Abstract
A uropathogenic Escherichia coli strain CFT073-specific DNA microarray that includes each open reading frame was used to analyze the transcriptome of CFT073 bacteria isolated directly from the urine of infected CBA/J mice. The in vivo expression profiles were compared to that of E. coli CFT073 grown statically to exponential phase in rich medium, revealing the strategies this pathogen uses in vivo for colonization, growth, and survival in the urinary tract environment. The most highly expressed genes overall in vivo encoded translational machinery, indicating that the bacteria were in a rapid growth state despite specific nutrient limitations. Expression of type 1 fimbriae, a virulence factor involved in adherence, was highly upregulated in vivo. Five iron acquisition systems were all highly upregulated during urinary tract infection, as were genes responsible for capsular polysaccharide and lipopolysaccharide synthesis, drug resistance, and microcin secretion. Surprisingly, other fimbrial genes, such as pap and foc/sfa, and genes involved in motility and chemotaxis were downregulated in vivo. E. coli CFT073 grown in human urine resulted in the upregulation of iron acquisition, capsule, and microcin secretion genes, thus partially mimicking growth in vivo. On the basis of gene expression levels, the urinary tract appears to be nitrogen and iron limiting, of high osmolarity, and of moderate oxygenation. This study represents the first assessment of any E. coli pathotype's transcriptome in vivo and provides specific insights into the mechanisms necessary for urinary tract pathogenesis.
Urinary tract infections (UTIs) are a serious health concern. Forty to 50% of women experience at least one UTI, leading to an estimated 8 million annual physician visits in the United States alone (39, 46). Uropathogenic Escherichia coli (UPEC) is by far the most common etiological agent of all UTIs. UPEC strain CFT073, derived from the clonal group O6:K2:H1 (26), was originally isolated from the blood and urine of a woman diagnosed with acute pyelonephritis (28). It is considered a prototype of the O6 serogroup, one of the most prevalent UPEC clonal lines (23, 24). The virulence of this strain was reproduced in the well-established CBA mouse model of ascending UTI (28). In addition to numerous virulence studies, the genome of E. coli CFT073 has recently been sequenced and compared to that of enterohemorrhagic E. coli EDL933 and the nonpathogenic laboratory strain E. coli MG1655 (42).
Mutations have been introduced into a number of candidate virulence genes in UPEC, leading to attenuated mutants in experimental UTI. These include fim, encoding type 1 fimbria (7, 13), sat, encoding secreted autotransporter toxin (14), cnf-1, encoding cytotoxic necrotizing factor (36), tonB, involved in iron transport (40), proP, involved in osmoprotectant transport (8), and degS (35). Large-scale screens for virulence factors of UPEC have also identified factors that aid UPEC during growth in urine (38) and have implicated capsule, lipopolysaccharide, iron acquisition systems, and the PhoU regulatory system in virulence (3, 35). Molecular Koch's postulates have been satisfied for type 1 fimbriae, DegS, and TonB (9).
In this report, we have quantified the gene expression for each open reading frame (ORF) of this uropathogen from organisms isolated directly from the urine of experimentally infected CBA/J mice. We have identified multiple virulence and metabolic factors that were upregulated to aid survival in the host. We also defined specific environmental conditions that appear to confront E. coli CFT073 during experimental UTI.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
For in vitro RNA preparation, UPEC strain CFT073 (28) was grown statically at 37°C in 100 ml of Luria broth (LB) until the optical density at 600 nm (OD600) reached 0.65. For RNA preparation from urine, this strain was grown statically at 37°C in 900 ml of pooled human urine until the OD600 reached 0.135 (mid-exponential phase for growth in urine). Urine was collected from healthy women volunteers aged 20 to 40 who had no history of UTI or antibiotic use in the prior 2 months. Each urine sample was immediately filter sterilized (0.2 μm pore size) and frozen at −80°C for use within 2 weeks. For each experimental replicate, a different pool of urine samples from three to five volunteers was used. The study was approved by the Institutional Review Board of the University of Maryland School of Medicine. LB- and urine-grown cultures were immediately treated with RNA Protect Bacterial Reagent (QIAGEN) to stabilize RNA according to the manufacturer's instructions and was harvested by centrifugation (8 min, 7,500 × g, 25°C). The bacterial pellet was frozen at −20°C until RNA extraction.
CBA/J mouse model of ascending UTI.
To establish a relevant source of in vivo-grown bacteria, 40 CBA/J mice were transurethrally inoculated as previously described (15, 21) with 5 × 109 CFU of E. coli CFT073. The infection was monitored daily by pooling urine from 10 mice at random and spiral plating on Luria agar to quantify CFU. Mice were reinoculated after 6 days to maintain infection at levels between 2 × 106 and 3 × 107 CFU/ml of urine, which allowed the collection of sufficient numbers of bacteria needed for RNA extraction and subsequent analysis. Urine was collected for 10 days, with the exception of the first 24 h following the inoculations. On average, 50 μl of urine was collected from each mouse every 45 min during an 8-h period. Urine was collected directly into 1.5-ml microcentrifuge tubes containing 650 μl of RNA Protect Bacterial Reagent (QIAGEN) and was pooled until the total volume reached approximately 1 ml. The RNA Protect-treated sample remained on ice until all samples had been collected for that day. A crystalline precipitate was allowed to settle before pipetting off the bacteria-containing supernatant into a 15-ml centrifuge tube for collection. Tubes were centrifuged (15 min, 2,500 × g, 4°C), the supernatant was decanted, and the bacterial pellet was frozen at −20°C until RNA extraction.
RNA isolation and cDNA synthesis.
RNA from both in vitro and in vivo samples was extracted using an RNeasy Mini kit with a 1-h on-column DNase digestion (QIAGEN) according to the RNeasy Mini handbook. Multiple RNA preparations of the same sample were pooled prior to cDNA synthesis. Up to 10 μg of RNA was mixed with 750 ng of random hexamers (Invitrogen) for each cDNA synthesis reaction according to a previously described protocol (37). SuperScript II reverse transcriptase (1,500 U; Invitrogen) was added to these reactions, along with First Strand buffer, dithiothreitol, and deoxyribonucleotides at concentrations recommended by the manufacturer (Invitrogen). The reactions were incubated at 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70°C for 10 min. Following RNaseH (Invitrogen) and RNaseA (Ambion) digestion, cDNA was purified with a QIAquick PCR Purification kit (QIAGEN) according to the QIAquick Spin handbook.
Quantitative real-time reverse transcription-PCR (qRT-PCR).
Primers designed to amplify papA_2 of E. coli CFT073 were 5′GTGCCTGCAGAAAATGCAGAT and 5′CCCGTTTTCCACTCGAATCA, and primers for gapA were 5′CATCGTTTCCAACGCATCCT and 5′ACCTTCGATGATGCCGAAGTT (forward and reverse primers, respectively). Thirty nanograms of cDNA and 300 nM (final concentration) each primer were mixed with 12.5 μl of 2× SYBR Green PCR Master Mix (ABI). Assays were performed in triplicate with the ABI Prism model 7900 instrument. All data were normalized to the internal standard gapA (encoding glyceraldehyde 3-phosphate dehydrogenase), and melting curve analysis demonstrated that the accumulation of SYBR Green-bound DNA was gene specific. The 2−ΔΔCT method (25) was used for analysis, and the data were transformed by log2 to obtain a fold change difference between growth conditions.
