Abstract
Type IV pili, filamentous surface appendages primarily composed of a single protein subunit termed pilin, play a crucial role in the initiation of disease by a wide range of pathogenic bacteria. Although previous electron microscopic studies suggested that pili might be present on the surface of Moraxella catarrhalis isolates, detailed molecular and phenotypic analyses of these structures have not been reported to date. We identified and cloned the M. catarrhalis genes encoding PilA, the major pilin subunit, PilQ, the outer membrane secretin through which the pilus filament is extruded, and PilT, the NTPase that mediates pilin disassembly and retraction. To initiate investigation of the role of this surface organelle in pathogenesis, isogenic pilA, pilT, and pilQ mutants were constructed in M. catarrhalis strain 7169. Comparative analyses of the wild-type 7169 strain and three isogenic pil mutants demonstrated that M. catarrhalis expresses type IV pili that are essential for natural genetic transformation. Our studies suggest type IV pilus production by M. catarrhalis is constitutive and ubiquitous, although pilin expression was demonstrated to be iron responsive and Fur regulated. These data indicate that additional studies aimed at elucidating the prevalence and role of type IV pili in the pathogenesis and host response to M. catarrhalis infections are warranted.
Moraxella catarrhalis is now recognized as an important human pathogen in both children and adults (12, 16, 19, 33, 49). This organism is a significant cause of otitis media and sinusitis in young children and also causes lower respiratory tract disease in adults, particularly those with chronic lung disease (12, 33, 34). In addition, a few reports have described nosocomial spread of this bacterium in respiratory wards (6-9, 35). The extremely high carriage rates reported in children, coupled with the fact that over 90% of M. catarrhalis clinical isolates are β-lactamase positive, suggest that infections with these organisms may increase (12, 48). Multiple studies, including those from our laboratory, have described specific bacterial components as potential virulence factors (for a recent review, refer to reference 49). Although little information is available regarding the actual steps involved in the pathogenesis of M. catarrhalis infections, it is clear that these organisms must attach to the human mucosal surface in order to establish colonization. Therefore, the identification of bacterial colonization factors and of new vaccine and treatment targets is a major focus of present research efforts. In this study, we describe the identification and characterization of the genes that are involved in the biosynthesis and assembly of M. catarrhalis pili.
Pili are homo- or heteropolymers composed of helically arranged subunits assembled and expressed on the surface of a broad spectrum of gram-negative bacteria and can be classified based on morphology and function. Type IV (MePhe) pili, classified based on amino acid similarities among their major pilin subunits and their assembly mechanisms, are expressed by a variety of important mammalian pathogens, including Neisseria gonorrhoeae, Neisseria meningitidis, Pseudomonas aeruginosa, Vibrio cholerae, Moraxella bovis, Eikenella corrodens, and Dichelobacter nodosus (13, 18). The highest homology between type IV pilins is located in the amino terminus, although a conserved disulfide bridge exists in the C terminus of the protein. The biogenesis and regulation of type IV pili are complex, involving a large number of different proteins that are related to the general type II secretion system of gram-negative bacteria, filamentous phage assembly, and bacterial DNA uptake systems (5, 18).
Type IV pili, usually polarly located rod-like fibers of variable length, are crucial factors for infectivity and disease manifestations of many pathogenic bacteria, as they play an essential role in colonization of host tissues (43). Besides mediating attachment to host cells, the type IV pili from a wide range of bacteria appear to be multifunctional organelles, since pilus expression may also be correlated with twitching motility, a form of bacterial translocation over moist surfaces that is a result of pili retraction, and biofilm formation and stability (31, 36). In addition, the expression of type IV pili is correlated with highly efficient transport of DNA across the bacterial membrane in many bacterial species. This mechanism of DNA transformation is a major contributor to the horizontal exchange of genetic information between naturally competent microorganisms (1, 24). Therefore, the molecular basis of pilus synthesis and assembly in M. catarrhalis and the phenotypic analysis of defined isogenic pil mutants is of considerable interest, as type IV pili have been shown to be involved in colonization and virulence in many important human mucosal pathogens.
Although M. catarrhalis has been previously reported to express type IV pili that mediate adherence to epithelial cells in vitro, those studies used electron microscopy (EM) and whole-cell trypsin treatments as a basis for their conclusions (2-4, 29, 38). In addition to the EM analyses, one study used genomic Southern hybridizations with an M. bovis pilin gene probe to identify a detectable band of hybridization to M. catarrhalis DNA; however, the authors concluded that their data did not discern whether type IV pili were expressed on the bacterial surface or if a complete pilin gene was even present within the genome (29). Therefore, definitive evidence for the expression of type IV pili by this bacterium was lacking, and the identification and characterization of the genes involved in the biogenesis and function of this surface structure by M. catarrhalis have not been reported to date.
In this report, we have identified M. catarrhalis type IV pilus biogenesis homologues and have cloned the genes encoding PilA, PilQ, and PilT. Comparative analyses of the wild type and the three isogenic pil mutants demonstrate that M. catarrhalis expresses type IV pili that are essential for natural competence. Our studies also indicate that pilin expression by M. catarrhalis is iron responsive and Fur regulated. In addition, we have detected the presence of pilin transcripts in a panel of clinical isolates from various geographic locations, indicating type IV pilus expression by M. catarrhalis may be ubiquitous in this human mucosal pathogen.
MATERIALS AND METHODS
Bacteria and culture conditions.
M. catarrhalis strains were routinely cultured at 35.5°C on brain heart infusion (BHI) agar in a 5% CO2 atmosphere as described previously (14, 25). Broth cultures in BHI were grown in a 37°C shaking water bath with constant agitation. The global panel of clinical strains, isolated from various geographic areas around the world from both adults and children, was kindly provided by Mark Achtman (Max Planck Institute, Berlin, Germany) and Timothy Murphy (Veterans Administration Research Center, Buffalo, N.Y.). The pediatric middle ear isolate M. catarrhalis 7169 (28) was used to construct the PilA-, PilT-, and PilQ-deficient mutants. The M. catarrhalis mutants were cultured as described elsewhere with the addition of kanamycin (25 μg per ml) or chloramphenicol (0.5 μg per ml) as required. Comparative growth and autoagglutination analyses of M. catarrhalis were performed as previously described (25, 37). Escherichia coli XL1-Blue was used as the host strain for plasmid DNA manipulations, and E. coli H1717, provided by Terry Connell (University at Buffalo, Buffalo, N.Y.), was used as the host strain for the Fur titration assays (FURTAs). E. coli strains were cultured at 37°C using Luria-Bertani agar plates and broth with antibiotic supplementation as required (100 μg of ampicillin per ml, 40 μg of kanamycin per ml, and 30 μg of streptomycin per ml).
General DNA manipulations.
