Abstract
The location and functional properties of antigen-specific memory T-cell populations in lymphoid and nonlymphoid compartments following DNA immunization or infection with Salmonella were investigated. Epitope-specific CD8+-T-cell expansion and retention during the memory phase were analyzed for DNA-immunized mice by use of a 5-h peptide restimulation assay. These data revealed that epitope-specific gamma interferon (IFN-γ)-positive CD8+ T cells occur at higher frequencies in the spleen, liver, and blood than in draining or peripheral lymph nodes during the expansion phase. Moreover, this distribution is maintained into long-term memory. The location and function of both CD4+ and CD8+ Salmonella-specific memory T cells in mice who were given a single dose of Salmonella enterica serovar Typhimurium was also quantitated by an ex vivo restimulation with bacterial lysate to detect the total Salmonella-specific memory pool. Mice immunized up to 6 months previously with S. enterica serovar Typhimurium had bacterium-specific CD4+ T cells that were capable of producing IFN-γ or tumor necrosis factor alpha (TNF-α) at each site analyzed. Similar findings were observed for CD8+ T cells that were capable of producing IFN-γ, while a much lower frequency and more restricted distribution were associated with TNF-α-producing CD8+ T cells. This study is the first to assess the frequencies, locations, and functions of both CD4+ and CD8+ memory T-cell populations in the same Salmonella-infected individuals and demonstrates the organ-specific functional compartmentalization of memory T cells after Salmonella infection.
An encounter with a previously unseen pathogen can lead to the expansion of antigen-specific T cells and the subsequent generation of long-lived CD4+ and CD8+ memory T cells (12, 26, 48, 51). The localization of antigen-specific T-cell populations during the progression from primary pathogen exposure to long-term memory can be visualized through the application of techniques such as major histocompatibility complex type I (MHC-I) tetramer staining and the adoptive transfer of naïve antigen-specific cells. In parallel with these studies, the functional attributes of distinct memory populations have also been examined, leading to the classifications of effector memory (TEM) and central memory (TCM) T cells (41, 53).
Large variations in the sizes of epitope-specific memory populations, but not in the kinetics of their generation, are observed for murine models of infection (9). These variations are dependent upon the extent of the initial “burst” of expansion from antigen-specific precursors, although not necessarily on the amount of antigen itself (5, 14, 19, 33, 35, 54). Epitope-specific CD8+ or CD4+ T cells generated following oral challenge with Listeria monocytogenes are present at higher frequencies in the liver and intestinal mucosa than in the spleen both during the peak of the primary response and once memory has been established (25, 32, 39). Moreover, epitope-specific CD8+ T cells generated in response to a viral or bacterial infection localize not only to lymphoid organs, but also to nonlymphoid tissues, including the kidneys, lungs, liver, and lamina propria (16, 29, 31, 32, 39). Indeed, higher frequencies of vesicular stomatitis virus-specific CD8+ memory T cells are present within these nonlymphoid tissues than within the spleen and other lymphoid sites, and few memory cells associate with lymph nodes (LN) (32). Furthermore, only a small minority of the total CD8+ memory cell population is retained within lymphoid tissues (32).
In contrast, a Salmonella infection model showed that the distributions of memory CD4+-T-cell populations generated after the stimulation of adoptively transferred cells were approximately equal for lymphoid and nonlymphoid sites (40). Infections with L. monocytogenes also revealed that bacterium-specific CD4+ T cells were localized to lymphoid as well as nonlymphoid tissues, although the route of bacterial administration influenced which organ contained the largest fraction of memory cells (25). Thus, the organ-specific distribution of antigen-specific T cells occurs during both the generation and the retention of memory.
Functionally, splenic epitope-specific CD8+ memory T cells produce gamma interferon (IFN-γ) and/or tumor necrosis factor alpha (TNF-α) after a peptide encounter (2-4, 46). Indeed, more lymphocytic choriomeningitis virus (LCMV)-specific CD8+ memory cells respond to peptides by producing both IFN-γ and TNF-α than during the primary response (15, 46, 47). Furthermore, CD8+ memory cells retain their cytolytic function (and express perforin) without the concomitant expression of IFN-γ or TNF-α ex vivo (45). Memory CD8+ T cells generated after bacterial infection and located in either the spleen or peripheral tissues were found to rapidly produce IFN-γ or TNF-α in response to peptide exposure (2-4, 39). However, memory CD8+ T cells from peripheral (nonlymphoid) sites exhibited direct cytotoxic activities ex vivo which were not present in similar populations taken from lymphoid tissues such as the spleen (32). Therefore, distinct fractions of memory T cells may be associated not only with a specific location, but also with a particular in vivo function. The identification of the TEM and TCM memory T-cell subsets further supports this notion (41, 53).