Microarrays and hybridization.
The E. coli CFT073-specific DNA microarray (NimbleGen Systems, Inc.) includes 5,611 ORFs and stable RNAs from version 17 of the compiled CFT073 genome sequence. Each ORF is represented on the glass slide by 17 unique probe pairs of 24-mer in situ-synthesized oligonucleotides. Each pair consists of a sequence perfectly matched to the ORF, and another adjacent sequence harbors two mismatched bases for determination of background and cross-hybridization. For each microarray, 3 to 5 μg of cDNA was fragmented using RQ1 DNaseI (Promega) partial digest and was then labeled with biotin-N6-ddATP (Perkin-Elmer Life Sciences) using terminal transferase (Roche) as described previously (37). Labeled cDNA samples were individually hybridized in triplicate to the CFT073-specific microarray according to the NimbleGen standard operating procedure. Following washes and labeling with a streptavidin-Cy3 complex according to the NimbleGen procedure, microarrays were scanned at 5-μm resolution using a GenePix 4000b scanner.
Data and statistical analysis.
For LB and in vivo microarrays, data were extracted using NimbleScan (NimbleGen) and an algorithm (courtesy of Yu Qiu, University of Wisconsin School of Medicine) applied to obtain a single measurement of signal intensity for each ORF. Data were normalized and converted to estimates of transcript abundance, using the total signal intensity to allow comparison of individual microarrays (1). A second set of LB-grown microarrays were processed along with the urine-grown microarray, where hybridization and normalization of the microarray were carried out by NimbleGen Systems, Inc. The signal intensity of an ORF was calculated by subtracting the mismatch probe intensity from the perfect match probe intensity for each of the 17 probe pairs, obtaining a mean difference value (excluding values greater than 3.0 standard deviations from the means). These urine- and LB-grown microarrays were normalized with the quantile normalization method and were analyzed with the RMA algorithm (6, 20). For all microarrays, a P value for each ORF was calculated by a two-tailed Welch's unpaired t test comparison of the three microarray replicates for each bacterial growth condition. Fold changes of an ORF between growth conditions were calculated by transformation of the following ratio: log2 [(average in vivo-grown or urine-grown signal intensity)/(average LB-grown signal intensity)]. Only fold changes of at least ±2 and P ≤ 0.05 were considered significant and are discussed in this report. Thus, ORFs characterized as upregulated (fold change, ≥2; P ≤ 0.05) or downregulated (fold change, ≤−2; P ≤ 0.05) during growth in vivo or during growth in urine are relative to growth in LB.
RESULTS
Alterations in the transcriptome of E. coli CFT073 during UTI.
To further understand the mechanisms UPEC employ in UTIs, we used E. coli CFT073-specific DNA microarrays to analyze the transcriptome of this strain grown under both in vivo and in vitro conditions. The source for in vivo-grown bacteria was the urine from infected mice, collected from 1 to 10 days postinoculation for use in RNA isolation. The sources for in vitro-grown RNA were cultures of E. coli CFT073 grown statically to mid-exponential phase at 37°C in either LB or filtered human urine. The expression level of each of 5,611 ORFs was determined for each condition. An ORF was considered differentially regulated if the statistically significant change in expression was also greater than twofold. All in vivo- and urine-derived data are described here relative to growth in LB.
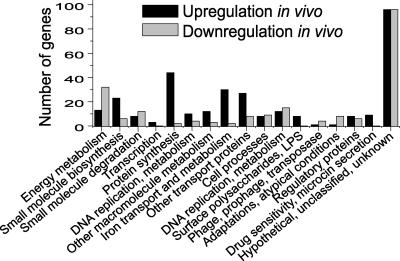
Overall, 313 genes were found upregulated (Table 1 lists the 50 most upregulated), and 207 genes were downregulated during growth in vivo. To identify candidate virulence genes specific to E. coli CFT073 that may play a role during UTI, all 313 genes upregulated in vivo were evaluated for the absence or presence of a homologue in the E. coli K-12 nonpathogenic strain (Table 1). This analysis revealed 45 genes that were not found in E. coli K-12 and 41 genes encoding hypothetical proteins that were unable to be confidently categorized. Of these 45 candidate virulence genes, 25 genes previously implicated in virulence encoded iron acquisition, capsule synthesis, or microcin secretion proteins and 13 encoded hypothetical proteins.
TABLE 1.
Top 50 genes upregulated in vivo during UTI
| Gene | Function | Fold changea | P value |
|---|---|---|---|
| rspA | Repression of stationary-phase gene sigma-S | 6.021 | 0.0059 |
| sitAb | Iron transport system SitA protein | 5.634 | <0.0001 |
| c1903 | Unknown | 5.556 | 0.0018 |
| rspB | Repression of stationary-phase gene sigma-S | 5.532 | <0.0001 |
| entE | Enterobactin synthetase component E (enterochelin) | 5.482 | 0.0032 |
| chuWb | Putative coproporphyrinogen III oxidase; hemin utilization | 5.417 | 0.0016 |
| yhjX | Putative drug resistance protein YhjX | 5.416 | 0.0312 |
| iucAb | Aerobactin protein IucA | 5.166 | 0.0025 |
| proV | Glycine betaine/l-proline transport ATP-binding protein ProV | 5.150 | 0.0025 |
| entA | Enterobactin system dehydrogenase | 5.115 | 0.0113 |
| entF | Enterobactin synthetase component F | 5.012 | 0.0134 |
| uxuB | d-mannonate oxidoreductase | 4.982 | 0.0049 |
| srlD | Sorbitol-6-phosphate 2-dehydrogenase | 4.912 | 0.0055 |
| uxuA | Mannonate dehydratase | 4.740 | 0.0026 |
| tyrA | T protein | 4.643 | 0.0002 |
| fdnI | Formate dehydrogenase, nitrate inducible | 4.623 | 0.0066 |
| deaD | Cold-shock DEAD-box protein A | 4.614 | 0.0024 |
| nirD | Nitrite reductase [NAD(P)H] small subunit | 4.601 | 0.0022 |
| yeaR | Hypothetical protein YeaR | 4.579 | 0.0018 |
| lpxB | Lipid-A-disaccharide synthase | 4.570 | 0.0006 |
| fimC | Type 1 fimbriae; chaperone protein FimC precursor | 4.553 | <0.0001 |
| c1905b | Hypothetical protein | 4.552 | 0.0081 |
| c5174b | Putative iron-regulated outer membrane virulence protein | 4.539 | 0.0024 |
| c1220 | Phospho-2-dehydro-3-deoxyheptonate aldolase, Trp sensitive | 4.529 | 0.0013 |
| yhfI | Hypothetical oxidoreductase YdfI | 4.523 | 0.0065 |
| iroNb | Siderophore receptor IroN | 4.491 | 0.0207 |