Restriction endonucleases and standard molecular biology reagents were obtained from New England Biolabs, Inc. (Beverly, Mass.), platinum Taq DNA polymerase High Fidelity was purchased from Invitrogen (Carlsbad, Calif.), and the pGEM-T Easy vector system was acquired from Promega (Madison, Wis.). Plasmid isolation and amplicon purification were performed using QiaPrep spin kits and MinElute kits, respectively (QIAGEN, Santa Clarita, Calif.). Restriction enzyme digestions, ligations, and transformations by electroporation were performed using standard methods. Chromosomal DNA was isolated as previously described (39). PCR amplifications of chromosomal DNA were performed for 25 cycles with the GeneAMP PCR system 9700 (PE Applied Biosystems, Foster City, Calif.), and annealing temperatures and extension lengths were primer set dependent. DNA nucleotide sequences of all constructs were obtained via automated DNA sequencing (RPCI Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, N.Y.) and analyzed with MacVector version 7.2 and the Wisconsin sequence analysis package (Genetics Computer Group, Madison, Wis.).
Identification and cloning of pilA, pilT, and pilQ.
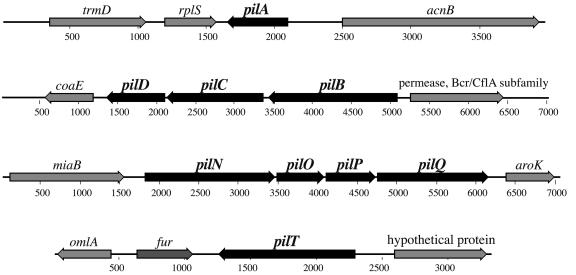
M. catarrhalis pil biogenesis homologues pilA, -B, -C, -D, -N, -O, -P, -Q, and -T were identified by BLAST searches of the patented M. catarrhalis genome (deposited under Incyte Genomics patent number WO0078968 located in the National Center for Biotechnology Information nucleotide database) using P. aeruginosa pilus-related gene sequences. PCR primers for cloning the 7169 pilA (primers 428 and 429), pilT (primers 413 and 414), and pilQ (primers 454 and 455) homologues were designed using the putative coding regions identified in the genome sequence (Table 1; Fig. 1). Sequence analysis of each amplicon indicated the identities between the genome sequence and strain 7169 sequence were 100% for both PilA and PilQ and 99.4% for PilT.
TABLE 1.
Nucleotide sequences of oligonucleotide primers used for PCR-based cloning procedures and RT-PCR analyses
| Primer | Sequence (5′-3′) |
|---|---|
| 413 | CTGTGATGTGTGCGGTAAG |
| 414 | CAACCAAGCGATTATCCC |
| 417 | AGATCTGGGTGACTAACTAGGAGGAATAAATGGCTAa |
| 424 | CGCTTCAGATTTACATTTATCCGC |
| 425 | ATTTGTTCGTTGGCAGTGG |
| 428 | TTCTGCCATTTGCCCAACCC |
| 429 | AACAAGCGTTACTTCTCCACTG |
| 430 | CGGTGTTTTGGCGATGTTTG |
| 431 | TTTTGGAGTTGCTTGGGATGTC |
| 454 | CATCTGATTGAAGCGGTC |
| 455 | AAATGTTTGGCAAGCAGC |
| 491 | CTCGAGGTCGACTCTAGAGGATCCCCGGGTCATTAb |
| 597 | GTTACCCAAACAGCAGAG |
| 598 | ATACCACCAAGAACGACG |
| 631 | CTCGAGGCTGTTTTTAGTGGCAATGb |
| 632 | AGATCTCCTTGTTGTGCTTTCATACGa |
| 633 | CATATGCCTCAATACCAAAAACGCc |
| 634 | GGATCCTAGTTAAGTTTTTGGAGTTGCd |
| 655 | CTCGAGGAGGTTATCTCAACACCAAAAGTCb |
| 656 | AGATCTGGTCATACTGCCTGTCACACTATCa |
| 657 | CTCGAGTGCCCCCAAAGAGCATTGACCb |
| 658 | AGATCTTACATTCACCCGAAAGCGAGCGa |
| 709 | ATGGCTGTCTCATCGCATC |
| 710 | ACTCCTCTGGTGTTGGTTCAC |
| 780 | GAAAGCACAACAAGGATTTACCTCTC |
| 781 | AAGCACATCTCAACAGCCCG |
| 782 | CCGATGATAGTGTGACAGGC |
| 783 | CCGTGATAATGGTGCTTCAG |
Engineered BglII site is underlined.
Engineered XhoI site is underlined.
Engineered NdeI site is underlined.
Engineered BamHI site is underlined.
FIG. 1.
Genetic organization of the M. catarrhalis pil biosynthesis-related homologues. Based on homologies to other systems, nine M. catarrhalis putative coding sequences with homology to genes involved in the biogenesis and assembly of type IV pili were identified (depicted in black). Thick arrows represent the size and orientation of the putative open reading frames identified. The pilA, pilT, and pilQ homologues were selected for further analysis.
Construction of isogenic mutants.
Our laboratory has previously used the nonpolar mutagenesis cassette aphA-3 from pUC18K to successfully construct isogenic mutants in M. catarrhalis (14, 25-28), and this resistance determinant was used to insertionally inactivate pilA, pilQ, and pilT in strain 7169 by an inverse PCR strategy. To briefly summarize, the strain 7169 pilA was cloned into the TA cloning vector pGEM-T Easy following PCR amplification with primers 428 and 429, resulting in pPilA10. Following sequence analysis, pPilA10 was used as the template in a PCR with primers 631 and 632, which contained engineered XhoI and BglII sites, respectively. This resulted in a deletion of 310 nucleotides (nt) internal to the 453-nt pilA coding region. aphA-3 was amplified from pUC18K using primers 417 and 491, which contained the corresponding restriction endonuclease sites for directional cloning. The amplicons from both reactions were purified and subjected to restriction digestion and ligation, resulting in pPILAk4. The same inverse PCR deletion-insertion mutagenesis strategy was employed to generate mutagenesis constructs for both pilQ and pilT, except that first an internal region of each coding sequence was amplified using primers 597 and 598 (1,178 nt of the 1,422-nt pilQ coding region) and primers 424 and 425 (984 nt of the 1,050-nt pilT coding region), respectively, and cloned in pGEM-T Easy. As described above, primers specific to either pilQ (primers 655 and 656) or pilT (primers 657 and 658) and containing engineered XhoI and BglII restriction sites were used in inverse PCRs with the internal pilQ and pilT plasmid constructs as templates, and the purified amplicons were restriction digested and ligated to aphA-3 as described above. This resulted in the generation of pPILQk6, which contained the 850-bp aphA-3 kanamycin resistance determinant within a 446-nt deletion of the pilQ coding region, and pPILTk3, with aphA-3 inserted within a 532-nt deletion of the pilT coding region. After verification by sequence analysis, the pPILAk4, pPILTk3, and pPILQk6 mutagenesis constructs (containing a nonpolar insertion of the aphA-3 resistance determinant within an internal deletion of the coding region, such that the ATG codon 3′ of aphA-3 was placed in frame with the remainder of the coding region for each gene) were amplified by PCR with gene-specific primers, and the resulting amplicons were purified and used to naturally transform M. catarrhalis 7169. In brief, a 100-μl aliquot of 7169 bacterial suspension (optical density at 600 nm [OD600] of 0.2) was plated onto BHI agar, and 20 ng of the purified DNA was spotted onto a portion of the bacterial lawn. After incubation for 5 h under standard growth conditions, the area of the bacterial lawn that had been inoculated with the mutagenesis construct was swabbed onto selective plates containing kanamycin. Insertional inactivation by the aphA-3 mutagenesis constructs was verified by sequence analysis of amplicons obtained from PCR analysis of chromosomal DNA prepared from several transformants from each mutagenesis event, with primers designed to flank the predicted site of cassette insertion. An insertionally inactivated pilA, pilQ, and pilT mutant from each transformation was selected for further study and termed 7169::pilAK4, 7169::pilQK6, and 7169::pilTK3, respectively.
