have not found homologues to the selected EST and GSS sequences in public databases, the availability of substantially more sequence information for the cloned ORFs resulted in the identification of homologues for TcG1-, TcG3-, TcG5-, and TcG8-encoded products. Among these, TcG1 and TcG8, identified as surface proteins by computational algorithms, were also recognized as membrane-associated proteins on the basis of significant homology to known surface proteins in public databases (Table 4). TcG1 shows significant similarity to Leishmania Ldp23 by BLAST (P > e−59) and ClustalW (73 and 70% homology at the DNA and protein levels, respectively) analyses. Motif search tools identified the MHC class II binding epitope (KVFDE) in TcG1 that was used to identify Ldp23 (7). Additionally, similar murine MHC class I and class II epitopes and HLA-1 binding motifs are detected in TcG1 and Ldp23. Recombinant Ldp23 protein is shown to stimulate proliferation and production of cytokine (IFN-γ) from T cells purified from the lymph nodes of Leishmania-infected mice (7). The homology of TcG1 to Ldp23 and the presence of similar MHC class I and class II epitopes are in themselves suggestive of surface expression and immunogenic properties of TcG1. In this study, several observations establish the surface expression of TcG1 in different developmental stages of T. cruzi and suggest its antigenic potential as a vaccine candidate. These include the detection of mRNA (Fig. 3) and binding of the antigen-specific antibodies to the plasma membrane (Fig. 4) in all parasite developmental stages, agglutination and trypanocidal activity of the TcG1 polyclonal serum (Table 5), and recognition of TcG1 by the antibodies induced in mice infected by T. cruzi.
TcG8 showed homology to ATP-dependent zinc metalloproteases, members of the AAA ATPase family (30). AAA ATPases function in selective ion transport events, actin-based motility, membrane traffic, and numerous biosynthetic pathways and exist as cytosolic, transmembrane, or membrane-associated proteins (30). The membrane localization of known AAA ATPases such as ABC transporter protein (21) and the significant homology of TcG8 to the AAA domain of members of the AAA ATPase family, however, support the possibility that TcG8-encoded antigen is expressed as a membrane protein in T. cruzi.
TcG3, on the basis of the presence of a DnaJ-binding domain, is identified as a homologue of the DnaJ family of heat shock proteins. In general, DnaJ family members are considered to be cytosolic proteins (20, 23), although expression of DnaJ homologues (Tcj2 and Tcj4) on cellular membranes in T. cruzi has been reported previously (42). The membrane association of TcG3 is suggested by a PSORT probability score of 0.6 for the plasma membrane. We detected immunostaining with anti-TcG3 antibody, albeit at low levels, at all three stages of parasite development (Fig. 4). Similarly, anti-TcG3 polyclonal serum showed marginal agglutination and trypanolytic activity (Table 5). Given the above observations, the inability to detect TcG3-specific mRNA at the trypomastigote stage (Fig. 3), and the low titer of anti-TcG3 antibodies (Fig. 5), we surmise that TcG3 is expressed at a very low level on the parasite surface and may not serve as an excellent antigenic target.
It is interesting that the cloned, full-length TcG5 gene showed significant homology to the gene encoding elongation factor 1α of trypanosomes, noted to be expressed as cytosolic or nuclear protein in different developmental stages (5). The substantial binding of the TcG5-specific antibodies to the surface of epimastigote and amastigote stages in flow cytometry analysis (Fig. 5) and the agglutination capacity against the epimastigote stage (Table 5) are therefore somewhat surprising. However, limited or no expression of TcG5 at mRNA (Fig. 3) and protein (Fig. 4) levels in the trypomastigote stage correlates with the lack of lytic activity of the TcG5-specific polyclonal serum (Table 5) and suggests that TcG5, even if surface expressed, may not be efficiently recognized by the host immune response.
The four genes TcG2, TcG4, TcG6, and TcG7 exhibited no clear paralogue in the public databases. However, for the purpose of assessing the antigenic potential of the products of these genes, our data show that the four genes are expressed, both at the mRNA and protein levels, in the trypomastigote and/or amastigote stage of T. cruzi (Fig. 3 and 4) (our unpublished data). The antigenic potential of TcG2, TcG4, and TcG6 is demonstrated by the observations that the encoded antigens were detected by CMS in Western blotting (unpublished results) and elicited antibodies that bound to the parasite surface (Fig. 4) and exhibited trypanolytic and agglutination activities (Table 5).