| dut | Deoxyuridine 5′-triphosphate nucleotidohydrolase | 4.488 | 0.0163 |
| chuSb | Putative heme/hemoglobin transport protein; hemin utilization | 4.473 | 0.0128 |
| iroBb | Putative glucosyltransferase; siderophore system | 4.466 | 0.0056 |
| iucDb | Aerobactin protein IucD | 4.433 | 0.0009 |
| sitBb | Iron transport system SitB protein | 4.404 | 0.0020 |
| yncE | Hypothetical protein YncE precursor | 4.341 | 0.0058 |
| rplI | 50S ribosomal protein L9 | 4.318 | 0.0122 |
| c4088 | Hypothetical protein | 4.271 | 0.0142 |
| yoaG | Hypothetical protein YoaG | 4.268 | <0.0001 |
| c3610b | Putative receptor | 4.242 | 0.0113 |
| c0672 | Conserved hypothetical protein | 4.234 | 0.0030 |
| c2203 | Hypothetical protein | 4.218 | 0.0001 |
| kpsEb | Capsule biosynthesis KpsE protein | 4.127 | 0.0126 |
| entC | Isochorismate synthase EntC, enterobactin system | 4.108 | 0.0011 |
| srlE | PTS system, glucitol/sorbitol-specific IIBC component | 4.096 | 0.0002 |
| rnpA | Ribonuclease P protein component | 4.094 | 0.0122 |
| fimF | Type 1 fimbriae; FimF protein precursor | 4.084 | 0.0001 |
| gidA | Glucose inhibited division protein A | 4.052 | 0.0021 |
| marR | Multiple antibiotic resistance protein MarR | 4.038 | 0.0002 |
| hmpA | Flavohemoprotein (hemoglobin-like protein) | 3.993 | 0.0148 |
| glnP | Glutamine transport system permease protein GlnP | 3.992 | 0.0016 |
| c4090 | Hypothetical protein | 3.966 | 0.0225 |
| srlB | PTS system, glucitol/sorbitol-specific IIA component | 3.956 | 0.0012 |
| chuTb | Putative periplasmic binding protein; hemin utilization | 3.956 | 0.0270 |
Fold change refers to growth in vivo relative to growth in LB.
This gene was not present in E. coli K-12.
By comparison to UPEC CFT073 grown in LB, bacteria coming from the urinary tract have upregulated expression of virulence and metabolic factors in the host (Fig. 1). Not only were a variety of factors upregulated, but these genes were presumably translated at a rapid rate. Thirty-five of the 50 most highly expressed genes in vivo encoded translational machinery (Table 2).
FIG. 1.
Functional categories of differentially expressed E. coli CFT073 genes. The number of genes that are significantly upregulated (black bars) or downregulated (grey bars) during growth in vivo (growth during UTI) relative to growth in static LB culture are categorized based on function.
TABLE 2.
Top 50 genes expressed by E. coli CFT073 in vivo during UTI
| Gene | Function | Signal intensity | Fold changea | P value | Among top 50 in urinec | Among top 50 in LBc |
|---|---|---|---|---|---|---|
| tufB | Protein chain elongation factor EF-Tu | 34.27 | 1.02 | 0.015 | + | + |
| rpsK | 30S ribosomal subunit protein S11 | 33.99 | 1.15 | <0.001 | + | + |
| rplE | 50S ribosomal subunit protein L5 | 32.91 | 1.73 | 0.004 | + | |
| fimAb | Major type 1 fimbrial subunit; fimbrin/pilin | 32.75 | 2.97 | 0.009 | ||
| rpsS | 30S ribosomal subunit protein S19 | 31.58 | 2.31 | 0.003 | ||
| rplX | 50S ribosomal subunit protein L24 | 31.27 | 1.71 | <0.001 | + | |
| rpsE | 30S ribosomal subunit protein S5 | 29.86 | 1.93 | <0.001 | + | |
| rpsN | 30S ribosomal subunit protein S14 | 29.28 | 1.65 | 0.006 | + | |
| rplR | 50S ribosomal subunit protein L18 | 29.01 | 1.87 | <0.001 | + | |
| rplJ | 50S ribosomal subunit protein L10 | 28.11 | 2.24 | 0.013 | ||
| rpmD | 50S ribosomal subunit protein L30 | 27.97 | 2.39 | 0.004 | + | |
| tufA | Protein chain elongation factor EF-Tu | 27.53 | 0.94 | 0.039 | + | |
| rplD | 50S ribosomal subunit protein L4 | 27.18 | 2.75 | 0.006 | ||
| rplC | 50S ribosomal subunit protein L3 | 27.00 | 2.88 | 0.003 | ||
| ompA | Outer membrane protein A (II*) | 26.05 | −1.23 | 0.016 | + | + |
| rplO | 50S ribosomal subunit protein L15 | 25.60 | 2.07 | 0.008 | + | |
| rpsD | 30S ribosomal subunit protein S4 | 25.49 | 0.97 | 0.028 | + | |
| rplF | 50S ribosomal subunit protein L6 | 25.42 | 2.17 | 0.012 | + | |
| rplL | 50S ribosomal subunit protein L7/L12 | 25.11 | 2.04 | 0.038 | ||
| prlA | Putative ATPase subunit of translocase | 24.74 | 1.98 | 0.011 | + | |
| rpsJ | 30S ribosomal subunit protein S10 | 24.11 | 2.59 | <0.001 | ||
| rplW | 50S ribosomal subunit protein L23 | 23.92 | 2.49 | 0.018 | ||
| rpsM | 30S ribosomal subunit protein S13 | 23.91 | 1.04 | 0.029 | + | |
| rpsH | 30S ribosomal subunit protein S8 | 23.27 | 1.96 | 0.013 | + | |
| rpsC | 30S ribosomal subunit protein S3 | 22.35 | 2.02 | 0.009 | ||
| gapA | Glyceraldehyde-3-phosphate dehydrogenase A | 22.31 | 0.04 | 0.178 | + | + |
| fusA | GTP-binding protein chain elongation factor EF-G | 21.96 | 1.76 | 0.002 | + | |
| rplB | 50S ribosomal subunit protein L2 | 21.83 | 2.30 | 0.007 | + | |
| rplP | 50S ribosomal subunit protein L16 | 21.09 | 1.72 | 0.018 | ||
| rplV | 50S ribosomal subunit protein L22 | 20.59 | 1.71 | 0.010 | ||
| rpoA | RNA polymerase, alpha subunit | 20.33 | 1.28 | 0.007 | + | |
| ptsH | PTS system protein HPr | 18.67 | 0.53 | <0.001 | ||
| rpsG | 30S ribosomal subunit protein S7 | 18.45 | 1.44 | 0.001 | + | |
| fimI | Type 1 fimbrial protein of unknown function | 17.97 | 3.65 | 0.007 | ||
| rpsL | 30S ribosomal subunit protein S12 | 17.71 | 1.28 | 0.008 | + | |
| rplN | 50S ribosomal subunit protein L14 | 17.40 | 1.70 | 0.006 | + | |
| ompC | Outer membrane protein C (porin) | 17.27 | −0.84 | 0.002 | + | + |
| rplK | 50S ribosomal subunit protein L11 | 16.78 | 2.09 | <0.001 | ||
| c1041 | Unknown | 16.08 | −1.01 | 0.016 | + | |
| rpsR | 30S ribosomal subunit protein S18 | 15.99 | 2.66 | <0.001 | ||
| rpsB | 30S ribosomal subunit protein S2 | 15.75 | 2.20 | 0.001 | ||
| uxuA | Mannonate hydrolase | 15.60 | 4.74 | 0.003 | ||
| rpsA | 30S ribosomal subunit protein S1 | 15.51 | 2.16 | <0.001 | ||
| c4086 | Hypothetical protein | 15.50 | 3.57 | 0.017 | ||
| ahpC | Alkyl hydroperoxide reductase, C22 subunit | 15.13 | 0.89 | 0.005 | ||
| rplA | 50S ribosomal subunit protein L1 | 14.91 | 2.74 | 0.020 | ||
| infC | Protein chain initiation factor IF-3 | 14.46 | −0.09 | 0.408 | + | + |
| c0287 | Hypothetical protein | 14.46 | 1.28 | 0.036 | ||
| aceE | Pyruvate dehydrogenase (decarboxylase) | 14.14 | 1.05 | 0.042 | ||
| acpP | Acyl carrier protein | 13.85 | 0.09 | 0.579 |
Fold change refers to growth in vivo relative to growth in LB.