Preparation of enriched surface-sheared pili and whole-cell lysates.
The shearing procedure for releasing cell surface components was performed as described elsewhere (40, 50) with the following modifications. In brief, bacteria were harvested after 16 to 18 h of growth on BHI agar plates, resuspended in 1.0 ml of sterile phosphate-buffered saline (PBS), vortexed vigorously for 1 min, and centrifuged at 12,000 × g for 10 min to separate the bacterial cells (pellet fraction) from the pilus-enriched supernatant (sheared fraction). The supernatants were collected, filter sterilized through a 0.2-μm-pore-size filter, and concentrated fivefold using 10,000 MWCO Amicon Centricon filtration devices (Millipore, Bedford, Mass.). Protein concentrations of the sheared surface components were quantitated using the Sigma Lowry protein assay kit and adjusted to equivalent amounts prior to electrophoresis. Whole-cell lysates were prepared concurrently by suspending plate-grown organisms in 10 ml of PBS to an OD600 of 0.3, collecting the bacteria by centrifugation, and resuspending the cell pellet with 100 μl of distilled H2O. Both preparations were analyzed by sodium dodecyl sulfate-14% polyacrylamide gel electrophoresis (SDS-PAGE) for immunoblot analyses, using our standard methods (10, 11).
RNA purification.
Total RNA was isolated using the RNeasy Mini kit (QIAGEN) following stabilization with RNAlater (Ambion) according to the manufacturer's recommendations. M. catarrhalis was inoculated to an OD600 of 0.06 in broth culture and incubated at 37°C with rotary shaking at 225 rpm. At 1 h (lag to early log), 3.5 h (early to mid-log), 5.5 h (mid- to late log), and 7.5 h (late log to early static) time points, or from agar-grown bacteria suspended to an OD600 of 0.3 in sterile PBS, aliquots were removed and processed either for RNA extraction or as whole-cell lysates for concomitant protein analysis. Purified RNA samples were subjected to RNase-free DNase treatment (Promega) to remove any residual contaminating chromosomal DNA, and the RNA was subjected to PCR analysis to verify purity. RNA concentrations were determined by measuring absorbance on an Eppendorf BioPhotometer, and 2 μg of total RNA from each sample was converted to cDNA by using the Applied Biosystems high-capacity cDNA archive kit according to the manufacturer's specifications in a final reaction volume of 100 μl; control reactions with no reverse transcriptase (RT) were also performed. cDNA was used for PCR analysis of mRNA transcripts at a concentration of 50 ng per reaction mixture, except for analysis of iron regulation of pilin expression, when 20 ng (final amount) was used. Primer sets, designed internal to the predicted coding sequence and used in RT-PCR analyses, included pilA-specific primers 430 and 431 and 780 and 781, pilQ-specific primers 782 and 783, and rpoD-specific primers 709 and 710.
Quantitative DNA transformation assay.
The quantitative transformation assay was based on our standard method for naturally transforming M. catarrhalis with the following modifications. Bacteria were grown on BHI agar plates for 16 to 18 h and suspended to an OD600 of 0.2 in PBS, and 100-μl aliquots were plated onto fresh plates. After 30 min at 35.5°C, 1 μl of a 100-ng/μl solution of purified linear DNA (M. catarrhalis omp103::CAT) was spotted onto the lawn in duplicate for each strain. Omp103 is a conserved membrane protein expressed by M. catarrhalis (unpublished data), and the omp103::CAT mutagenesis construct (containing the chloramphenicol resistance determinant from pACYC184 inserted into a 1.4-kb deletion within the 2.7-kb omp103 coding region) was kindly provided by Kristin Furano. A 3-mm-diameter agar plug was removed from the site of DNA inoculation after 5 h of incubation under standard growth conditions and transferred to 1 ml of sterile PBS. After vortexing, serial dilutions were plated onto agar plates with and without chloramphenicol supplementation. After overnight incubation, CFU were determined for both the selective and nonselective media. Transformation frequencies are expressed as the transformant CFU per total CFU and represent the average of five highly reproducible independent assays.
Construction and purification of rPilA.
The pET expression system (Novagen, Madison, Wis.) was used according to the manufacturer's recommendations to generate recombinant PilA (rPilA). In brief, gene-specific primers 633 and 634 containing engineered NdeI and BamHI restriction sites were used to amplify nt 82 through 453 (corresponding to amino acids 123 to 150) of the pilA coding sequence, using strain 7169 genomic DNA as the template in PCRs. The N terminus of the pilin was not included, to prevent oligomerization and optimize solubility of monomeric pilin fusion proteins as previously described for P. aeruginosa (20). Purified amplicons were double digested with NdeI and BamHI and cloned in frame with the N-terminal His6 tag by ligation into the expression vector pET16b digested with the same endonucleases. Following transformation into competent E. coli BL21(DE3)pLysS, recombinant plasmids were isolated and the orientation of the insert was confirmed by DNA sequence analysis. Specific overexpression and purification of the His6-tagged fusion proteins were performed according to the recommended protocol (Novagen), summarized as follows: transformed bacteria were grown to mid-exponential phase (OD600 of ∼0.6) at 37°C with vigorous aeration, and then recombinant protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h. The cells were harvested by centrifugation, and the resulting pellet was resuspended in BugBuster protein extraction reagent with benzonase nuclease (Novagen) using 5 ml of reagent per gram of cells according to the manufacturer's specifications. The extract was clarified by centrifugation to separate the soluble protein fraction (supernatant) and insoluble fraction (pellet), and both the pellet and supernatant were analyzed for the solubility and expression of the recombinant protein by SDS-PAGE analysis. Soluble His6-tagged rPilA was purified by metal exchange chromatography using the Ni2+-charged His · Bind purification kit (Novagen) according to the manufacturer's instructions. Eluate sample fractions were spectrophotometrically measured at an absorbance of 280 nm, and protein-containing fractions were pooled, concentrated using Amicon Centricon filtration devices (10,000 MWCO), and analyzed by SDS-PAGE for purity.