The present study assessed the locations, numbers, and functions of antigen-specific T-cell populations in lymphoid and nonlymphoid compartments following DNA immunization or infection with Salmonella enterica serovar Typhimurium. A polyclonal restimulation of cells from Salmonella-infected mice allowed the numbers and cytokine production capacities of CD4+ and CD8+ memory T-cell responses to the whole bacterium to be assessed. This study is the first function- and location-specific analysis of both CD4+ and CD8+ memory T-cell populations from the same Salmonella-infected animals and demonstrates the organ-specific functional compartmentalization of Salmonella-specific memory T-cell responses.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were between 6 and 8 weeks of age at the time of immunization. Age-matched naïve controls were included in all experiments. The mice were bred and housed at the animal facilities at Lund University or were purchased from Taconic M&B (Ry, Denmark) and housed at the Laboratory for Experimental Biomedicine at Göteborg University. All animals were provided with food and water ad libitum.
Plasmid immunization and peptides.
The eukaryotic expression plasmids pCI/C and pCI/S, encoding the hepatitis B virus core and small surface antigens (HBsAg), respectively, were described previously (24, 42) and were prepared on a large scale by Qiagen Ltd., Düsseldorf, Germany. The DNAs were resuspended at a final concentration of 1 mg/ml in sterile phosphate-buffered saline (PBS), and mice were immunized with 50 μg in a single injection at the base of the tail. Immunization with pCI/S generates a Kb-restricted CD8+-T-cell response to the peptide ILSPFLPL (KbS), corresponding to bases 208 to 215 of HBsAg (43). The Kb-restricted SIINFEKL (KbOVA) peptide, corresponding to bases 257 to 264 of ovalbumin, was used as a control.
Salmonella immunization.
The S. enterica serovar Typhimurium χ4550 strain used for these studies was described previously (55). This SR-11 derivative contains deletions in the genes encoding adenylate cyclase and the cyclic AMP receptor protein (Δcya-1 Δcrp-1) that reduce its virulence relative to the wild-type strain. Whereas mice infected with even low doses of wild-type S. enterica serovar Typhimurium succumb to the infection in 7 to 10 days, mice given 108 to 109 χ4550 cells survive, clear the bacteria ∼4 weeks after infection, generate both CD4+- and CD8+-T-cell responses, and are immune to a lethal challenge with virulent S. enterica serovar Typhimurium (23, 55). Like fully virulent S. enterica serovar Typhimurium, χ4550 acquired orally replicates in Peyer's patches, mesenteric LN (MLN), the spleen, and the liver. The kinetics of χ4550 growth in these organs are delayed relative to those after infection with virulent bacteria (our unpublished data), and unlike the case with virulent bacteria, the mice control χ4550 and survive. Although additional studies would be necessary to directly compare the memory T cells generated after infection with fully virulent S. enterica serovar Typhimurium (given in nonfatal doses) to those generated after infection with χ4550, χ4550 provides a model for assessing long-term memory in mice who are immune to a lethal challenge.
The bacteria were cultured overnight with constant agitation in Luria-Bertani broth. The concentration was then estimated spectrophotometrically, and the bacteria were washed and resuspended in sterile PBS. For intragastric immunization, mice were given 0.1 ml of 0.1 M NaHCO3 solution followed 5 min later by 109 bacteria in a volume of 0.1 ml. Mice immunized intraperitoneally were given 106 bacteria in a volume of 0.1 ml. The actual bacterial dose administered was determined by counting colonies after the plating of serial dilutions of the inoculum onto Luria-Bertani agar plates. Mice were sacrificed for analysis 9 to 28 weeks after immunization. No differences in the parameters analyzed were observed, for any site examined, as a result of the route of immunization or the time elapsed between immunization and analysis.
In vitro restimulation and flow cytometry.