Genes statistically upregulated in vivo at least twofold are in boldface.
A plus sign indicates this gene was also present among the 50 most expressed genes in human urine or LB.
Adhesins.
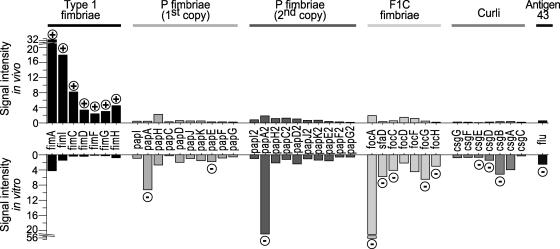
Colonization is a major challenge facing UPEC in the urinary tract. Thus, adherence factors that aid in this process are essential to survival. The genome sequence of E. coli CFT073 revealed an abundance of fimbrial adhesins relative to levels in both enterohemorrhagic and nonpathogenic laboratory E. coli strains. Indeed, as many as 12 fimbrial gene clusters have been predicted or demonstrated (4, 21, 42). Our in vivo microarray data revealed that the fim genes encoding type 1 fimbriae were preferentially expressed 12 to 72 times more highly than each of the 11 other fimbrial gene clusters. Additionally, fimA, encoding the major structural subunit, was the fourth most highly expressed gene in vivo overall; the first three were related to translation. fim genes were the only genes previously implicated as virulence factors among the 50 most highly expressed genes in vivo (Table 2). The 11 remaining fimbrial gene clusters were minimally expressed, if at all.
Not only were type 1 fimbriae highly expressed, but they were also highly upregulated (three- to fivefold) (Fig. 2) compared to levels for the LB culture grown statically, conditions known to enhance type 1 fimbrial production (33). Both of the pap gene clusters of strain CFT073 encoding P fimbriae were downregulated two- to fourfold, and the foc/sfa gene cluster and sfaB recombinase of F1C fimbriae were also downregulated two- to fivefold. The csg genes, which encode thin aggregative curli fibers, were downregulated two- to fourfold in vivo. The flu gene, encoding antigen 43 and implicated in autoaggregation and biofilm formation, was also downregulated twofold in vivo (Fig. 2). Besides type 1 fimbriae, only one other adhesion-related gene (ydeS, encoding a hypothetical fimbria-like structural protein) was upregulated during UTI.
FIG. 2.
Expression of adhesins in E. coli CFT073. The signal intensity, corresponding to the relative expression of a gene, is shown for selected adhesin genes in vivo (growth during UTI) or in vitro (growth in static LB culture). Genes upregulated (encircled plus signs) or downregulated (encircled minus signs) in vivo relative to in vitro are indicated.
Iron acquisition systems.
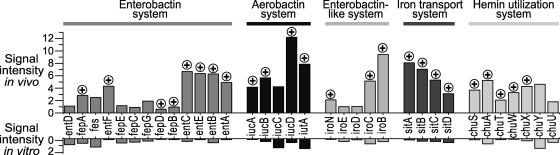
Iron is essential for growth of E. coli, and five iron uptake and assimilation systems were upregulated during bacterial growth in the urinary tract. Siderophore systems were upregulated in vivo two- to fivefold, providing evidence that the urinary tract is an iron-limiting environment (31). These systems include ent genes encoding enterobactin, iuc genes encoding aerobactin, and iro genes encoding an ent-like system. Additionally, iutA, exbD, fhuA, fhuC, and the fep genes, all involved in the transport of these ferrisiderophore complexes; the chu genes, required for hemin utilization; and sit genes, encoding an iron transport system, were upregulated two- to fivefold (Fig. 3). The expression of tonB, necessary for iron and siderophore uptake, remained unchanged between in vivo and LB growth.
FIG. 3.
Expression of iron acquisition systems in E. coli CFT073. The signal intensity, corresponding to the relative expression of a gene, is shown for selected iron acquisition systems in vivo (growth during UTI) or in vitro (growth in static LB culture). Genes upregulated (encircled plus signs) in vivo relative to the same genes in vitro are indicated.
Motility.
E. coli CFT073 is motile, as is characteristic of the species. There has been speculation that flagella assist UPEC in the ascent from the bladder to the kidneys through the ureters (29). The role of flagella in colonization is unknown, but epidemiological data indicate that nonmotile strains may be overrepresented among UTI isolates (10, 16). Our in vivo microarray data demonstrated that several genes involved in flagellation were downregulated in comparison to these genes during LB growth (Table 3), including flgL and flgK, class III genes encoding hook-filament flagellar structural components, and cheW, encoding a positive regulator of chemotaxis. Importantly, the fliC gene, encoding flagellin, was downregulated fivefold. Although not meeting statistical criteria, all 11 remaining class III flagellar and chemotaxis genes demonstrated a trend toward downregulation in vivo. Overall, these data do not support a primary role for motility during infection of the urinary tract.
TABLE 3.
