Abstract
Haemophilus ducreyi is a strict human pathogen and the causative agent of the sexually transmitted disease chancroid. The genome of the human-passaged strain of H. ducreyi (35000HP) contains two homologous genes whose protein products have estimated molecular masses of 46 and 43 kDa. A comparative analysis of the deduced amino acid sequences revealed that these proteins share 27 to 33% identity to the outer membrane protein P2 (OmpP2), a major porin of Haemophilus influenzae. Therefore, these proteins have been designated OmpP2A and OmpP2B, respectively. The detection of ompP2A and ompP2B transcripts by reverse transcriptase PCR indicated that these genes were independently transcribed in H. ducreyi 35000HP. Western blot analysis of outer membrane proteins isolated from a geographically diverse collection of H. ducreyi clinical isolates revealed that OmpP2A and OmpP2B were differentially expressed among these strains. Although PCR analysis suggested that ompP2A and ompP2B were conserved among the strains tested, the differential expression observed was due to nucleotide additions and partial gene deletions. Purified OmpP2A and OmpP2B were isolated under nondenaturing conditions, and subsequent analysis demonstrated that these two proteins exhibited porin activity. OmpP2A and OmpP2B are the first porins described for H. ducreyi.
The gram-negative bacterium Haemophilus ducreyi is the causative agent of the sexually transmitted genital ulcerative disease chancroid (23, 34, 37, 42). Chancroid is endemic to underdeveloped countries of Africa, Asia, Latin America, and the Caribbean (37, 42). Although relatively uncommon in developed countries, it is estimated that over 7 million cases of chancroid occur each year (19, 23, 37). H. ducreyi is a strict human pathogen, and infection with this bacterium has been implicated as a cofactor in the transmission of human immunodeficiency virus (20, 37). In fact, chancroid is a common infection in all 18 countries where the adult human immunodeficiency virus prevalence surpasses 8% of the population (37). Protective immunity is not induced after infection, and the only effective treatment involves the use of antibiotics (23, 34, 37). As a result, antibiotic-resistant isolates have emerged, thus complicating the standard therapeutic regimen (19, 23).
The bacterial outer membrane (OM) represents the interface between the organism and the extracellular environment. Thus, the OM and its constituents play a pivotal role in cell survival and the host-pathogen interaction. Characterization of OM proteins (OMPs) of H. ducreyi has led to the identification of several proteins with important biological functions. Some of these include a hemolysin, the hemoglobin binding receptor HgbA, a TonB-dependent heme receptor, TdhA, cytolethal distending toxin, fimbria-like protein cluster, the large supernatant proteins LspA1 and LspA2, and ducreyi serum resistance antigen DsrA (10, 11, 22, 30, 35, 38, 46). Despite research efforts focusing on the characterization of OMPs, the biology of H. ducreyi remains poorly understood. Interestingly, to date there have been no reports describing H. ducreyi porins. Porins not only facilitate nutrient acquisition and provide membrane stability but have also been shown to act as virulence determinants, stimulate immune responses, and influence antibiotic susceptibility (1, 2, 8, 14, 15, 17, 26, 27, 45). Previous studies suggested that the H. ducreyi major outer membrane protein (MOMP) and OmpA2 were putative porins, but subsequent data revealed these proteins did not exhibit porin activity (21, 40).
In this study we have identified two proteins, designated OmpP2A and OmpP2B, that share partial protein identity to the major porin OmpP2 of Haemophilus influenzae (6, 44). Western blot analysis, using antibodies specific to each protein, revealed that OmpP2A and OmpP2B are differentially expressed among clinical isolates from various geographic regions. Sequence analysis of ompP2A and ompP2B from this panel of H. ducreyi isolates indicated that the differential expression was due to nucleotide additions and partial gene deletions. In addition, functional analysis of purified OmpP2A and OmpP2B in black lipid bilayer assays revealed these proteins exhibited porin activity in 1 M KCl and thus represent the first porins identified for H. ducreyi.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in these studies are shown in Table 1. H. ducreyi 35000 was isolated from a patient in Winnipeg, Canada (16). H. ducreyi 35000HP was isolated from a biopsy of a human subject who was experimentally infected with the original 35000 isolate for 13 days, and this strain has been described previously (36). This strain has been sequenced to completion (GenBank accession no. NC_002940). H. ducreyi strain MF35000 is a laboratory variant of 35000 that was recovered after a single passage through humans in the original description of the human challenge model, and this strain has been routinely cultured in our laboratory (36). This variant is uncharacterized as to the similarities and differences with 35000HP, and this strain was primarily used as a tool to initially identify OmpP2B. The other clinical isolates were selected in order to represent a diverse collection. All H. ducreyi strains were routinely cultured at 35°C in 5% CO2 on chocolate agar plates containing gonococcal agar (36 g/liter; Becton-Dickinson, Cockeysville, Md.) and bovine hemoglobin (10 g/liter; U.S. Biochemicals, Cleveland, Ohio) supplemented with 1% (vol/vol) IsoVitaleX (Becton-Dickinson). Escherichia coli was cultured at 37°C on Luria-Bertani agar or broth supplemented with antibiotics when necessary (ampicillin at 100 μg/ml and kanamycin at 30 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| H. ducreyi strains | ||
| 35000HP | Human-passaged 35000 | 36 |
| MF35000 | Derivative of 35000HP | This study |
| 35000 | Clinical isolate, Winnipeg, Canada | 16 |
| A75 | Clinical isolate, London, England | CDCa |
| A76 | Clinical isolate, London, England | CDC |
| CIP542 | Clinical isolate, Pasteur Institute | CDC |
| HD41 | Clinical isolate, Tampa, Fla. | CDC |
| HD151 | Clinical isolate, New Orleans, La. | CDC |
| HD293 | Clinical isolate, Thailand | CDC |
| HD346 | Clinical isolate, Kenya | CDC |
| NY8 | Clinical isolate, New York, N.Y. | CDC |
| R3 | Clinical isolate, Rochester, N.Y. | CDC |
| E. coli strains | ||
| XL-1 Blue MRF′ | Cloning host | Stratagene |
| BL21(DE3) | Recombinant protein expression host | Novagen |
| Plasmids | ||
| pGEM-T Easy | Cloning vector | Promega |
| pET-30a | Expression vector | Novagen |
| pDTP113 | pET-30a + 1.3 kb ompP2B fragment | This study |
| pDTP114 | pET-30a + 1.2 kb ompP2A fragment | This study |
CDC, Centers for Disease Control and Prevention.