EM.
To analyze piliation phenotype, bacteria were grown for 18 to 20 h on agar plates, and poly-l-lysine treated (1 μg per ml) Formvar-coated grids (Ladd Research Industries, Inc.) were used to lift cells from colonies. The grids were then fixed, negatively stained, and viewed as described previously (21, 23).
MAb 4G9.
Monoclonal antibody (MAb) 4G9 was developed by injecting BALB/C mice twice, at a 2-week interval, with 37.5 μg of purified rPilA and performing a fusion using standard methods as previously described (10).
FURTAs.
The FURTA technique, performed essentially as described by Stojiljkovic et al., employs a Fur-regulated lacZ fusion as a reporter gene that allows the detection of transformants carrying multicopy Fur binding sites as Lac+ colonies on MacConkey agar plates (45). In brief, pPILAk6 (the mutagenesis construct used to develop the pilA isogenic mutant which contains the 282 nt of sequence 5′ to the pilA ATG) and paphA-3 (primers 417 and 491 were used to amplify aphA-3 from pUC18K,and the resulting amplicon was cloned into pGEM-T easy and used as a negative control) were introduced into E. coli H1717 carrying the chromosomal Fur-repressible fhuF::lacZ fusion. After 16 h of growth at 37°C, ampicillin-resistant transformants were screened for the Lac+ phenotype on MacConkey lactose agar plates (Difco) in the presence and absence of 50 μM ferrous ammonium sulfate supplementation. In the presence of iron, Fur binds to the fhuF promoter region and, therefore, the lacZ reporter gene is not expressed and colonies are white on MacConkey lactose agar plates. However, Fur will not bind to the fhuF promoter region in the absence of iron or in the presence of both iron and Fur binding sequences carried on a multicopy plasmid that will titrate out the Fur protein from binding sites within the reporter construct. In both of these latter cases, the lacZ reporter gene is expressed and the colonies appear red on MacConkey lactose plates. Three independent growth assays were performed in duplicate.
Nucleotide sequence accession number.
The nucleotide sequences of the M. catarrhalis 7169 pilA, pilQ, and pilT genes have been deposited with GenBank under accession numbers AY647185, AY647186, and AY647187, respectively.
RESULTS
Identification of pil biosynthesis homologues.
Using genes characterized to encode the proteins essential for pilin filament assembly and function in P. aeruginosa as queries for BLAST searches of the patented M. catarrhalis nonannotated genome, deposited through the National Center for Biotechnology Information, we identified two separate gene clusters (pilBCD and pilNOPQ) and two noncontiguous coding sequences (pilA and pilT) potentially encoding components of type IV pilin assembly located at the inner and outer membrane (Fig. 1). To begin a defined phenotypic and genetic investigation of pilus expression by M. catarrhalis, we initially selected the putative pilA, pilT, and pilQ homologues for further analyses. Primers designed to the regions flanking the putative coding sequences were used to amplify the corresponding regions from M. catarrhalis strain 7169, and the resulting amplicons were subjected to DNA sequence analysis.
Similar to the major pilin subunit-encoding genes of the gonococci and the meningococci, and in contrast to the V. cholerae and P. aeruginosa homologues, the M. catarrhalis pilA was not located within a pilus assembly gene cluster. The deduced 150-amino-acid PilA polypeptide exhibited significant sequence similarity to the major type IV pilin structural subunit of other type IV pilus-expressing bacteria (data not shown). The type IV pilins from other mucosal pathogens, including P. aeruginosa, N. gonorrhoeae, M. bovis, and D. nodus, comprise 145 to 160 amino acids and have a similar predicted structure (18). Consistent with other type IV pilin subunits, the N terminus of the M. catarrhalis PilA is predicted to be highly hydrophobic and contained the consensus short polar signal sequence followed by a phenylalanine residue that is predicted to be methylated following proteolytic processing. In addition, the +5 glutamate is conserved in the M. catarrhalis PilA, a residue previously shown to be essential for both methylation of the mature pilin and subunit oligomerization into the helical pilin filament. The C terminus of the protein contains a putative disulfide bridge that spans 13 amino acids. The disulfide bridge of pilin subunits, typically spanning 12 to 15 amino acids, has been shown to be essential for pilin-mediated adherence and interactions with host tissues (31). To facilitate analysis of type IV pilus expression by M. catarrhalis, an rPilA fusion protein was constructed that lacked the hydrophobic N-terminal oligomerization domain and was used to generate a PilA-specific MAb termed 4G9.
The M. catarrhalis PilT has 64 to 72% similarity to other bacterial PilT homologues. PilT is a nucleotide binding protein (ATPase) that mediates pilin filament disassembly and is homologous to PilB, which has the opposite effect and is required for pilin filament assembly (53). Although PilB homologues exist in other systems, PilT is unique to type IV pilus biogenesis (32). The M. catarrhalis PilQ (50 to 63% similarity to other PilQ homologues) is located within an operon of genes involved in functions attributed to pilin expression, as observed in other organisms expressing type IV pili (31). PilQ is a member of a large family of proteins termed secretins, which form gated pores in the bacterial outer membrane. In type IV pilus systems, the pilus filament is extruded through a dodecameric donut-shaped outer membrane complex comprised of PilQ (31).
Comparative analysis of the wild-type and pil mutant M. catarrhalis strains.
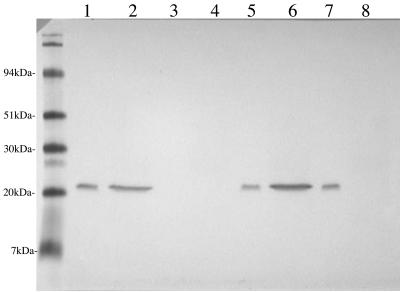
In order to begin functional analysis of type IV pilus expression by M. catarrhalis, isogenic mutants deficient in PilA, PilT, or PilQ expression (termed 7169::pilAK4, 7169::pilTK3, and 7169::pilQK6, respectively) were constructed. To investigate the localization of PilA in the wild-type and mutant strains, whole bacterial lysates and sheared surface components were analyzed by SDS-PAGE and MAb 4G9-probed immunoblotting. Comparative analysis of the mutants indicated that whereas wild-type 7169 contained both pilin subunits within the cell as well as on the surface (Fig. 2, lanes 1 and 2), 7169::pilAK4 exhibited the expected PilA-null phenotype (lanes 3 and 4). 7169::pilTK3 appeared to have slightly more pilin in the sheared surface components than the wild-type strain (lane 6), and 7169::pilQK6 contained pilin subunits within the cell but not on the cell surface (lanes 7 and 8). This result was consistent with the expected phenotypes of the three mutant strains based on the predicted homology of these proteins to essential components of type IV pilus systems. Additional comparative analyses of the wild-type and three isogenic mutant strains revealed that the lack of either PilA, PilQ, or PilT expression had no discernible effect on bacterial growth characteristics or autoagglutination properties (data not shown).