Expression of E. coli CFT073 class III genes of motility and chemotaxis
| Gene | Function | Fold changea | P value |
|---|---|---|---|
| flgLb | Flagellar biosynthesis; hook-filament junction protein | −3.19 | 0.054 |
| flgK | Flagellar biosynthesis, hook-filament junction protein 1 | −2.36 | 0.004 |
| fliD | Flagellar biosynthesis; filament capping protein; enables filament assembly | −1.31 | 0.018 |
| fliS | Flagellar biosynthesis; repressor of class 3a and 3b operons (RflA activity) | −2.53 | 0.206 |
| fliT | Flagellar biosynthesis; repressor of class 3a and 3b operons (RflA activity) | −1.04 | 0.441 |
| fliC | Flagellar biosynthesis; flagellin, filament structural protein | −4.65 | 0.006 |
| tar | Methyl-accepting chemotaxis protein II, aspartate sensor receptor | −3.88 | 0.060 |
| cheZ | Chemotactic response; CheY protein phophatase | −3.49 | 0.107 |
| cheY | Transmits chemoreceptor signals to flagellar motor components | −3.73 | 0.146 |
| cheB | Response regulator for chemotaxis (CheA sensor); protein methylesterase | −0.89 | 0.397 |
| cheR | Response regulator for chemotaxis; protein glutamate methyltransferase | −0.35 | 0.678 |
| cheW | Positive regulator of CheA protein activity | −3.35 | 0.012 |
| cheA | Sensory transducer kinase between signal receptors and CheB and CheY | −2.03 | 0.117 |
| motB | Enables flagellar motor rotation | −1.36 | 0.186 |
| motA | Proton conductor component of motor | −0.14 | 0.903 |
Fold change refers to growth in vivo relative to growth in LB.
Genes statistically downregulated in vivo at least two-fold are in boldface.
Capsule and LPS.
Capsular polysaccharide and lipopolysaccharide (LPS) O side chains may be involved in UPEC virulence, perhaps by conferring resistance to serum and phagocytosis (18). These microarray data confirmed the importance of extracellular polysaccharides. The genes kpsE, kpsD, kpsC, and kpsS, involved in the transport of polysaccharide to the cell surface, were upregulated two- to fourfold during infection. lpxA and lpxB, both involved in lipid A biosynthesis, were upregulated three- to fivefold. nanA, encoding an enzyme involved in capsular sialic acid catabolism, and wzzE, encoding a putative transport protein involved in LPS biosynthesis, were also upregulated in vivo.
Antibiotic resistance.
Genes that confer antibiotic resistance were found upregulated during bacterial growth in vivo. These include marAB (two- to three fold), encoding multiple antibiotic resistance proteins, yhjX (fivefold), encoding a putative drug resistance protein, and emrA (twofold), encoding a multidrug resistance secretion protein. Microcins are secreted antibiotic peptides that may be used to compete with surrounding bacteria. mchCDEF, encoding microcin H47, was upregulated two- to fourfold in the urinary tract.
E. coli CFT073 transcriptome as a probe for nutrient levels and conditions within the urinary tract.
E. coli senses its environment and modulates gene expression to best utilize available resources. By examining the transcriptome of E. coli CFT073 isolated from the urine of infected mice, we established that these bacteria can act as sensitive bioprobes to reveal the composition of this environment.
Oxygen level.
E. coli, a facultative anaerobe, can adapt to growth in various redox environments. Based on the pattern of gene expression of E. coli CFT073, the mouse urinary tract is more oxygen rich than the LB static culture. Many genes indicative of anaerobiosis were downregulated under in vivo conditions. Most interestingly, frdABCD (encoding fumarate reductase), glpABC (encoding sn-glycerol-3-phosphate dehydrogenase), and aspA (encoding aspartase), all Fnr-regulated genes downregulated during growth in the urinary tract, were all found to be upregulated in Vibrio cholerae during in vivo growth in the intestinal rabbit ileal loop model (45). tdcABC (encoding genes for anaerobic threonine usage), tdcE (encoding formate acetyltransferase), and Fnr-regulated genes ansB (encoding l-asparaginase II) and hypC (affects hydrogenase activity) were also downregulated during growth in the urinary tract. Accordingly, cyoAB and cyoE, cytochrome o genes indicative of aerobic growth, were upregulated twofold in vivo. On the other hand, the fdnGHI operon encoding anaerobic formate dehydrogenase-N was upregulated under in vivo conditions, thus implying oxygen limitation. arcA and fnr, the key regulators of respiration and indicators of oxygenation, remained unchanged between in vivo and LB growth. Thus, bacteria were growing neither strictly anaerobically nor strictly aerobically. The urinary tract is likely a combination of these conditions, as bacteria isolated from urine may have been growing in different niches of the urinary tract with different levels of oxygenation.
Nitrogen limitation.
Although nitrogen is abundant in urine (e.g., urea is present at ∼0.5 M [11]), it is a limiting resource in the urinary tract for E. coli, which typically lacks the urease enzyme required to catalyze the hydrolysis of urea to ammonia and CO2. Nitrogen limitation induces nitrogen-regulated (Ntr) genes, such as glnA, which encodes glutamine synthetase. While growing in the urinary tract, glnA was upregulated fourfold. To provide an exogenous nitrogen source, glutamine importers glnP and glnQ were also upregulated threefold. gadA and gadB (encoding glutamate decarboxylases), for the conversion of l-glutamate to γ-aminobutyrate, and ybaS (encoding a probable glutaminase), for the degradation of glutamine, were downregulated 7- to 10-fold, perhaps as a coordinated response to maximize the amount of ammonia assimilated. These data indicate that E. coli CFT073 faces nitrogen limitation in the urinary tract.
Iron limitation.
E. coli CFT073 faces iron limitation in the urinary tract as indicated by the uniform upregulation of iron acquisition systems as described above.
High osmolarity and osmotic stress.
The cytosolic concentration of osmoprotectants is elevated under high osmolarity, as measured here with the upregulation of pro gene expression of E. coli CFT073 in the urinary tract. proP and proVWX, encoding systems for the transport of osmoprotectants proline and glycine betaine, were upregulated two- to fivefold in vivo.
Carbon utilization and starvation response.
E. coli CFT073 utilized different sources of carbon depending on whether growth was in vivo or in LB. While in the urinary tract, metabolism shifted so that hexuronates and hexanates could be exploited to support growth. uxuA and uxuB, encoding mannonate dehydratase and oxidoreductase, and uxaA and uxaB, encoding altronate hydrolase and oxidoreductase, respectively, were upregulated two- to fivefold. Genes encoding glucitol and fructose uptake and catabolism were upregulated three- to fivefold. The genes encoding the uptake and catabolism of glycerol, sn-glycerol-3-phosphate, and trehalose were downregulated three- to sevenfold in vivo. Carbon sources that were used equally during in vivo and LB growth conditions include glucose, mannose, and mannitol. Genes encoding carbon starvation proteins were not consistently up- or downregulated. Thus, E. coli CFT073 was not limited for carbon sources during acute infection of the urinary tract.
Conditions unchanged between in vivo and LB growth.
Groups of genes that were not differentially expressed between in vivo and LB growth conditions provide insight into the similarity between these environments. There was no measurable change in genes that normally respond to pH, oxidative stress, DNA damage, heat or cold shock, cell density dependence, sulfur availability, or phosphate availability (data not shown).