OMP isolation and analysis.
Sarkosyl-insoluble OMPs were isolated as previously described (12, 21). Protein samples were diluted 1:1 in sample buffer (0.5 M Tris [pH 6.8], 10% sodium dodecyl sulfate [SDS], 20% glycerol, 0.1% bromophenol blue) and boiled at 100°C for 10 min. SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed with standard procedures previously used in our laboratory. Broad-range prestained molecular weight standards were purchased from Bio-Rad (Hercules, Calif.).
DNA manipulations.
Standard molecular biology techniques were employed. Restriction endonucleases were obtained from New England Biolabs (Beverly, Mass.). Plasmid isolation and DNA extraction from agarose gels were performed using the QIAprep Spin Miniprep and MinElute gel extraction kits, respectively, from QIAGEN (Santa Clarita, Calif.). Chromosomal DNA was isolated as previously described (32). DNA sequence analysis, primer design, and protein analysis were performed using MacVector 7.1.1 and AssemblyLign (Accelyrs Inc., San Diego, Calif.). Primers used in this study are shown in Table 2. Standard 50-μl volume PCRs were performed for 30 cycles using a GeneAMP 9700 series PCR system (PE Applied Biosystems, Foster City, Calif.). DNA was amplified using Pfu polymerase from Stratagene (La Jolla, Calif.).
TABLE 2.
Primers used in the amplification of ompP2A and ompP2B
| Primer | Sequence (5′→3′)c | Region (nt) |
|---|---|---|
| 237b | GGTAGCCGTATTGGTTTATCAGCC | 3207-3230 |
| 238b | GGTTCATCTGCTTTTCACCCATTG | 3723-3700 |
| 254b | ATGAAAAAAACATTTGTAGCATTAG | 2991-3015 |
| 255b | TGTGAAGACTGATACGGGCG | 4704-4685 |
| 290b | GGATCCTCAGCAatgGTTACGCTTTATG | 3042-3060 |
| 291b | CTCGAGTCAACATACACCTTACCTTAT | 4340-4320 |
| 330a | GGATCCATGGTTGTTTATGAAGCAGATG | 1400-1421 |
| 331a | CTCGAGTTACCAAGACACCCGGACACCA | 2617-2596 |
| 364a | GGTAGTCGTTTGGGTATTACTGC | 1631-1653 |
| 365a | CGCTTGAGTTTTATCCGTATTGTC | 2116-2093 |
| 400a | GCGAATACCTCGTAAACTTGAAAG | 2512-2535 |
| 401b | CCAAAACCAACAAAACCACG | 3349-3330 |
| 635a | AGAAATGTTGTCTTATGTCTGG | 1276-1297 |
| 636a | CCATCTATGTCAACACGAGC | 2886-2867 |
| 702a | CCCGATAAGGAAGAAAAATAGC | 787-808 |
Used in the amplification of ompP2A.
Used in the amplification of ompP2B.
Engineered restriction endonuclease sites are in bold letters, BamHI sites are in italics, and XhoI sites are underlined. Nucleotides engineered within the primer are indicated as lowercase letters.
Protein sequence analysis.
OMPs isolated from H. ducreyi MF35000 were separated on an SDS-PAGE gel and stained with Coomassie brilliant blue as described above, and the band corresponding to OmpP2B was excised. Tryptic digest and amino acid analysis were performed by Borealis Biosciences Inc. (Toronto, Canada).
DNA sequencing of ompP2A and ompP2B.
To evaluate genetic conservation, primers 635 and 636 were used to amplify the entire ompP2A open reading frame (ORF) and flanking sequence from strains 35000HP, MF35000, 35000, CIP542, HD41, HD151, HD293, HD346, NY8, and R3. Primers 702 and 636 were used to amplify ompP2A from strains A75 and A76. The entire ompP2B ORF was amplified by PCR from all H. ducreyi strains using primers 254 and 255. Plasmid constructs and PCR products were sequenced at the Roswell Park Cancer Institute Biopolymer Facility (Buffalo, N.Y.).
RNA isolation and RT-PCR.
H. ducreyi strain 35000HP was harvested and resuspended in 3 ml of sterile phosphate-buffered saline (pH 7.4) to an optical density at 600 nm (OD600) of 0.3. Total cellular RNA was isolated using the RNeasy Mini kit (QIAGEN). Residual chromosomal DNA was digested with RQ1 DNase (Promega, Madison, Wis.) as recommended by the manufacturer. Reverse transcriptase PCR (RT-PCR) was performed using the OneStep RT-PCR kit following the manufacturer's suggested parameters (QIAGEN). Primers 364 and 365 were used to amplify an internal 486-bp ompP2A fragment, and primers 237 and 238 were used to amplify an internal 517-bp ompP2B fragment. These primers were determined to be specific for each respective gene (data not shown). Primers 400 and 401, specific for the 3′ end of ompP2A and the 5′ end of ompP2B, were used to evaluate cotranscription. Control reactions were performed in the absence of RT activation and in the absence of a nucleic acid template.
Cloning ompP2A and ompP2B.
Putative signal sequences of OmpP2A and OmpP2B were identified with SignalP Server (Center for Biological Sequence Analysis, Lyngby, Denmark). A 1.2-kb DNA fragment containing mature ompP2A was amplified by PCR from 35000HP chromosomal DNA using primers 330 and 331. The same method was used to amplify a 1.3-kb DNA fragment containing mature ompP2B by using primers 290 and 291. Adenines were added to blunt-ended amplicons as recommended by the pGEM-T Easy vector manual (Promega), and the resulting products were cloned into pGEM-T Easy (Promega). The inserts were subsequently subcloned into the BamHI and XhoI sites within pET30a (Novagen, Madison, Wis.) to create individual His-S-tagged fusion proteins. Plasmids pDTP113 (ompP2B) and pDTP114 (ompP2A) were independently electroporated into the E. coli expression strain BL21(DE3).