FIG. 2.
To investigate the localization of PilA in the wild-type and mutant strains, whole bacterial lysates (WCL; odd lanes) or sheared surface components (SS; even lanes) were analyzed by SDS-PAGE and immunoblotting. Comparative analysis of the MAb 4G9-probed immunoblot indicated that whereas the wild-type 7169 contained pilin subunits both within the cell and on the surface (lanes 1 and 2), pilAK4 exhibited the expected PilA-null phenotype (lanes 3 and 4), pilTK3 appeared to have slightly more pilin in the SS (lanes 5 and 6), and pilQK6 contained pilin subunits within the cell but not on the cell surface (lanes 7 and 8). Molecular size standards are shown in kilodaltons.
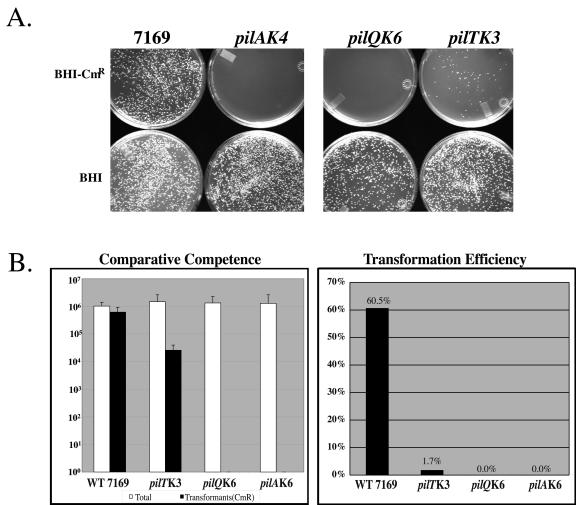
Expression of type IV pili is correlated with natural genetic transformation in many bacterial species, although not all type IV pilus-expressing organisms are competent for DNA uptake (31). Although the mechanism by which type IV pili mediate natural transformation remains unknown, nonpiliated or loss-of-function mutants have been shown to be severely impaired in transformation frequencies, with an approximately 1,000-fold reduction in DNA uptake (31, 47). The contribution of M. catarrhalis pili to DNA competence was investigated in a quantitative transformation assay. As summarized in Fig. 3, the pil mutants were defective in DNA uptake compared to the wild-type strain. Whereas 60% of DNA-exposed wild-type 7169 cells were consistently transformed by the selectable marker (M. catarrhalis omp103::CAT), less than 2% of the pilT mutants were competent for DNA uptake, and both the PilA- and PilQ-null mutants were completely nontransformable (Fig. 3B).
FIG. 3.
(A) The contribution of M. catarrhalis pili to DNA competence was investigated in a quantitative transformation assay (a representative set of plates is shown). (B) These data indicate that the pil mutants are defective in DNA uptake compared to the wild-type 7169.
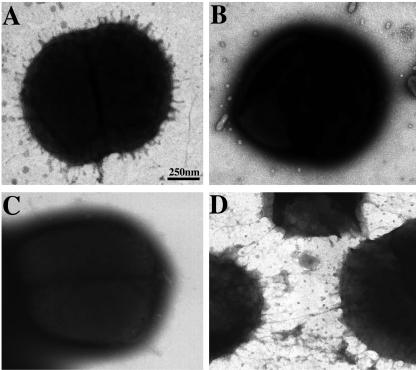
EM analysis confirmed the differential piliation phenotypes of the wild-type 7169, 7169::pilAK4, 7169::pilQK6, and 7169::pilTK3 strains (Fig. 4). In contrast to 7169 (Fig. 4A), both 7169::pilAK4 (Fig. 4B) and 7169::pilQK6 (Fig. 4C) lacked detectable pili on the surface of the cell. 7169::pilTK3 (Fig. 4D), on the other hand, appeared to be hyperpiliated, with observably longer and denser pili.
FIG. 4.
Piliation phenotypes as assessed by EM studies of the wild-type and pilA,-Q, and -T mutant strains. (A) EM analysis revealed short, peritrichous pili on the wild-type 7169. In contrast, whereas both the PilA-deficient (B) and PilQ-deficient (C) strains lacked detectable pili on the surface, the PilT-null mutant (D) appeared hyperpiliated with longer, tangled pili.
Investigation of pilus expression by M. catarrhalis.
Type IV pilus expression has been shown to be tightly growth phase or growth condition regulated in other bacterial systems. Therefore, to investigate expression of pili by M. catarrhalis, total RNA was isolated from M. catarrhalis 7169 at various time points during broth-based growth, as well as from plate-grown organisms, and subjected to RT-PCR analysis (Fig. 5). These analyses detected mRNA transcripts during all phases of broth-based growth, although the level of pilA transcript was reduced as the broth cultures reached late log and early stationary phases. Corresponding immunoblot analysis of whole-cell lysates, prepared simultaneously with RNA isolation, demonstrated PilA expression occurred under all conditions evaluated (data not shown).
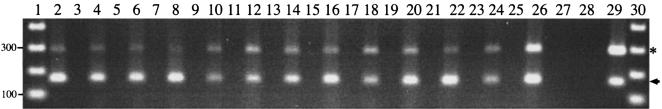
FIG. 5.
RT-PCR analysis of pilin expression by M. catarrhalis 7169 at various time points during broth-based growth (lane 2, 1.5 h; lane 3, 3.0 h; lane 4, 5.0 h, lane 5, 7.5 h) as well as from plate-grown organisms (lane 6). pilA transcript was detected during all phases of growth (amplicons denoted by arrowhead). rpoD served as the internal control (amplicons denoted by asterisk). One representative set of controls is depicted, including an amplification reaction that included RNA but lacked RT activation (lane 7), 7169 chromosomal DNA (lane 8), and a no-template control (lane 9). Samples were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide. Molecular size standards (lanes 1 and 10) are depicted in 100-bp increments.