Verification of microarray.
qRT-PCR was used to independently verify the levels of transcript for an example gene, papA_2, which was found downregulated during growth in vivo 3.5-fold by microarray analysis. gapA, encoding glyceraldehyde 3-phosphate dehydrogenase, was used as the normalizing internal standard. Microarray analysis demonstrated that gapA expression remained unchanged between in vivo and in vitro growth, thus confirming the validity of this standard. qRT-PCR analysis demonstrated a greater transcript level sensitivity than microarray analysis, as papA_2 was downregulated 6.24-fold ± 0.15-fold during growth in vivo relative to growth in LB.
To verify the biological reproducibility of our in vivo microarray analysis, the results presented here were compared to those of an analogous microarray of an E. coli CFT073 dsdA (d-serine deaminase) mutant isolated from the urine of infected CBA/J mice. The upregulation of specific genes encoding type 1 fimbriae, several iron acquisition systems, capsular polysaccharide, and microcin during the in vivo growth of E. coli CFT073 dsdA demonstrated the biological reproducibility of our major findings (our unpublished data). Pearson's correlation coefficient (r2) between wild-type E. coli CFT073 grown in vivo and CFT073 dsdA grown in vivo was 0.7583. By comparison, the correlation coefficient between wild-type E. coli CFT073 grown in vivo and grown in vitro in LB was 0.4059, suggesting that the wild type and dsdA mutant have similar overall gene expression patterns when grown in vivo.
Alterations in the transcriptome of E. coli CFT073 during growth in human urine.
E. coli CFT073 grown to mid-exponential phase in filtered human urine partially mimics growth in the urinary tract. The transcriptome of E. coli CFT073 grown in vitro in human urine, relative to growth in LB, demonstrated the upregulation of genes encoding two iron acquisition systems (iuc and iro gene clusters), capsular sialic acid catabolism genes (nanA and nanT), and a microcin secretion gene (mchB). Overall, 54 genes were found upregulated (Table 4 lists the 50 most upregulated) and 88 genes were found downregulated during growth in human urine. Many of the most highly expressed genes in vivo are also among the most highly expressed in human urine, including ompA, ompC, and many ribosomal protein genes (Table 2). However, in sharp contrast to growth in vivo, papA_2 (encoding the major P fimbriae subunit), focA (encoding the major F1C fimbriae subunit), and fliC (encoding flagellin) were highly expressed in human urine (data not shown).
TABLE 4.
Top 50 genes upregulated during in vitro growth in human urine
| Gene | Function | Fold changea | P value | Also upregulated in vivob |
|---|---|---|---|---|
| yeiC | Hypothetical sugar kinase YeiC | 6.646 | 0.0022 | + |
| rbsD | High affinity ribose transport protein | 4.987 | 0.0021 | |
| artJ | Arginine-binding periplasmic protein 2 precursor | 4.730 | 0.0195 | + |
| c0336 | PTS system, mannitol (cryptic)-specific IIA component | 4.721 | 0.0032 | |
| ompF | Outer membrane protein F precursor | 4.686 | 0.0048 | |
| iucD | Aerobactin protein IucD | 4.587 | 0.0047 | + |
| c0335 | Hypothetical protein | 4.513 | 0.0032 | + |
| nanA | N-acetylneuraminate lyase subunit | 4.225 | 0.0374 | + |
| leuC | 3-Isopropylmalate dehydratase large subunit | 4.180 | 0.0219 | |
| iroB | Siderophore glucosyltransferase | 4.144 | 0.0138 | + |
| leuB | 3-Isopropylmalate dehydrogenase | 4.141 | 0.0232 | |
| dppA | Periplasmic dipeptide transport protein precursor | 4.124 | 0.0131 | + |
| c0088 | Hypothetical protein | 4.030 | 0.0377 | |
| uxuA | Mannonate dehydratase | 3.967 | 0.0170 | + |
| c0334 | Putative integral membrane protein | 3.889 | 0.0079 | |
| ilvC | Ketol-acid reductoisomerase | 3.887 | 0.0343 | |
| yncE | Hypothetical protein YncE precursor | 3.849 | 0.0225 | + |
| ybiW | Putative formate acetyltransferase 3 | 3.728 | 0.0491 | |
| yeaR | Hypothetical protein YeaR | 3.709 | 0.0202 | + |
| argC | N-acetyl-gamma-glutamyl-phosphate reductase | 3.682 | 0.0267 | |
| ilvE | Branched-chain amino acid aminotransferase | 3.594 | 0.0274 | + |
| narK | Nitrite extrusion protein 1 | 3.568 | 0.0422 | |
| ybeJ | Glutamate/aspartate periplasmic binding protein precursor | 3.370 | 0.0262 | + |
| argB | Acetylglutamate kinase | 3.363 | 0.0320 | |
| ygbJ | Hypothetical oxidoreductase YgbJ | 3.263 | 0.0257 | |
| nanT | Putative sialic acid transporter | 3.194 | 0.0241 | |
| fdnH | Formate dehydrogenase-N beta subunit | 3.155 | 0.0427 | + |
| galE | UDP-glucose 4-epimerase | 3.039 | 0.0235 | |
| narG | Respiratory nitrate reductase 1 alpha chain | 3.014 | 0.0287 | |
| leuL | Leu operon leader peptide | 3.003 | 0.0167 | |
| fdnI | Formate dehydrogenase-N gamma subunit | 2.953 | 0.0207 | + |
| mglA | Galactoside transport ATP-binding protein | 2.925 | 0.0226 | |
| galK | Galactokinase | 2.895 | 0.0455 | |
| hmpA | Flavohemoprotein (hemoglobin-like protein) | 2.843 | 0.0302 | + |
| iutA | Aerobactin protein IutA | 2.796 | 0.0349 | + |
| serA | d-3-phosphoglycerate dehydrogenase | 2.793 | 0.0444 | |
| ytfE | Hypothetical protein YtfE | 2.753 | 0.0395 | + |
| c3835 | Hypothetical protein | 2.582 | 0.0401 | |
| livG | High-affinity branched-chain amino acid transport ATP-binding protein | 2.479 | 0.0493 | |
| ilvH | Acetolactate synthase isozyme III small subunit | 2.443 | 0.0166 | |
| uxaC | Uronate isomerase | 2.436 | 0.0392 | |
| livJ | Leu/lle/Val-binding protein precursor | 2.416 | 0.0291 | |
| hisJ | Histidine-binding periplasmic protein precursor | 2.385 | 0.0117 | |
| yjiY | Hypothetical protein YjiY | 2.278 | 0.0479 | |
| mchB | Microcin H47 secretion protein | 2.228 | 0.0462 | |
| asnB | Asparagine synthetase B (glutamine hydrolyzing) | 2.204 | 0.0395 | |
| cyoD | Cytochrome o ubiquinol oxidase protein | 2.184 | 0.0458 | |
| narI | Respiratory nitrate reductase 1 gamma chain | 2.176 | 0.0278 | |
| cyoE | Protoheme IX farnesyltransferase | 2.176 | 0.0370 | + |
| argE | Acetylornithine deacetylase | 2.088 | 0.0199 |
Fold change refers to growth in human urine relative to growth in LB.