Recombinant protein expression and purification.
Recombinant OmpP2A (rOMP2A) and rOmpP2B expression and purification were performed as recommended by the manufacturer (Novagen). In brief, 100-ml cultures were inoculated to an OD600 of 0.05 with E. coli BL21(DE3) harboring either pDTP113 or pDTP114. Cells were grown with vigorous shaking to an OD600 of 0.6, and recombinant protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. Cells were harvested 3 h postinduction. Soluble and insoluble cellular fractions were obtained using the Bugbuster extraction reagent (Novagen) and analyzed by SDS-PAGE. Inclusion bodies, containing rOmpP2A or rOmpP2B, were solubilized in 6 M urea, and the fusion proteins were purified by Ni+ affinity chromatography. Protein samples were dialyzed in 10 mM Tris (pH 8.0) containing 100 mM NaCl and 1 mM EDTA to remove urea.
Polyclonal antibody development.
Polyclonal antisera to rOmpP2A and rOmpP2B were developed by Proteintech Group Inc. (Chicago, Ill.) using their 56-day standard immunization protocol. Briefly, a preimmune bleed was collected on day zero, and two sets of rabbits (two per group) were immunized with 50 μg of either rOmpP2A or rOmpP2B on days 1, 14, 28, and 35. The titers of the antisera were evaluated on day 42 by enzyme-linked immunosorbent assays versus each respective recombinant protein, and each respective antiserum was harvested.
Affinity purification of antibodies.
OmpP2A- and OmpP2B-specific antibodies were purified from the polyclonal antisera using the Amino-Link Plus immobilization kit (Pierce, Rockford, Ill.). Protein coupling and blocking were carried out as recommended by the manufacturer. Approximately 1.5 mg of rOmpP2A and rOmpP2B was independently coupled to the 4% beaded agarose support at pH 10. Antibodies were eluted in 1-ml fractions with 100 mM glycine (pH 2.5). Eluted fractions were neutralized by the addition of 100 μl of 1 M Tris (pH 7.5). Samples were concentrated in a Centricon YM-30 concentrator (Millipore, Bedford, Mass.). Western blotting assays were performed to assess the purity of the antibodies.
OmpP2A and OmpP2B purification.
Sarkosyl-insoluble OMPs isolated from H. ducreyi strain 35000HP were prepared from approximately 300 chocolate agar plates and used to purify OmpP2A and OmpP2B. Unheated protein samples were solubilized in sample buffer and separated on an SDS-PAGE gel (described above). A section of the unstained gel corresponding to either OmpP2A or OmpP2B was excised. Gel slices were eluted overnight at 4°C in 10 mM Tris (pH 8.0) containing 1 mM EDTA and 100 mM NaCl (5). Eluted proteins were concentrated with a Centricon YM-10 concentrator (Millipore). To confirm the purity of OmpP2A and OmpP2B, samples heated (100°C for 10 min) or unheated in sample buffer were analyzed by SDS-PAGE and Western blotting.
Planar lipid bilayer experiments.
Analysis of the pore-forming ability of OmpP2A and OmpP2B was performed using the planar lipid bilayer technique as previously described (18). Briefly, the apparatus consisted of a Teflon chamber with two compartments connected by a small hole (0.1 to 0.2 mm2). A membrane was formed across the hole by painting on a solution of 1.5% (wt/vol) oxidized cholesterol in n-decane. Bilayer formation was indicated by the membrane turning optically black to incident light. Purified protein was diluted into 0.1% (vol/vol) Triton X-100 prior to the addition of a salt solution bathing the planar bilayer membrane. Conductance through the pores was measured after the application of voltage, using a pair of Ag-AgCl electrodes inserted into the aqueous solutions on both sides of the membrane. The current through the pores was boosted by a preamplifier, monitored by a storage oscilloscope, and recorded on a chart recorder. The experiments were repeated at least five times, and the standard deviations represent all of the conductance increments assessed from a single experiment and more than 100 channels.
RESULTS
Identification of the ompP2A and ompP2B loci.
Analysis of OMPs isolated from H. ducreyi MF35000 routinely cultivated in our lab revealed a prominent OMP with an estimated molecular mass of 43 kDa (data not shown). Partial amino acid sequence was obtained by tryptic digest, and subsequent database searches revealed that the amino acid sequence was identical to a translated putative ORF in the H. ducreyi 35000HP genome (GenBank accession no. NC_002940) (data not shown). The 1.2-kb ORF encoded a mature protein with an estimated molecular mass of 43 kDa and a pI of 8.79. A second 1.3-kb ORF 374 nucleotides upstream was also identified in the 35000HP sequence. This latter ORF encoded a mature protein with an estimated molecular mass of 46 kDa and a pI of 9.28. Based on the fact that the 35000HP chromosome had been completely sequenced and annotated, the remainder of the studies outlined in this report were performed with this well-characterized strain.