In conclusion, we demonstrate the power of bioinformatic analysis and the experimental system reported here in the identification of novel parasite-specific proteins that have potential utility as vaccine candidates. Specifically, our data show that the EST, but not the GSS, database is an efficient resource for the identification of potential ORFs. Screening of the 924 ESTs identified 348 sequences for putative novel proteins of unknown function, of which 17 were identified to encode putative secreted or membrane-associated GPI proteins by our selection criteria. In comparison, screening of 1,493 GSS sequences resulted in the identification of only two ORFs for putative secreted or membrane-associated GPI proteins. Among the selected candidates screened by in vitro studies, TcG1, TcG2, and TcG4 elicited maximally potent trypanolytic antibodies, which is in agreement with the intensity of the surface expression of these antigens in infective and intracellular stages. Accordingly, these candidates are being evaluated to determine their potential to induce protective immunity against T. cruzi infection and disease.
Acknowledgments
This work was supported in part by grants from the American Heart Association (0160074Y), the Sealy Memorial Foundation (2546-01), and the National Institutes of Health (AI053098-01).
Our thanks are due to Ajay Bhatia for computational analysis of the GSS database and Mardelle Susman for critical editing of the manuscript.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel, R. D., A. Bairoch, and D. F. Hochstrasser. 1994. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem. Sci. 19:258-260. [DOI] [PubMed] [Google Scholar]
- 4.Aslund, L., L. Carlsson, J. Henriksson, M. Rydaker, G. C. Toro, N. Galanti, and U. Pettersson. 1994. A gene family encoding heterogeneous histone H1 proteins in Trypanosoma cruzi. Mol. Biochem. Parasitol. 65:317-330. [DOI] [PubMed] [Google Scholar]
- 5.Billaut-Mulot, O., R. Fernandez-Gomez, M. Loyens, and A. Ouaissi. 1996. Trypanosoma cruzi elongation factor 1-α: nuclear localization in parasites undergoing apoptosis. Gene 174:19-26. [DOI] [PubMed] [Google Scholar]
- 6.Brener, Z., and R. T. Gazzinelli. 1997. Immunological control of Trypanosoma cruzi infection and pathogenesis of Chagas' disease. Int. Arch. Allergy Immunol. 114:103-110. [DOI] [PubMed] [Google Scholar]
- 7.Campos-Neto, A., L. Soong, J. L. Cordova, D. Sant'Angelo, Y. A. Skeiky, N. H. Ruddle, S. G. Reed, C. Janeway, Jr., and D. McMahon-Pratt. 1995. Cloning and expression of a Leishmania donovani gene instructed by a peptide isolated from major histocompatibility complex class II molecules of infected macrophages. J. Exp. Med. 182:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Costa, F., G. Franchin, V. L. Pereira-Chioccola, M. Ribeirao, S. Schenkman, and M. M. Rodrigues. 1998. Immunization with a plasmid DNA containing the gene of trans-sialidase reduces Trypanosoma cruzi infection in mice. Vaccine 16:768-774. [DOI] [PubMed] [Google Scholar]
- 10.Crossman, A., J. S. Brimacombe, M. A. Ferguson, and T. K. Smith. 1999. Synthesis of some second-generation substrate analogues of early intermediates in the biosynthetic pathway of glycosylphosphatidylinositol membrane anchors. Carbohydr. Res. 321:42-51. [DOI] [PubMed] [Google Scholar]
- 11.Di Noia, J. M., D. O. Sanchez, and A. C. Frasch. 1995. The protozoan Trypanosoma cruzi has a family of genes resembling the mucin genes of mammalian cells. J. Biol. Chem. 270:24146-24149. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Presas, A. M., J. T. Zavala, I. B. Fauser, M. T. Merchant, L. R. Guerrero, and K. Willms. 2001. Ultrastructural damage of Trypanosoma cruzi epimastigotes exposed to decomplemented immune sera. Parasitol. Res. 87:619-625. [DOI] [PubMed] [Google Scholar]
- 13.Frasch, A. C. 2000. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol. Today 16:282-286. [DOI] [PubMed] [Google Scholar]
- 14.Garg, N., V. L. Popov, and J. Papaconstantinou. 2003. Profiling gene transcription reveals a deficiency of mitochondrial oxidative phosphorylation in Trypanosoma cruzi-infected murine hearts: implications in chagasic myocarditis development. Biochim. Biophys. Acta 1638:106-120. [DOI] [PubMed] [Google Scholar]
- 15.Garg, N., and R. L. Tarleton. 1998. Elicitation of protective cellular and humoral immune responses to Trypanosoma cruzi infection using DNA vaccines can be augmented with cytokines p. 1421-1426. Proceedings of the 10th International Congress of Immunology, New Delhi, India.