A plus sign indicates this gene was also upregulated during growth in vivo relative to growth in LB.
DISCUSSION
This study represents the first quantification of the genome-wide transcriptional profile of the most common human uropathogen, E. coli, during colonization of the urinary tract. These measurements, taken with bacteria collected from the urine of infected mice, allow us to assess the conditions in the urinary tract as seen from the point of view of the pathogen. We describe the first use of microarray technology, based on the sequence of a urinary tract pathogen, to globally examine the induction of specific virulence genes and the coordinate regulation among different virulence and metabolic gene clusters during a UTI. Expression patterns were verified by an example gene by qRT-PCR and by a comparison of major findings to those of in vivo microarray analysis of an E. coli CFT073 dsdA mutant. In addition, this microarray analysis represents the first assessment of the transcriptome of any E. coli pathotype in vivo.
The transcriptional analysis of E. coli CFT073 during colonization of the murine urinary tract was feasible because the urinary tract is naturally sterile and because the amount of host nucleic acid contamination from exfoliated epithelial cells (12, 30) was insignificant. This allowed us to collect the urine from experimentally infected mice and directly extract CFT073-specific RNA. We experienced limited complications in isolating ample RNA, provided a sufficient volume of urine was collected from infected animals. Previous studies demonstrated that individual CBA/J mice are consistently and relatively uniformly infected with E. coli CFT073 (13). Additionally, urine from 20 mice individually cultured at the conclusion of these microarray experiments demonstrated that all mice remained infected (data not shown). Urine was sampled and pooled over a range of time (1 to 10 days) from a large collection of infected mice (n = 40). Bacteria expelled in the urine likely represent an appropriate sample of the bacteria that had recently infected the kidneys, ureters, bladder, and urethra. Therefore, we assume that the RNA used for this in vivo microarray analysis provides a representation of the responses of a uropathogen to a summation of these environmental situations encountered in the urinary tract.
Urine was not collected within the first 24 h of the experimental UTI. We desired to exclude noncolonizing bacteria that were present in the inoculum that may have washed out following transurethral challenge. Instead we focused on a bacterial population that had colonized and adapted to the murine urinary tract. Admittedly, this analysis likely excluded the transient transcription of genes encoding colonization factors and other virulence determinants required for the transition from the in vitro LB culture to growth in the urinary tract. Additionally, any gene transiently expressed during the ongoing urine collection would be diluted out in the process of pooling urine samples. Thus, our analysis emphasizes genes that are efficiently expressed throughout space and time during UTI.
Microarray data presented here are internally consistent and compatible with previous studies. There is coordinated regulation of genes within an operon, and transcript levels reflect the expected abundance. For example, the entire fimAICDFGH gene cluster of type 1 fimbriae is upregulated in vivo compared to that in LB growth (Fig. 2), and as expected there is significantly more fimA (major structural subunit gene) transcript than fimI, more fimI than fimC, more fimC than fimD, and more fimD than fimF. This is consistent with the prediction of a transcript-stabilizing stem-loop structure at the end of fimA (34) as well as in vitro expression studies of other chaperone-usher fimbrial gene clusters (2). Genes within iron acquisition operons are also uniformly upregulated (Fig. 3), consistent with a previous study of E. coli CFT073 that observed the induction of three siderophore systems during murine peritonitis (35).
We affirm the importance of several known virulence genes by reporting the upregulation of these genes in vivo, such as those encoding type 1 fimbriae, siderophores, capsule, drug resistance, and microcin. We have also identified 13 new candidate virulence genes, encoding hypothetical proteins, which were upregulated during in vivo growth and were simultaneously not found in the nonpathogenic E. coli K-12 strain. Additionally, these analyses provide valuable information on the expression of surface proteins in vivo for the purpose of vaccine-directed research.
Several other microarray analyses found that genes encoding ribosomal proteins were highly expressed when cells were exponentially growing in rich media (5, 41, 45). This suggests that the high level of in vivo expression of genes involved in translation indicates a rapid rate of exponential growth of pyelonephritogenic E. coli CFT073 in the urinary tract. This is somewhat surprising, given that urine is an incomplete and relatively poor growth medium in vitro (19, 38). Although bacteria may acquire some nutrients via contact with the urinary tract epithelium, our experiments indicate that nitrogen and iron remain limiting. In addition, bacteria are expending additional energy responding to osmotic stress and producing virulence and metabolic factors specific for survival in the host. Despite these specific nutrient deprivations and energy expenditures, rapid growth is sustained in the urinary tract. This is in contrast to observations of V. cholerae during growth in the intestine, where fewer genes for protein synthesis tended to be found among the 300 most highly expressed genes compared to that of growth in LB (45).
Microarray analysis demonstrates a strikingly high level of type 1 fimbrial expression during growth in vivo, as fimA expression was the fourth highest gene expressed overall. In addition, type 1 fimbrial genes are highly upregulated compared to that of growth in LB. These data support the findings of studies using signature-tagged mutagenesis and phase-locked mutants (3, 13). Type 1 fimbrial expression in vivo contrasts sharply with the near lack of expression of the 11 other fimbrial types previously demonstrated (4, 21) or predicted by the E. coli CFT073 genome (42). Coordinate regulation among fimbrial operons has been described previously and may provide an explanation for this observation (17, 44). P fimbriae, argued as being virulence factors during UTI (43), are found downregulated here in vivo despite the presence of P-fimbrial Gal-Gal receptors in mice (22, 32). Our microarray experiments suggest that a better way to study the importance of other adherence factors may be use of a Fim-deficient UPEC mutant.
The signals necessary for transcriptional alterations in UPEC during transition from growth in the intestine to growth in the urinary tract remain undisclosed. However, this microarray analysis of bacteria responding to the urinary tract can be compared to the two other true in vivo microarray analyses, where V. cholerae was isolated from rice-water stools of infected patients (5, 27). By using bacteria as bioprobes for their respective environments, differences in these niches are revealed by patterns of bacterial gene expression. For example, the urinary tract is nitrogen and iron limiting for E. coli, of moderate oxygenation, and of higher osmolarity and pH than the anaerobic, nitrogen-rich gastrointestinal tract.
Differences in gene expression between growth in vivo and growth in vitro in human urine may be partially due to species-specific differences in urine composition. This use of human urine as a growth medium thus examines the important relationship between in vivo murine studies and human infection. This work also suggests that in vitro growth of UPEC in human urine, rather than in LB, is a useful tool to more closely imitate growth conditions encountered during UTI. The upregulation of important virulence factors, including iron acquisition systems, capsule, and microcin, was observed. However, the most highly expressed virulence factor during UTI, type 1 fimbriae, was not induced in urine. The examination of gene expression in vivo by using the experimental murine model of ascending UTI thus remains essential.
Acknowledgments
We thank Jim Kaper for critical reading of the manuscript and the Genome Expression Center at the University of Wisconsin—Madison.