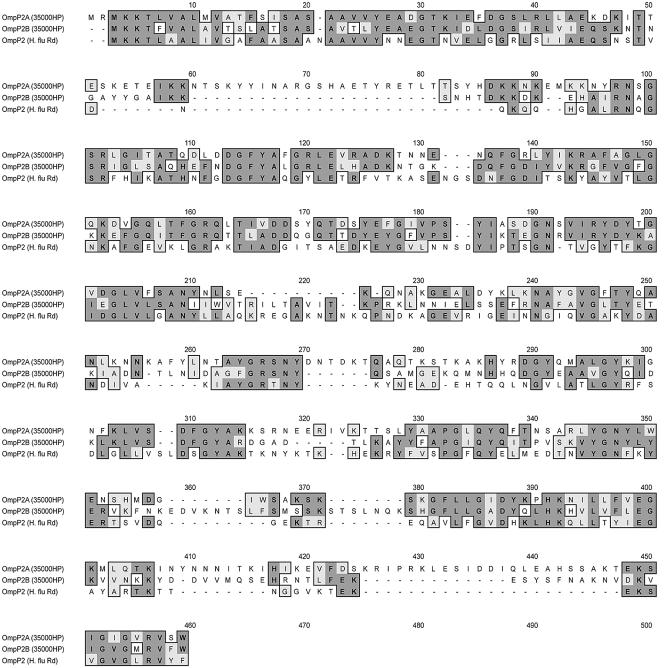
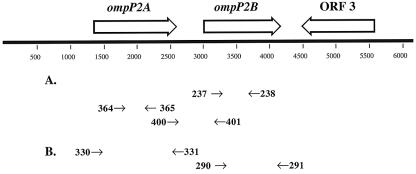
A comparative analysis revealed that the two tandem H. ducreyi ORFs were 54% homologous at the nucleotide level and their predicted gene products were 40% identical and 58% similar at the amino acid level. An NCBI BLAST search revealed that these proteins shared 27 to 33% identity with the major porin OmpP2 of H. influenzae. A comparative Clustal alignment of the amino acid sequence is shown in Fig. 1. Based on these data, the 46- and 43-kDa proteins of H. ducreyi 35000HP were designated OmpP2A and OmpP2B, respectively. In addition, located 600 bp downstream of OmpP2B, an uncharacterized 1.1-kb ORF transcribed in the opposite orientation was identified. The deduced amino acid sequence of this ORF shared 64% similarity to the general secretory pathway component PulD of Aeromonas hydrophila (39). The arrangement of the ompP2A and ompP2B genetic loci is shown in Fig. 2.
FIG. 1.
Clustal alignment of the amino acid sequences of OmpP2A and OmpP2B of H. ducreyi 35000HP versus OmpP2 of H. influenzae Rd (typeable). Identical residues are denoted by dark boxes, and similar residues are denoted by light boxes.
FIG. 2.
Genetic organization of the ompP2A and ompP2B loci from H. ducreyi 35000HP. ORF 3 is a putative PulD homolog. Large arrows indicate the direction of transcription. Primers used in RT-PCR are represented by the arrows in panel A, and primers used for cloning are denoted by the arrows in panel B.
Detection of ompP2A and ompP2B transcripts by RT-PCR.
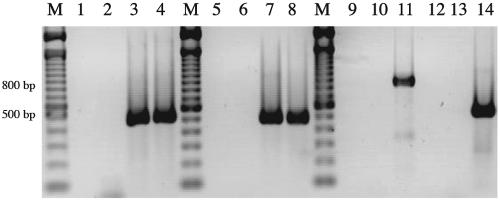
As a preliminary analysis of expression, internal primers to ompP2A and ompP2B were designed and used in RT-PCR. Figure 3 demonstrates that the predicted ompP2A (486 bp; lane 8) and ompP2B (517 bp; lane 4) RT-PCR products were amplified using the primers specific for each internal region. In addition, primers spanning the 3′ end of ompP2A and the 5′ end of ompP2B amplified an 838-bp amplicon in the presence of DNA template (lane 11). However, no RT-PCR product was amplified, using the same primers, in the presence of RNA template (lane 12). These analyses indicated that ompP2A and ompP2B are expressed as independent transcripts in H. ducreyi 35000HP.
FIG. 3.
Detection of ompP2A and ompP2B transcripts by RT-PCR. Total cellular RNA and chromosomal DNA were isolated from H. ducreyi 35000HP. Lanes 1 to 4, 13, and 14, primers 237 and 238, internal to ompP2B; lanes 5 to 8, primers 364 and 365, internal to ompP2A; lanes 9 to 12, primers 400 and 401, spanning the ompP2A-ompP2B intergenic region. Lanes 1, 5, 9, and 13, no nucleic acid template; lanes 2, 6, and 10, no RT; lanes 3, 7, and 11, chromosomal DNA; lanes 4, 8, 12, and 14, total RNA. Lanes marked M indicate molecular size standards in 100-bp increments. Lanes 13 and 14 are controls for RNA integrity.
Cloning and recombinant protein expression of OmpP2A and OmpP2B.
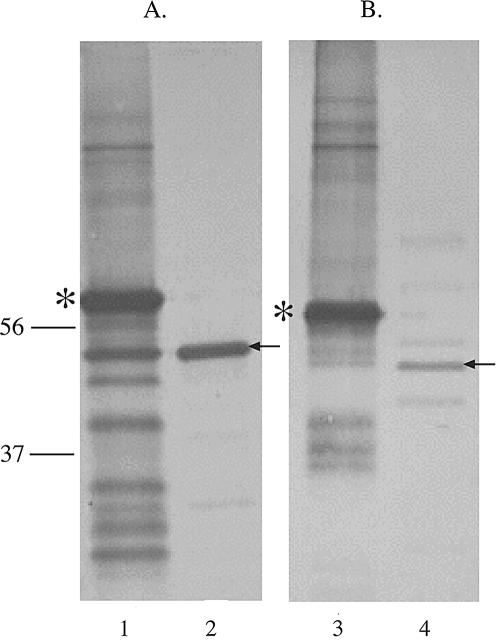
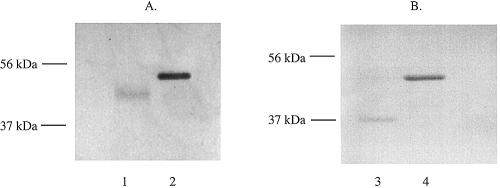
Both ompP2A and ompP2B were cloned from H. ducreyi 35000HP lacking their signal sequence, expressed as fusion proteins in E. coli BL21(DE3), and used to develop antibodies. Figure 4 shows Western blot assays of rOmpP2A, rOmpP2B, and OMPs isolated from 35000HP. Figure 4A shows that the polyclonal antibodies developed against rOmpP2A reacted with the fusion protein as contained in inclusion bodies (lane 1), and the results shown in Fig. 4B confirmed that the antibodies to rOmpP2B also reacted with the respective fusion protein (lane 3). The data also showed that the antibodies recognized OmpP2A (Fig. 4A, lane 2) and OmpP2B (Fig. 4B, lane 4) in the OMPs of H. ducreyi 35000HP. The increased molecular mass of the recombinant proteins was due to the presence of a 5-kDa His-S tag.