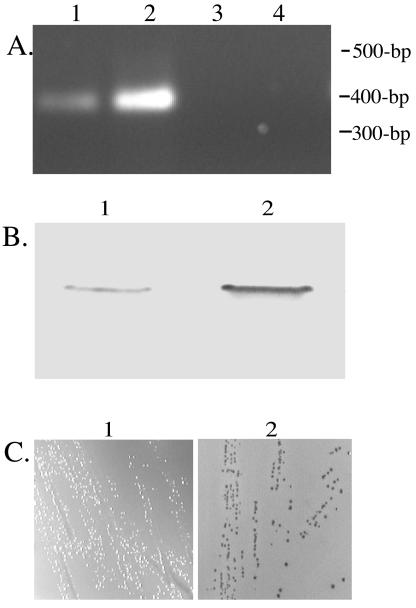
Furthermore, additional RT-PCR analyses investigating the effects of various growth conditions on type IV pilus expression indicated increased levels of pilus expression under iron-stressed growth conditions (data not shown). Recent studies have identified the M. catarrhalis fur homologue, which was demonstrated to encode the pleotrophic iron-responsive transcriptional regulator Fur (14). The M. catarrhalis Fur-deficient mutant (7169fur1) has been shown to constitutively express Fur-responsive, iron-regulated proteins on the surface of the bacterium irrespective of iron availability (14). This mutant was used to confirm the initial observations suggesting that pilA transcription was iron regulated. Figure 6A demonstrates the increased level of pilA transcript observed in 7169fur1 (lane 2) compared to that in the wild-type 7169 (lane 1). Equivalent amounts of RNA were subjected to RT-PCR analysis with pilA-specific primers. The corresponding increase in pilin expression on the surface of the organism is depicted in Fig. 6B, demonstrating an increased amount of pili produced by M. catarrhalis cells displaying an iron-stressed phenotype. The finding that pilin expression in M. catarrhalis is iron regulated and Fur responsive was further supported by the positive results of the FURTA (Fig. 6C). When strain H1717 was transformed with the control plasmid paphA-3, no LacZ expression was observed and the colonies remained white in the presence of iron (Fig. 6C, left). This indicated that the sequence carried by this recombinant plasmid did not titrate out the Fur repressor from binding to fhu binding sites. In contrast, colonies of strain H1717 transformed with pPILAk6 were red on MacConkey iron plates (Fig. 6C, right). This observation indicated that the transcription of the M. catarrhalis pilA was under the control of Fur, since pPILAk6 was capable of titrating the Fur protein from binding sites within fhuF, causing expression of the reporter gene. Consistent with these results, although the M. catarrhalis Fur consensus binding sequence has not yet been identified, a putative Fur box matching the E. coli Fur box consensus was identified (13 of 19 bp identity) in the upstream region of pilA located on the FURTA-positive clone pPILAk6 (data not shown).
FIG. 6.
Analysis of iron-regulated expression of M. catarrhalis pilin. (A) Comparison of wild-type 7169 (lane 1) and the isogenic Fur mutant 7169fur1(lane 2) pilA transcripts by agarose gel analysis of RT-PCR amplicons. Control reactions, lacking RT activation, are depicted in lanes 3 and 4 for RNA isolated from 7169 and 7169fur1, respectively. (B) MAb 4G9 was used in immunoblot analysis of surface pilin expression by comparing the sheared surface components from 7169 (lane 1) and 7169fur1 (lane 2). (C) Representative depiction of the regulation of pilA expression by Fur, as demonstrated by the FURTA-positive phenotype of H1717 cells transformed with pPILAk4 (right panel) compared to the FURTA-negative phenotype of the H1717 cells transformed with the control plasmid paphA-3 (left panel). Red colonies appear dark gray and white colonies appear pale gray, as imaged by the monochromatic AlphaImager 2200 documentation and analysis system (Imgen Technologies, Alexandria, Va.).
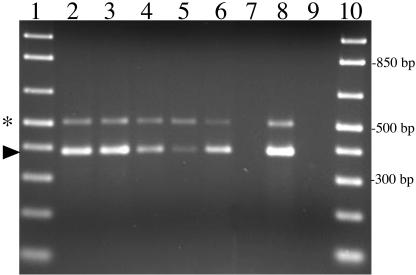
The prevalence of type IV pilus expression by M. catarrhalis was ascertained by investigating the expression of pilA and pilQ by RT-PCR analyses of RNA isolated from a geographically and physiologically diverse panel of clinical isolates. As shown in Fig. 7, transcription of the type IV pilin genes corresponding to the major pilin, PilA, and the outer membrane secretin component, PilQ, was detected in every strain analyzed. These data suggest the expression of type IV pili by M. catarrhalis may be ubiquitous within this bacterial species and represents an important virulence factor in the pathogenesis of M. catarrhalis infections.
FIG. 7.
RT-PCR analysis of pilA and pilQ expression by a panel of M. catarrhalis clinical isolates. RT-PCR amplicons, corresponding to pilA (bottom band, arrow) and pilQ (top band, asterisk) mRNA transcripts, were analyzed by agarose gel electrophoresis and ethidium bromide staining. Even lanes (2 to 26) represent reactions in the presence of RT, and odd lanes (3 to 27) represent control reactions lacking RT activation to verify RNA purity and absence of DNA contamination. Total RNA was isolated and analyzed from the following M. catarrhalis clinical isolates: lanes 2 and 3, 43617 (American Type Culture Collection); lanes 4 and 5, 7169 (Buffalo, N.Y.); lanes 6 and 7, KSA (Buffalo, N.Y.); lanes 8 and 9, BC40 (Buffalo, N.Y.); lanes 10 and 11, sk633 (Buffalo, N.Y.); lanes 12 and 13, O35E (Houston, Tex.); lanes 14 and 15, Tal1 (Philadelphia, Pa.); lanes 16 and 17, Af218 (England); lanes 18 and 19, 27335 (France); lanes 20 and 21, 27479 (Japan); lanes 22 and 23, 210044 (Angola); lanes 24 and 25, 27325 (Germany); lanes 26 and 27, 27512 (Belgium). Controls are depicted in lane 28, no-template control reaction, and lane 29, 7169 chromosomal DNA. Molecular size standards (lanes 1 and 30) are depicted in 100-bp increments.
DISCUSSION
In this report, we describe the identification and characterization of the genes that are involved in the biosynthesis and assembly of M. catarrhalis pili. While previous EM studies have suggested that some strains of Moraxella spp. express pili, or other putative surface appendages such as Hag or UspA1 (2-4, 29, 37), definitive data demonstrating pilus expression by M. catarrhalis have been lacking until now, and these structures have been essentially ignored as putative virulence factors for the past decade. The data we present in this study demonstrate that we have identified multiple M. catarrhalis genes with homology to genes of the type IV pilus systems described for other important gram-negative human pathogens.
Type IV pili are composed of pilin subunits which share several distinctive common features, including a modified N-terminal residue resulting from the posttranslational methylation of a conserved Phe residue found in the +1 position following cleavage of the short positively charged signal sequence, which is dependent on the presence of a Gly residue at amino acid position −1 and a Glu residue at +5. In addition to the hydrophobic N-terminal domain that is highly conserved and essential for both pilin secretion and subunit-subunit interactions within the pilus filament, a pair of C-terminal cysteine residues that form a functionally critical disulfide bridge have also been described (5, 42, 44). Analysis of the deduced amino acid sequence of the M. catarrhalis pilin homologue indicates PilA contains identically positioned amino acid residues as identified among the previously characterized members of type IV pilin-expressing bacteria. Furthermore, analysis of the PilA-deficient mutant 7169::pilAK4 indicates the disruption of pilA prevents pilus expression on the surface of M. catarrhalis, in accordance with the monomeric nature of type IV pilin filaments.