This work was supported by National Institutes of Health (NIH) grants AI43363 (H.L.T.M.), DK49720 (H.L.T.M. and M.S.D), and DK63250 (R.A.W.) and by NIH National Research Service award A0T32GM072125 (B.J.H.). Custom NimbleGen microarrays were supported by NIH SBIR grant R44-HG-02193 to NimbleGen Systems with a subcontract to the Application Development Center at the University of Wisconsin—Madison.
Editor: A. D. O'Brien
REFERENCES
- 1.Allen, T. E., and B. O. Palsson. 2003. Sequence-based analysis of metabolic demands for protein synthesis in prokaryotes. J. Theor. Biol. 220:1-18. [DOI] [PubMed] [Google Scholar]
- 2.Baga, M., M. Goransson, S. Normark, and B. E. Uhlin. 1985. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EMBO J. 4:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani-Mougeot, F. K., S. Pancholi, M. Daoust, and M. S. Donnenberg. 2001. Identification of putative urovirulence genes by subtractive cloning. J. Infect. Dis. 183(Suppl. 1):S21-S23. [DOI] [PubMed] [Google Scholar]
- 5.Bina, J., J. Zhu, M. Dziejman, S. Faruque, S. Calderwood, and J. Mekalanos. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. USA 100:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 7.Connell, I., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 93:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culham, D. E., C. Dalgado, C. L. Gyles, D. Mamelak, S. MacLellan, and J. M. Wood. 1998. Osmoregulatory transporter ProP influences colonization of the urinary tract by Escherichia coli. Microbiology 144:91-102. [DOI] [PubMed] [Google Scholar]
- 9.Falkow, S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10(Suppl. 2):S274—S276. [DOI] [PubMed] [Google Scholar]
- 10.Green, C. P., and V. L. Thomas. 1981. Hemagglutination of human type O erythrocytes, hemolysin production, and serogrouping of Escherichia coli isolates from patients with acute pyelonephritis, cystitis, and asymptomatic bacteriuria. Infect. Immun. 31:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith, D. P., D. M. Musher, and C. Itin. 1976. Urease. The primary cause of infection-induced urinary stones. Investig. Urol. 13:346-350. [PubMed] [Google Scholar]
- 12.Gunther, N. W. I., V. Lockatell, D. E. Johnson, and H. L. Mobley. 2001. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect. Immun. 69:2838-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunther, N. W. I., J. A. Snyder, V. Lockatell, I. Blomfield, D. E. Johnson, and H. L. Mobley. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 70:3344-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guyer, D. M., S. Radulovic, F. E. Jones, and H. L. Mobley. 2002. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect. Immun. 70:4539-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagberg, L., I. Engberg, R. Freter, J. Lam, S. Olling, and C. Svanborg Eden. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrmann, B., and L. G. Burman. 1985. Pathogenesis of Escherichia coli cystitis and pyelonephritis: apparent lack of significance of bacterial motility and chemotaxis towards human urine. Infection 13:4-7. [DOI] [PubMed] [Google Scholar]
- 17.Holden, N. J., B. E. Uhlin, and D. L. Gally. 2001. PapB paralogues and their effect on the phase variation of type 1 fimbriae in Escherichia coli. Mol. Microbiol. 42:319-330. [DOI] [PubMed] [Google Scholar]
- 18.Horwitz, M. A., and S. C. Silverstein. 1980. Influence of the Escherichia coli capsule on complement fixation and on phagocytosis and killing by human phagocytes. J. Clin. Investig. 65:82-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hull, R. A., and S. I. Hull. 1997. Nutritional requirements for growth of uropathogenic Escherichia coli in human urine. Infect. Immun. 65:1960-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, D. E., C. V. Lockatell, R. G. Russell, J. R. Hebel, M. D. Island, A. Stapleton, W. E. Stamm, and J. W. Warren. 1998. Comparison of Escherichia coli strains recovered from human cystitis and pyelonephritis infections in transurethrally challenged mice. Infect. Immun. 66:3059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, J. R., T. Berggren, D. S. Newburg, R. H. McCluer, and J. C. Manivel. 1992. Detailed histopathological examination contributes to the assessment of Escherichia coli urovirulence. J. Urol. 147:1160-1166. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, J. R., A. L. Stell, P. Delavari, A. C. Murray, M. Kuskowski, and W. Gaastra. 2001. Phylogenetic and pathotypic similarities between Escherichia coli isolates from urinary tract infections in dogs and extraintestinal infections in humans. J. Infect. Dis. 183:897-906. [DOI] [PubMed] [Google Scholar]
- 25.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 26.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 27.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mobley, H. L., M. D. Island, and G. Massad. 1994. Virulence determinants of uropathogenic Escherichia coli and Proteus mirabilis. Kidney Int. Suppl. 47:S129-S136. [PubMed] [Google Scholar]
- 30.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 31.Neidhardt, F. C., and R. Curtiss. 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 32.O'Hanley, P., D. Lark, S. Falkow, and G. Schoolnik. 1985. Molecular basis of Escherichia coli colonization of the upper urinary tract in BALB/c mice. Gal-Gal pili immunization prevents Escherichia coli pyelonephritis in the BALB/c mouse model of human pyelonephritis. J. Clin. Investig. 75:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Old, D. C., and J. P. Duguid. 1970. Selective outgrowth of fimbriate bacteria in static liquid medium. J. Bacteriol. 103:447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen, P. B., and P. Klemm. 1994. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett. 116:95-100. [DOI] [PubMed] [Google Scholar]
- 35.Redford, P., P. L. Roesch, and R. A. Welch. 2003. DegS is necessary for virulence and is among extraintestinal Escherichia coli genes induced in murine peritonitis. Infect. Immun. 71:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rippere-Lampe, K. E., A. D. O'Brien, R. Conran, and H. A. Lockman. 2001. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf1) attenuates the virulence of uropathogenic Escherichia coli. Infect. Immun. 69:3954-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo, T. A., S. T. Jodush, J. J. Brown, and J. R. Johnson. 1996. Identification of two previously unrecognized genes (guaA and argC) important for uropathogenesis. Mol. Microbiol. 22:217-229. [DOI] [PubMed] [Google Scholar]
- 39.Schappert, S. M. 1999. Ambulatory care visits to physician offices, hospital outpatient departments, and emergency departments: United States Series 13, no. 143. National Center for Health Statistics, Centers for Disease Control and Prevention, Atlanta, Ga. [PubMed]
- 40.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei, Y., J. M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wullt, B., G. Bergsten, M. Samuelsson, N. Gebretsadik, R. Hull, and C. Svanborg. 2001. The role of P fimbriae for colonization and host response induction in the human urinary tract. J. Infect. Dis. 183(Suppl. 1):S43—S46. [DOI] [PubMed] [Google Scholar]
- 44.Xia, Y., D. Gally, K. Forsman-Semb, and B. E. Uhlin. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 19:1450-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zielske, J. V., K. N. Lohr, R. H. Brook, and G. A. Goldberg. 1981. Conceptualization and measurement of physiologic health for adults. Urinary tract infection, vol. 16. R-2262/16-HHS. The Rand Corporation, Washington, D.C.