- 16.Garg, N., and R. L. Tarleton. 2002. Genetic immunization elicits antigen-specific protective immune responses and decreases disease severity in Trypanosoma cruzi infection. Infect. Immun. 70:5547-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg, N., R. L. Tarleton, and K. Mensa-Wilmot. 1997. Proteins with glycosylphosphatidylinositol (GPI) signal sequences have divergent fates during a GPI deficiency. GPIs are essential for nuclear division in Trypanosoma cruzi. J. Biol. Chem. 272:12482-12491. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Munain, C., J. L. De Diego, P. Bonay, N. Girones, and M. Fresno. 1993. GP 50/55, a membrane antigen of Trypanosoma cruzi involved in autoimmunity and immunosuppression. Biol. Res. 26:209-218. [PubMed] [Google Scholar]
- 19.Higuchi, M. 1999. Human chronic chagasic cardiopathy: participation of parasite antigens, subsets of lymphocytes, cytokines and microvascular abnormalities. Mem. Inst. Oswaldo Cruz 94:263-267. [DOI] [PubMed] [Google Scholar]
- 20.Houry, W. A. 2001. Chaperone-assisted protein folding in the cell cytoplasm. Curr. Protein Pept. Sci. 2:227-244. [DOI] [PubMed] [Google Scholar]
- 21.Jones, P. M., and A. M. George. 2004. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol. Life Sci. 61:682-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, J. M., H. M. Ward, M. A. Miles, and G. Kendall. 1992. A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucleic Acids Res. 20:3963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konieczny, I., and M. Zylicz. 1999. Role of bacterial chaperones in DNA replication. Genet. Eng. (New York) 21:95-111. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., and R. L. Tarleton. 1998. The relative contribution of antibody production and CD8+ T cell function to immune control of Trypanosoma cruzi. Parasite Immunol. 20:207-216. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. G., and L. H. Van der Ploeg. 1997. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu. Rev. Microbiol. 51:463-489. [DOI] [PubMed] [Google Scholar]
- 26.Medina-Acosta, E., and G. A. Cross. 1993. Rapid isolation of DNA from trypanosomatid protozoa using a simple ′mini-prep' procedure. Mol. Biochem. Parasitol. 59:327-329. [DOI] [PubMed] [Google Scholar]
- 27.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 29.Ouaissi, A., E. Guilvard, Y. Delneste, G. Caron, G. Magistrelli, N. Herbault, N. Thieblemont, and P. Jeannin. 2002. The Trypanosoma cruzi Tc52-released protein induces human dendritic cell maturation, signals via Toll-like receptor 2, and confers protection against lethal infection. J. Immunol. 168:6366-6374. [DOI] [PubMed] [Google Scholar]
- 30.Patel, S., and M. Latterich. 1998. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8:65-71. [PubMed] [Google Scholar]
- 31.Planelles, L., M. C. Thomas, C. Alonso, and M. C. Lopez. 2001. DNA immunization with Trypanosoma cruzi HSP70 fused to the KMP11 protein elicits a cytotoxic and humoral immune response against the antigen and leads to protection. Infect. Immun. 69:6558-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plata, F., F. G. Pons, and H. Eisen. 1984. Antigenic polymorphism of Trypanosoma cruzi: clonal analysis of trypomastigote surface antigens. Eur. J. Immunol. 14:392-399. [DOI] [PubMed] [Google Scholar]
- 33.Rassi, A., Jr., A. Rassi, and W. C. Little. 2000. Chagas' heart disease. Clin. Cardiol. 23:883-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos, M. A., N. Garg, and R. L. Tarleton. 1997. The identification and molecular characterization of Trypanosoma cruzi amastigote surface protein-1, a member of the trans-sialidase gene super-family. Mol. Biochem. Parasitol. 86:1-11. [PubMed] [Google Scholar]
- 35.Santos-Buch, C. A., and A. M. Acosta. 1985. Pathology of Chagas' disease, p. 145-183. In I. Tizard (ed.), Immunology and pathology of trypanosomiasis. CRC Press, Boca Raton, Fla.