FIG. 4.
Western blots probed with antisera against rOmpP2A (A) and rOmpP2B (B) as expressed in E. coli BL21(DE3). Lanes 1 and 3, inclusion bodies containing rOmpP2A and rOmpP2B, respectively; lanes 2 and 4, 35000HP OMPs. The asterisk denotes recombinant protein; arrows denote the positions of protein in the OM. Molecular mass (in kilodaltons) is shown on left.
Analysis of OmpP2A and OmpP2B expression among clinical isolates of H. ducreyi.
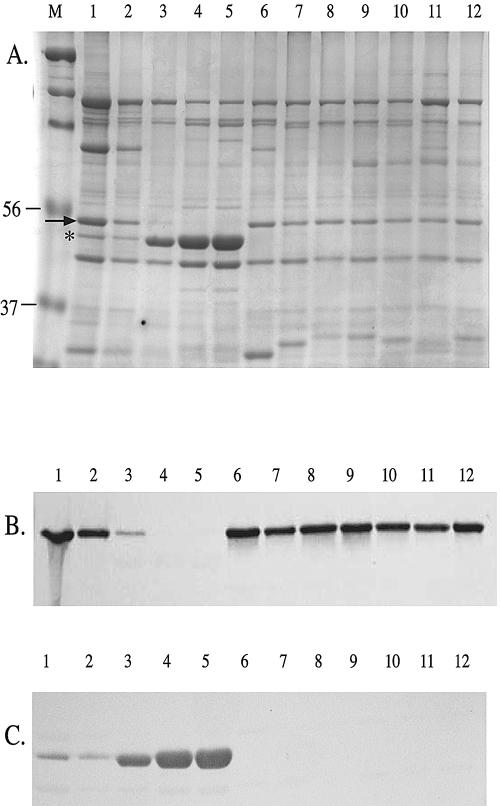
Western blot analysis was used to assess the conservation of OmpP2A and OmpP2B expression in a panel of H. ducreyi strains. OMPs isolated from 35000HP, MF35000, and 10 H. ducreyi isolates selected from various geographic regions were separated on an SDS-PAGE gel (Fig. 5A). Immunoblots were probed with affinity-purified polyclonal antibodies specific to OmpP2A (Fig. 5B) or OmpP2B (Fig. 5C). H. ducreyi 35000, 35000HP, and MF35000 expressed both OmpP2A and OmpP2B (Fig. 5B and C, lanes 1, 2, and 3, respectively). However, H. ducreyi strains CIP542, HD41, HD151, HD293, HD346, NY8, and R3 expressed only OmpP2A (Fig. 5B, lanes 6 to 12). In contrast, isolates A75 and A76 exclusively expressed OmpP2B (Fig. 5C, lanes 4 and 5).
FIG. 5.
Evaluation of OmpP2A and OmpP2B expression by SDS-PAGE (A) and Western blotting (B and C). Lanes: 1, 35000; 2, 35000HP; 3, MF35000; 4, A75; 5, A76; 6, CIP542; 7, HD41; 8, HD151; 9, HD293; 10, HD346; 11, NY8; 12, R3. Corresponding immunoblots were probed with OmpP2A antibodies (B) and OmpP2B antibodies (C). The arrow denotes OmpP2A; the asterisk denotes OmpP2B. Molecular mass (in kilodaltons) is shown on the left.
Genetic analysis of ompP2A and ompP2B.
To begin to understand the differential expression of OmpP2A and OmpP2B, the genes (including their flanking sequences) were independently amplified from each strain of H. ducreyi evaluated above. PCR analysis of ompP2A and ompP2B yielded a consistent product from all of the H. ducreyi strains tested, with the exception of strains A75 and A76. The ompP2A amplicon from these two isolates was approximately 250 bp shorter than the expected product size of 2.1 kb (data not shown). The sequences of ompP2A from strains MF35000, 35000, CIP542, HD41, HD151, HD293, HD346, and R3 were identical to that of the ompP2A from the sequenced strain 35000HP. The ompP2A gene from strain NY8 contained a 6-nucleotide deletion at positions 198 to 203 and a cytosine-to-thymine substitution at position 212 (data not shown). Sequence analysis also revealed that ompP2A from strains A75 and A76 contained a 261-bp deletion encompassing the 5′ upstream sequence, the entire promoter region, and 103 bp of the coding region (data not shown). This resulted in the lack of OmpP2A expression in these strains.
Interestingly, the above analysis for ompP2B demonstrated a consistently similar PCR product among all of the H. ducreyi strains tested (data not shown), yet not all strains expressed OmpP2B. Sequence analysis revealed that the lack of OmpP2B expression in strains CIP542, HD41, HD151, HD293, HD346, NY8, and R3 was due to the addition of two adenines at position 556 within the coding region. The additional adenines created a frameshift that resulted in premature termination of OmpP2B. A truncated OmpP2B product was not observed by Western blotting in these strains (data not shown). Interestingly, strains A75 and A76 also contained two additional adenines at position 556. However, a guanine insertion 15 bp upstream of the two adenines restored the proper reading frame. The deduced amino acid sequences of OmpP2B from strains A75 and A76 were identical to those of the other isolates expressing OmpP2B with the exception of a 5-amino-acid change at positions 182 to 186 (data not shown).
Purification of OmpP2A and OmpP2B.