In addition to pilA, we have also located eight additional genes that have homology to components of the type IV pilus biosynthetic apparatus. The pilBCD locus contains genes predicted to encode proteins that are directly involved in the export and biosynthesis of type IV pili. In other type IV pilus systems, it has been demonstrated that PilB is a nucleotide binding protein required for energizing pilin extrusion, PilC is an inner membrane component of the pilus biosynthetic machinery, and PilD is the prepilin peptidase and methylase (31). The pilNOPQ operon is predicted to encode inner and outer membrane components of pilus assembly machinery. In addition to being required for pilin biogenesis, these genes also have homology to genes required for DNA uptake and twitching motility. PilQ is a member of a large family of proteins termed secretins, which form gated pores in the bacterial outer membrane. In type IV pilus systems, the pilus filament is extruded through a dodecameric donut-shaped outer membrane complex comprised of PilQ (31). Consistent with this function, our data demonstrated that the 7169::pilQK6 mutant lacked pili on the bacterial surface and was no longer competent for natural genetic transformation, even though pilin monomers were still produced within the cell.
It is interesting that in M. catarrhalis, this gene cluster consists solely of pilNOPQ and lacks the pilM homologue present in this operon identified in other type IV piliated species studied to date. Although the significance of this difference is not yet known, pilM has been hypothesized to link the production of polar type IV pili to cell shape and division (5, 30, 31). As shown in the EM studies, M. catarrhalis appears to express peritrichous pili, and it is possible that lack of classical polar type IV pilus production by this organism may be due, at least in part, to the absence of pilM. However, additional studies are needed to investigate this hypothesis.
pilT was identified downstream of the M. catarrhalis fur homologue. PilT is a nucleotide binding protein that is required for pilin retraction, disassembly, and degradation. Consistent with the phenotype of the PilT-deficient mutant 7169::pilTK3 described in this study, pilT mutants are unable to retract their pili, resulting in hyperpiliation (31). PilT is also required for twitching motility, a form of flagellum-independent bacterial surface motility that is mediated by polar type IV pili. It is also of interest that we were unable to demonstrate that M. catarrhalis exhibits twitching motility, and it is tempting to speculate that this phenomenon may also be related to the fact that M. catarrhalis does not express polarly located type IV pili.
One of the most striking observations made while undertaking comparative analyses of the isogenic PilA-, PilT-, and PilQ-deficient mutants was the absolute correlation between pilus expression and competence for DNA uptake. The DNA transformation experiments revealed that the loss of piliation completely inhibited the competence of M. catarrhalis for DNA uptake. Previous studies of the pathogenic Neiserria spp. indicated the frequency of DNA transformation was reduced by 1,000- to 10,000-fold in nonpiliated mutants (47, 54). In contrast, natural competence in M. catarrhalis appears to be absolutely correlated with pilus expression on the bacterial surface, as transformation was totally inhibited in both the PilA- or PilQ-deficient mutants, both of which were completely deficient in surface-expressed pili.
Furthermore, our studies indicated that M. catarrhalis appears to constitutively express a single pilin. This is in stark contrast to the highly variable pilus expression displayed by N. gonorrhoeae, which not only switches from piliated to nonpiliated states but also undergoes high-frequency antigenic variation of the pilin monomer with the capacity of producing up to 107 pilin variants as a result of intragenic recombination with transcriptionally silent pilS loci (reviewed in references 15 and 22). N. meningitidis, M. bovis, Moraxella lacunata, and E. corrodens also exhibit antigenic and phase variations of their type IV pili (17, 51, 52). We have not identified either putative silent loci of variant pilin sequences or more than one major pilin gene encoded within the M. catarrhalis genome.
In addition, to address the disparity in the literature surrounding the relative frequency and occurrence of pilus expression by M. catarrhalis as assessed by EM analyses (reviewed in reference 49), we performed RT-PCR experiments to investigate the transcription of pilA and pilQ in a panel of global isolates. Our data suggest type IV pilus expression by M. catarrhalis may be ubiquitous in this human mucosal pathogen.
The RT-PCR analyses also led to the interesting observation that pilus expression by M. catarrhalis is iron responsive and Fur regulated. When cultured under iron-depleted conditions or when using the Fur-deficient mutant 7169fur1, M. catarrhalis showed both increased expression of pilA transcript as well as concomitant increased expression of pili on the bacterial cell surface. Furthermore, the FURTA-positive phenotype of sequence upstream of pilA is consistent with the iron regulation of pilin through binding of the pleotrophic iron-responsive transcriptional regulator Fur. Recently, it has been demonstrated that limited iron availability results in an increase in gonococcal pilin antigenic variation, DNA transformation, and DNA repair (41). In addition, iron limitation was shown to induce expression of the Burkholderia cenocepacia cable pilus (46). Iron is a critical element essential for the growth of many organisms, and there is little free iron available in the human body (26). The fact that iron limitation has an inducing effect on pilus expression supports the speculation that bacteria are primed to colonize host surfaces and initiate infection by responding to low-iron conditions within the host by upregulating pilus expression. It will be interesting to ascertain whether iron regulation of pilus expression is a common occurrence in other type IV pilus-expressing mammalian pathogens.
In summary, we have conclusively demonstrated that M. catarrhalis expresses type IV pili. These surface filaments are absolutely essential for natural transformation, and this study represents the first definitive demonstration of Fur-regulated pilin expression. RT-PCR analyses demonstrated expression of the pilA and pilQ genes involved in type IV pilus biogenesis in all strains of M. catarrhalis evaluated to date. Taken together, these data indicate additional studies investigating the contribution of type IV pilus expression to the virulence and pathogenicity of M. catarrhalis are warranted.
Acknowledgments
This research was supported by Public Health Service research grants AI46422 and DC005827 awarded to A.A.C.