- 36.Schenkman, S., M. A. Ferguson, N. Heise, M. L. de Almeida, R. A. Mortara, and N. Yoshida. 1993. Mucin-like glycoproteins linked to the membrane by glycosylphosphatidylinositol anchor are the major acceptors of sialic acid in a reaction catalyzed by trans-sialidase in metacyclic forms of Trypanosoma cruzi. Mol. Biochem. Parasitol. 59:293-303. [DOI] [PubMed] [Google Scholar]
- 37.Schnapp, A. R., C. S. Eickhoff, D. Sizemore, R. Curtiss III, and D. F. Hoft. 2002. Cruzipain induces both mucosal and systemic protection against Trypanosoma cruzi in mice. Infect. Immun. 70:5065-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sepulveda, P., M. Hontebeyrie, P. Liegeard, A. Mascilli, and K. A. Norris. 2000. DNA-based immunization with Trypanosoma cruzi complement regulatory protein elicits complement lytic antibodies and confers protection against Trypanosoma cruzi infection. Infect. Immun. 68:4986-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh, H., and G. P. S. Raghava. 2001. ProPred: prediction of HLA-DR binding sites. Bioinformatics 17:1236-1237. [DOI] [PubMed] [Google Scholar]
- 40.Stoppani, A. O. 1999. The chemotherapy of Chagas disease. Medicina (Buenos Aires) 59:147-165. (In Spanish.) [PubMed] [Google Scholar]
- 41.Tarleton, R. L. 1996. Immunity to Trypanosoma cruzi, p. 227-247. In S. Kaufmann (ed.), Host response to intracellular pathogens. R.G. Landes Company, Austin, Tex.
- 42.Tibbetts, R. S., J. L. Jensen, C. L. Olson, F. D. Wang, and D. M. Engman. 1998. The DnaJ family of protein chaperones in Trypanosoma cruzi. Mol. Biochem. Parasitol. 91:319-326. [DOI] [PubMed] [Google Scholar]
- 43.Tomlinson, S., F. Vandekerckhove, U. Frevert, and V. Nussenzweig. 1995. The induction of Trypanosoma cruzi trypomastigote to amastigote transformation by low pH. Parasitology 110:547-554. [DOI] [PubMed] [Google Scholar]
- 44.Umekita, L. F., and I. Mota. 2000. How are antibodies involved in the protective mechanism of susceptible mice infected with T. cruzi? Braz. J. Med. Biol. Res. 33:253-288. [DOI] [PubMed] [Google Scholar]
- 45.Vazquez, M., H. Lorenzi, A. G. Schijman, C. Ben-Dov, and M. J. Levin. 1999. Analysis of the distribution of SIRE in the nuclear genome of Trypanosoma cruzi. Gene 239:207-216. [DOI] [PubMed] [Google Scholar]
- 46.Wizel, B., N. Garg, and R. L. Tarleton. 1998. Vaccination with trypomastigote surface antigen 1-encoding plasmid DNA confers protection against lethal Trypanosoma cruzi infection. Infect. Immun. 66:5073-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization. 1999. Chagas disease: tropical diseases progress in research, 1997-1998. WHO Tech. Rep. Ser. 14:31-42. [Google Scholar]
- 48.World Health Organization. 1997. Tropical disease research progress 1995-1996. WHO Tech. Rep. Ser. 1:108-139. [Google Scholar]