OmpP2A and OmpP2B were isolated from OMP preparations of H. ducreyi 35000HP in multiple rounds of gel elution as described in Materials and Methods. The proteins were analyzed by SDS-PAGE and Western blotting. Figure 6 shows two immunoblots containing purified OmpP2A (Fig. 6A) and OmpP2B (Fig. 6B) under nondenaturing conditions (lanes 1 and 3, respectively) and denaturing conditions (lanes 2 and 4, respectively). The Western blots were probed with either antibodies to OmpP2A (Fig. 6A) or antibodies to OmpP2B (Fig. 6B). These studies showed that the heated form of OmpP2A (lane 2) and OmpP2B (lane 4) migrated at a slower rate than the unheated form of each protein (lanes 1 and 3, respectively), demonstrating that these proteins are heat modifiable. This is an important observation, as this is a characteristic that is consistent with a subset of other known porins (1). In addition, it was apparent that the antibodies exhibited stronger reactivities to the denatured form of each protein. It seems likely that this difference in reactivity was due to the fact that the polyclonal antibodies were developed against recombinant proteins; therefore, a majority of the antibodies produced would likely be reactive towards linear epitopes.
FIG. 6.
Purification and heat modifiability of native OmpP2A and OmpP2B. Immunoblots were probed with affinity-purified antibodies developed against either OmpP2A (A) or OmpP2B (B). Lanes: 1, unheated; 2, heated; 3, unheated; 4, heated. Heated samples were boiled for 10 min at 100°C.
Porin function analysis.
Addition of either OmpP2A or OmpP2B in small amounts to either solution bathing a planar lipid bilayer membrane led to an increase in conductance that was resolved at higher amplification into discrete conductance increments. By analogy with many similar experiments, the stepwise increases in conductance represent the stepwise entry of single porin channels into the membrane, creating water-filled conductance pathways. Averaged over more than 100 discrete events, the mean size of OmpP2A conductance increment in the presence of 1 M KCl was 0.86 ± 0.04 nS (mean ± standard error, where a siemen is a unit of conductance equivalent to 1 ohm−1). For OmpP2B, the mean size of the conductance increment in the presence of 1 M KCl was 1.20 ± 0.05 nS (data not shown).
DISCUSSION
In this report we identified and characterized two homologous genes in H. ducreyi 35000HP. These tandem genes encode distinct proteins that have homology to OmpP2 of H. influenzae; thus, we have designated these genes ompP2A and ompP2B. OmpP2 is the major porin of H. influenzae, and there have been numerous studies describing the critical importance of this OMP both in the biology of the species and in the pathogenesis of Haemophilus infections (7, 14, 26, 31, 33). Previous studies have demonstrated that antibodies directed to OmpP2 elicit bactericidal activity (25, 26). Although OmpP2 expression is conserved, previous data have also demonstrated the heterogeneous nature of this protein in nontypeable H. influenzae at both the nucleotide and amino acid levels (3, 13, 24, 26). Despite these observations, the surface-exposed loops 5 and 6 of this major outer membrane porin are under consideration as potential vaccine antigens versus H. influenzae (26). Based on the significance of OmpP2 to H. influenzae and the similarities between this porin and OmpP2A and OmpP2B of H. ducreyi, studies were performed analyzing the conservation and expression of these latter proteins among various clinical isolates. It is also important to note that to our knowledge there have been no studies to date describing OMPs of H. ducreyi that exhibit porin activity.
To determine if OmpP2A and OmpP2B expression was conserved, a panel of H. ducreyi clinical isolates from various geographic regions were analyzed. Affinity-purified polyclonal antibodies were developed to rOmpP2A and rOmpP2B and used in Western blot studies. These data demonstrated that OmpP2A and OmpP2B were differentially expressed in the OM of the H. ducreyi strains tested. Indeed, the majority of the strains tested in these studies expressed OmpP2A exclusively. It was somewhat surprising that H. ducreyi 35000 and its derivative strains were the only isolates that expressed both OmpP2A and OmpP2B. While our genetic studies identified possible explanations for the lack of expression in other strains, the significance of this observation is unexplained and remains a current focus of our work.
A second interesting observation was that strains A75 and A76 expressed OmpP2B but did not appear to express OmpP2A. Previous studies have described H. ducreyi strains A75 and A76 as extremely sensitive to the bactericidal activity of normal human sera, while other studies have used strain A75 as an avirulent control in various animal systems (9, 11, 29, 41, 47). However, these strains have been shown to be defective in expression of other putative virulence determinants, including lipooligosaccharides, hemolysin activity, and DsrA; therefore, no correlation can be drawn between the expression of OmpP2A and/or OmpP2B and virulence (11, 29, 41).
Further inspection of the Western blotting data indicated that the relative levels of OmpP2A and OmpP2B expression varied among strains. Whereas OmpP2B was the MOMP in strains MF35000, A75, and A76, this protein appeared to be a minor component in the membranes of strains 35000 and 35000HP. In contrast, the relative levels of OmpP2A expression remained fairly consistent in the clinical isolates expressing this protein. Previous studies using H. ducreyi 35000HP detected an apparent increase in the expression of an OMP in a defined MOMP mutant (40). Although the nature of this protein was unknown at the time of the study (40), it now appears that this protein may have been OmpP2B. Despite these interesting observations, more detailed genetic analysis must be performed to effectively determine the significance of these apparent differences in protein expression.
Molecular studies comparing ompP2A and ompP2B among the evaluated strains demonstrated a high degree of homology at the nucleotide sequence level. This is in direct contrast to the allelic polymorphism that is exhibited by the ompP2 of nontypeable H. influenzae (13, 24, 31). Two distinct patterns emerged as putative mechanisms controlling the expression of OmpP2A and OmpP2B. A partial gene deletion was evident in the strains that did not express OmpP2A. This defect, identical in both strains A75 and A76, involved the 5′ region of ompP2A, including the upstream flanking sequence, the initial start site, and a portion of the coding region. Further studies are needed to assess the frequency of this expression pattern among clinical isolates and the mechanism leading to this disruption.