Editor: D. L. Burns
REFERENCES
- 1.Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, K. 1992. Fimbriae of Branhamella catarrhalis as possible mediators of adherence to pharyngeal epithelial cells. APMIS 100:1066-1072. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, K., N. Rikitomi, and K. Matsumoto. 1992. Fimbriation, hemagglutination and adherence properties of fresh clinical isolates of Branhamella catarrhalis. Microbiol. Immunol. 36:1009-1017. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed, K., N. Rikitomi, T. Nagatake, and K. Matsumoto. 1990. Electron microscopic observation of Branhamella catarrhalis. Microbiol. Immunol. 34:967-975. [DOI] [PubMed] [Google Scholar]
- 5.Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. [DOI] [PubMed] [Google Scholar]
- 6.Berk, S. L. 1990. From Micrococcus to Moraxella. The reemergence of Branhamella catarrhalis. Arch. Intern. Med. 150:2254-2257. [PubMed] [Google Scholar]
- 7.Berk, S. L., J. H. Kalbfleisch, et al. 1996. Antibiotic susceptibility patterns of community-acquired respiratory isolates of Moraxella catarrhalis in western Europe and in the USA. J. Antimicrob. Chemother. 38(Suppl. A):85-96. [DOI] [PubMed] [Google Scholar]
- 8.Berk, S. L., and A. Verghese. 1989. Emerging pathogens in nosocomial pneumonia. Eur. J. Clin. Microbiol. Infect. Dis. 8:11-14. [DOI] [PubMed] [Google Scholar]
- 9.Berner, R., R. F. Schumacher, M. Brandis, and J. Forster. 1996. Colonization and infection with Moraxella catarrhalis in childhood. Eur. J. Clin. Microbiol. Infect. Dis. 15:506-509. [DOI] [PubMed] [Google Scholar]
- 10.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campagnari, A. A., K. L. Shanks, and D. W. Dyer. 1994. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect. Immun. 62:4909-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright, M. C., and H. McKenzie. 1997. Moraxella (Branhamella) catarrhalis—clinical and molecular aspects of a rediscovered pathogen. J. Med. Microbiol. 46:360-371. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, L. A., and J. Berenguer. 2000. Secretion and assembly of regular surface structures in gram-negative bacteria. FEMS Microbiol. Rev. 24:21-44. [DOI] [PubMed] [Google Scholar]
- 14.Furano, K., and A. A. Campagnari. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect. Immun. 71:1843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Rodriguez, J. A., and M. J. Fresnadillo Martinez. 2002. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 50(Suppl. S2):59-73. [DOI] [PubMed] [Google Scholar]
- 17.Heinrich, D. W., and A. C. Glasgow. 1997. Transcriptional regulation of type 4 pilin genes and the site-specific recombinase gene, piv, in Moraxella lacunata and Moraxella bovis. J. Bacteriol. 179:7298-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 19.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 20.Keizer, D. W., C. M. Slupsky, M. Kalisiak, A. P. Campbell, M. P. Crump, P. A. Sastry, B. Hazes, R. T. Irvin, and B. D. Sykes. 2001. Structure of a pilin monomer from Pseudomonas aeruginosa: implications for the assembly of pili. J. Biol. Chem. 276:24186-24193. [DOI] [PubMed] [Google Scholar]
- 21.Ketterer, M. R., J. Q. Shao, D. B. Hornick, B. Buscher, V. K. Bandi, and M. A. Apicella. 1999. Infection of primary human bronchial epithelial cells by Haemophilus influenzae: macropinocytosis as a mechanism of airway epithelial cell entry. Infect. Immun. 67:4161-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kline, K. A., E. V. Sechman, E. P. Skaar, and H. S. Seifert. 2003. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R's of molecular genetics of two human-specific bacterial pathogens. Mol. Microbiol. 50:3-13. [DOI] [PubMed] [Google Scholar]
- 23.Long, C. D., R. N. Madraswala, and H. S. Seifert. 1998. Comparisons between colony phase variation of Neisseria gonorrhoeae FA1090 and pilus, pilin, and S-pilin expression. Infect. Immun. 66:1918-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luke, N. R., S. Allen, B. W. Gibson, and A. A. Campagnari. 2003. Identification of a 3-deoxy-d-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant. Infect. Immun. 71:6426-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luke, N. R., R. J. Karalus, and A. A. Campagnari. 2002. Inactivation of the Moraxella catarrhalis superoxide dismutase SodA induces constitutive expression of iron-repressible outer membrane proteins. Infect. Immun. 70:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luke, N. R., T. A. Russo, N. Luther, and A. A. Campagnari. 1999. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect. Immun. 67:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrs, C. F., and S. Weir. 1990. Pili (fimbriae) of Branhamella species. Am. J. Med. 88:36S-40S. [DOI] [PubMed] [Google Scholar]
- 30.Martin, P. R., A. A. Watson, T. F. McCaul, and J. S. Mattick. 1995. Characterization of a five-gene cluster required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 16:497-508. [DOI] [PubMed] [Google Scholar]
- 31.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 32.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy, T. F. 1998. Lung infections. 2. Branhamella catarrhalis: epidemiological and clinical aspects of a human respiratory tract pathogen. Thorax 53:124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, T. F., and S. Sethi. 2002. Chronic obstructive pulmonary disease: role of bacteria and guide to antibacterial selection in the older patient. Drugs Aging 19:761-775. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rikitomi, N., B. Andersson, K. Matsumoto, R. Lindstedt, and C. Svanborg. 1991. Mechanism of adherence of Moraxella (Branhamella) catarrhalis. Scand. J. Infect. Dis. 23:559-567. [DOI] [PubMed] [Google Scholar]
- 39.Russo, T. A., J. E. Guenther, S. Wenderoth, and M. M. Frank. 1993. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol. Microbiol. 9:357-364. [DOI] [PubMed] [Google Scholar]
- 40.Sauvonnet, N., G. Vignon, A. P. Pugsley, and P. Gounon. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serkin, C. D., and H. S. Seifert. 2000. Iron availability regulates DNA recombination in Neisseria gonorrhoeae. Mol. Microbiol. 37:1075-1086. [DOI] [PubMed] [Google Scholar]
- 42.Shaw, C. E., and R. K. Taylor. 1990. Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect. Immun. 58:3042-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, W., and H. Sun. 2002. Type IV pilus-dependent motility and its possible role in bacterial pathogenesis. Infect. Immun. 70:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smyth, C. J., M. B. Marron, J. M. Twohig, and S. G. Smith. 1996. Fimbrial adhesins: similarities and variations in structure and biogenesis. FEMS Immunol. Med. Microbiol. 16:127-139. [DOI] [PubMed] [Google Scholar]
- 45.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 46.Tomich, M., and C. D. Mohr. 2004. Transcriptional and posttranscriptional control of cable pilus gene expression in Burkholderia cenocepacia. J. Bacteriol. 186:1009-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tonjum, T., and M. Koomey. 1997. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships—a review. Gene 192:155-163. [DOI] [PubMed] [Google Scholar]
- 48.Vaneechoutte, M., G. Verschraegen, G. Claeys, B. Weise, and A. M. Van den Abeele. 1990. Respiratory tract carrier rates of Moraxella (Branhamella) catarrhalis in adults and children and interpretation of the isolation of M. catarrhalis from sputum. J. Clin. Microbiol. 28:2674-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vignon, G., R. Kohler, E. Larquet, S. Giroux, M. C. Prevost, P. Roux, and A. P. Pugsley. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 185:3416-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villar, M. T., J. T. Helber, B. Hood, M. R. Schaefer, and R. L. Hirschberg. 1999. Eikenella corrodens phase variation involves a posttranslational event in pilus formation. J. Bacteriol. 181:4154-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villar, M. T., R. L. Hirschberg, and M. R. Schaefer. 2001. Role of the Eikenella corrodens pilA locus in pilus function and phase variation. J. Bacteriol. 183:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 54.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]