The second pattern that emerged from these genetic studies involved the addition or deletion of specific nucleotides that ultimately affected the expression of OmpP2B. For example, the sequences of ompP2B from strains CIP542, HD41, HD151, HD293, HD346, NY8, and R3 were completely identical to that of the ompP2B from the sequenced strain 35000HP, with the exception of a two-adenine addition at position 556. These additional nucleotides created a frameshift that resulted in premature termination of OmpP2B. It is important to note that these adenine substitutions occurred at the exact same position in the gene, suggesting that they are clonal in nature. Interestingly, strains A75 and A76 also contained the additional two adenines at position 556 and yet, as stated above, these strains expressed OmpP2B. Further analysis of ompP2B from these two isolates revealed a single guanine addition at position 541 of the nucleotide sequence, which restored the correct reading frame for expression of the complete protein. The fact that the addition of these two adenines was detected in the majority of the strains selected for these studies suggests that this insertion event may be more prevalent among H. ducreyi isolates, although again further analysis is needed.
Functional studies indicated that both purified OmpP2A and OmpP2B formed channels in planar bilayers, indicating that they are porins. Previous studies have established that monomeric OmpP2 from H. influenzae type b formed channels in planar bilayers with a single channel conductance of 1.1 nS (43). This compares favorably with the average value (1.2 nS) observed for OmpP2B. In contrast, OmpP2A demonstrated a smaller single channel conductance of 0.86 nS. By comparison, the E. coli trimeric porins OmpF, OmpC, and PhoE have single channel conductance values ranging from 0.5 to 0.7 nS (trimers, 1.5 to 2.1 nS) (5). It is known that the rather modest differences in conductance between OmpC (1.5 nS) and OmpF (1.9 nS) (4) of E. coli lead to a five- to sixfold difference in rates of diffusion of, e.g., β-lactams (28), and these porins vary in antibiotic-resistant clinical isolates. Thus, it is tempting to speculate that the variations in the expression of OmpP2A and OmpP2B have been selected for by antibiotic treatment of patients infected with H. ducreyi, although this is speculation at this point. We are focusing future experiments on evaluating the importance of OmpP2A and OmpP2B expression to the general biology and pathogenesis of H. ducreyi infections.
Acknowledgments
Funding for this research was provided by a grant from the John R. Oishei Foundation, Buffalo, N.Y. (A.A.C.) and by a Canadian Institutes for Health Research grant to R.E.W.H. D.T.P. was supported in part as a graduate student by the Arthur A. Schomburg Graduate Fellowship. R.E.W.H. was the holder of a Canada Research Chair.
We thank Nicole R. Luke (University at Buffalo) for valuable assistance in the preparation of the manuscript and Robert S. Munson, Jr. (Children's Institute and the Ohio State University) for assistance with the H. ducreyi genome.
Editor: D. L. Burns
REFERENCES
- 1.Achouak, W., T. Heulin, and J. M. Pages. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Alberti, S., G. Marques, S. Hernandez-Alles, X. Rubires, J. M. Tomas, F. Vivanco, and V. J. Benedi. 1996. Interaction between complement subcomponent C1q and the Klebsiella pneumoniae porin OmpK36. Infect. Immun. 64:4719-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, J., S. Grass, D. Jeanteur, and R. S. Munson, Jr. 1994. Diversity of the P2 protein among nontypeable Haemophilus influenzae isolates. Infect. Immun. 62:2639-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benz, R., A. Schmidt, and R. E. W. Hancock. 1985. Ion selectivity of gram-negative bacterial porins. J. Bacteriol. 2:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bina, J., M. Bains, and R. E. Hancock. 2000. Functional expression in Escherichia coli and membrane topology of porin HopE, a member of a large family of conserved proteins in Helicobacter pylori. J. Bacteriol. 182:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns, J. L., and A. L. Smith. 1987. A major outer-membrane protein functions as a porin in Haemophilus influenzae. J. Gen. Microbiol. 133:1273-1277. [DOI] [PubMed] [Google Scholar]
- 7.Cope, L. D., S. E. Pelzel, J. L. Latimer, and E. J. Hansen. 1990. Characterization of a mutant of Haemophilus influenzae type b lacking the P2 major outer membrane protein. Infect. Immun. 58:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowley, B., V. J. Benedi, and A. Domenech-Sanchez. 2002. Expression of SHV-2 β-lactamase and of reduced amounts of OmpK36 porin in Klebsiella pneumoniae results in increased resistance to cephalosporins and carbapenems. Antimicrob. Agents Chemother. 46:3679-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutro, S. M., G. E. Wood, and P. A. Totten. 1999. Prevalence of, antibody response to, and immunity induced by Haemophilus ducreyi hemolysin. Infect. Immun. 67:3317-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins, C., C. J. Chen, and C. E. Thomas. 1995. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect. Immun. 63:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes, K. J., K. D. Bruce, A. Ball, and T. H. Pennington. 1992. Variation in length and sequence of porin (ompP2) alleles of non-capsulate Haemophilus influenzae. Mol. Microbiol. 6:2107-2112. [DOI] [PubMed] [Google Scholar]
- 14.Galdiero, M., M. D'Amico, F. Gorga, C. Di Filippo, M. D'Isanto, M. Vitiello, A. Longanella, and A. Tortora. 2001. Haemophilus influenzae porin contributes to signaling of the inflammatory cascade in rat brain. Infect. Immun. 69:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galdiero, M., M. D'Isanto, M. Vitiello, E. Finamore, and L. Peluso. 2001. Porins from Salmonella enterica serovar Typhimurium induce TNF-alpha, IL-6 and IL-8 release by CD14-independent and CD11a/CD18-dependent mechanisms. Microbiology 147:2697-2704. [DOI] [PubMed] [Google Scholar]
- 16.Hammond, G. W., C. J. Lian, J. C. Wilt, and Alan R. Ronald. 1978. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob. Agents. Chemother. 13:608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock, R. E., and F. S. Brinkman. 2002. Function of pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17-38. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, R. E. W., and R. Benz. 1986. Demonstration and chemical modification of a specific phosphate binding site in the phosphate-starvation-inducible outer membrane porin protein P of Pseudomonas aeruginosa. Biochim. Biophys. Acta 860:699-707. [DOI] [PubMed] [Google Scholar]
- 19.Haydock, A. K., D. H. Martin, S. A. Morse, C. Cammarata, K. J. Mertz, and P. A. Totten. 1999. Molecular characterization of Haemophilus ducreyi strains from Jackson, Mississippi, and New Orleans, Louisiana. J. Infect. Dis. 179:1423-1432. [DOI] [PubMed] [Google Scholar]
- 20.Humphreys, T. L., C. T. Schnizlein-Bick, B. P. Katz, L. A. Baldridge, A. F. Hood, R. A. Hromas, and S. M. Spinola. 2002. Evolution of the cutaneous immune response to experimental Haemophilus ducreyi infection and its relevance to HIV-1 acquisition. J. Immunol. 169:6316-6323. [DOI] [PubMed] [Google Scholar]
- 21.Klesney-Tait, J., T. J. Hiltke, I. Maciver, S. M. Spinola, J. D. Radolf, and E. J. Hansen. 1997. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J. Bacteriol. 179:1764-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, D. A., M. K. Stevens, J. L. Latimer, C. K. Ward, K. Deng, R. Blick, S. R. Lumbley, C. A. Ison, and E. J. Hansen. 2001. Characterization of Haemophilus ducreyi cdtA, cdtB, and cdtC mutants in in vitro and in vivo systems. Infect. Immun. 69:5626-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montero, J. A., L. L. Zaulyanov, S. H. Houston, and J. T. Sinnot. 2002. Chancroid: an update. Infect. Med. 21:174-178. [Google Scholar]
- 24.Munson, R., Jr., C. Bailey, and S. Grass. 1989. Diversity of the outer membrane protein P2 gene from major clones of Haemophilus influenzae type b. Mol. Microbiol. 3:1797-1803. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, T. F., and L. C. Bartos. 1988. Human bactericidal antibody response to outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 56:2673-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neary, J. M., K. Yi, R. J. Karalus, and T. F. Murphy. 2001. Antibodies to loop 6 of the P2 porin protein of nontypeable Haemophilus influenzae are bactericidal against multiple strains. Infect. Immun. 69:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido, H. 1994. Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 269:3905-3908. [PubMed] [Google Scholar]
- 28.Nikaido, H., E. Y. Rosenberg, and J. Foulds. 1983. Porin channels in Escherichia coli: studies with β-lactams in intact cells. J. Bacteriol. 153:232-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1985. Role of lipopolysaccharide and complement in the susceptibility of Haemophilus ducreyi to human serum. Infect. Immun. 50:495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer, K. L., S. Grass, and R. S. Munson, Jr. 1994. Identification of a hemolytic activity elaborated by Haemophilus ducreyi. Infect. Immun. 62:3041-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regelink, A. G., D. Dahan, L. V. M. Moller, J. W. Coulton, P. Eijk, P. Van Ulsen, J. Dankert, and L. V. Van Alphen. 1999. Variation in the composition and pore function of major outer membrane pore protein P2 of Haemophilus influenzae from cystic fibrosis patients. Antimicrob. Agents Chemother. 43:226-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo, T. A., J. E. Guenther, S. Wenderoth, and M. M. Frank. 1993. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol. Microbiol. 9:357-364. [DOI] [PubMed] [Google Scholar]
- 33.Sanders, J. D., L. D. Cope, G. P. Jarosik, I. MacIver, J. L. Latimer, G. B. Toews, and E. J. Hansen. 1993. Reconstitution of a porin-deficient mutant of Haemophilus influenzae type B with a porin gene from nontypeable H. influenzae. Infect. Immun. 61:3966-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spinola, S. M., M. E. Bauer, and R. S. Munson, Jr. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 70:1667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 71:7178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spinola, S. M., L. M. Wild, M. A. Apicella, A. A. Gaspari, and A. A. Campagnari. 1994. Experimental human infection with Haemophilus ducreyi. J. Infect. Dis. 169:1146-1150. [DOI] [PubMed] [Google Scholar]
- 37.Steen, R. 2001. Eradicating chancroid. Bull. W. H. O. 79:818-826. [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas, C. E., B. Olsen, and C. Elkins. 1998. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect. Immun. 66:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, S. R., and T. J. Trust. 1995. A specific PulD homolog is required for the secretion of paracrystalline surface array subunits in Aeromonas hydrophila. J. Bacteriol. 177:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Throm, R. E., J. A. Al-Tawfiq, K. R. Fortney, B. P. Katz, A. F. Hood, C. A. Slaughter, E. J. Hansen, and S. M. Spinola. 2002. Evaluation of an isogenic major outer membrane protein-deficient mutant in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:2602-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Totten, P. A., D. A. Norn, and W. A. Stamm. 1995. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect. Immun. 63:4409-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vachon, V., R. Laprade, and J. W. Coulton. 1986. Properties of the porin of Haemophilus influenzae type b in planar lipid bilayer membranes. Biochim. Biophys. Acta 86:74-82. [DOI] [PubMed] [Google Scholar]
- 44.Vachon, V. D., J. Lyew, and J. W. Coulton. 1985. Transmembrane permeability channels across the outer membrane of Haemophilus influenzae. J. Bacteriol. 162:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel, U., and M. Frosch. 1999. Mechanisms of neisserial serum resistance. Mol. Microbiol. 32:1133-1139. [DOI] [PubMed] [Google Scholar]
- 46.Ward, C. K., J. L. Latimer, J. Nika, M. Vakevainen, J. R. Mock, K. Deng, R. J. Blick, and E. J. Hansen. 2003. Mutations in the lspA1 and lspA2 genes of Haemophilus ducreyi affect the virulence of this pathogen in an animal model system. Infect. Immun. 71:2478-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood, G. E., S. M. Dutro, and P. A. Totten. 2001. Haemophilus ducreyi inhibits phagocytosis by U-937 cells, a human macrophage-like cell line. Infect. Immun. 69:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]