Animals were sacrificed, peripheral blood was collected into heparinized tubes, and organs were removed aseptically. Livers were perfused with PBS (Gibco BRL Life Technologies, Paisley, United Kingdom) before removal to eliminate contaminating peripheral blood. Single-cell suspensions of lymphoid organs were prepared by physical disruption and resuspension in RPMI medium (Gibco BRL) supplemented with 10% fetal calf serum. Livers were mashed through a cell filter, and hepatic leukocytes were purified over a Percoll gradient as described previously (50). Erythrocytes in peripheral blood samples were lysed by hypotonic shock before use. For peptide restimulation, 106 cells in 200 μl of RPMI plus fetal calf serum were plated in 96-well plates. The KbOVA or KbS peptide was added to a final concentration of 5 μg/ml, and the cells were incubated for 1 h at 37°C. Negative control wells received medium only. Brefeldin A (BFA; Sigma, St. Louis, Mo.) was then added to all wells to a final concentration of 5 μg/ml, and the cells were incubated for a further 4 h. For polyclonal restimulation, cells were seeded at 5 × 106 cells/well in 24-well plates, and a lysate of S. enterica serovar Typhimurium χ4550 was added at a dilution of 1:100. For experiments to examine the role of interleukin-12 (IL-12) in T-cell activation, the anti-IL-12 clone C17.8 (Pierce Biotechnology, Rockford, Ill.) or an isotype control (rat anti-immunoglobulin G2a; BD Pharmingen, San Diego, Calif.) was used at 10 μg/ml in addition to the bacterial lysate (17, 27). To address the contribution of lipopolysaccharide (LPS) to T-cell activation, we added LPS purified from S. enterica serovar Typhimurium (L2262; Sigma), instead of the bacterial lysate, at the same concentration as that in the bacterial lysate (1 to 1.25 μg/ml). The plates were incubated for 18 h, with BFA added to a final concentration of 5 μg/ml for the final 4 h. At the end of the culture period, the cells were harvested, stained, and analyzed by flow cytometry.
Flow cytometry for intracellular cytokines was performed as described previously (22). Briefly, Fc receptors were blocked with antibodies purified from the hybridoma 2.4.G2, and cells were stained for their surface phenotype by the use of fluorescein isothiocyanate, phycoerythrin (PE), allophycocyanin, or PE-Cy7 conjugates of anti-CD4 (GK1.5), anti-CD8α (YTS.169), and anti-T-cell receptor beta (anti-TCRβ) (H57-597) (all from BD Biosciences, San Diego, Calif.). The cells were then fixed and permeabilized, and intracellular cytokines were detected with fluorescein isothiocyanate or biotin conjugates of anti-TNF-α or anti-IFN-γ, followed by streptavidin-allophycocyanin as required (BD Biosciences). Isotype control antibodies (BD Biosciences) with the appropriate conjugates were used. Samples were acquired with a FACSCalibur or LSRII flow cytometer (BD Biosciences) and then analyzed with CellQuest (BD Biosciences) or FloJo (Tree Star Inc., Ashland, Oreg.) software, respectively. The frequencies of specifically responding T cells were calculated with the following equation: (CT/T)test − (CT/T)control, where CT equals the number of cytokine-positive TCRβ+ CD4+ or TCRβ+ CD8+ T cells and T equals the total number of TCRβ+ CD4+ or TCRβ+ CD8+ T cells. The control was either restimulation with medium alone and staining with an anticytokine antibody or restimulation with lysate or LPS and staining with an isotype-matched control antibody.
RESULTS
Detection of low frequencies of epitope-specific CD8+ T cells following DNA immunization.
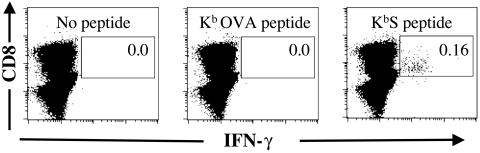
C57BL/6 mice immunized with pCI/S generated a KbS-specific CD8+-T-cell response that was detectable by a 5-h ex vivo peptide restimulation assay followed by flow cytometric analysis of intracellular IFN-γ (Fig. 1). For a naïve splenocyte population that was restimulated with the KbS peptide, no IFN-γ-positive events were detected when over 100,000 gated CD8+ TCRαβ+ cells were collected (n = 6; data not shown). Furthermore, splenocytes from either naïve or pCI/S-immunized animals that were restimulated with medium only or the irrelevant KbOVA peptide gave less than one IFN-γ-positive event per 100,000 CD8+ T cells (Fig. 1). By analyzing large numbers of CD8+ T cells (50,000 to 150,000), we determined specific responding frequencies as low as 1:10,000 with confidence. For samples such as liver and peripheral blood samples, it was often not possible to analyze these large numbers of cells, which resulted in a higher limit of detection for these samples than for other organs. However, KbS epitope-specific frequencies for any organ taken from naïve or pCI/C-immunized animals were always 1:10,000 or lower.
FIG. 1.
Detection of low frequencies of KbS-specific CD8+ T cells. Single-cell suspensions of splenocytes were incubated for 5 h in medium alone or with either the KbOVA peptide or KbS peptide, as indicated in the dot plots. BFA was included for the final 4 h of incubation. The cells were gated for the expression of TCRβ, and IFN-γ expression within the TCRβ+ CD8+ population was examined. The data shown are for a mouse that was immunized 14 days previously with plasmid pCI/S. Similar results were obtained in seven independent experiments in which a total of over 60 pCI/S-immunized and 12 control (6 pCI/C-immunized and 6 naïve mice) mice were examined. Over 100,000 TCRβ+ CD8+ cells are shown in each case. The percentage of cells within the indicated gate is shown for each restimulation. All axes represent log fluorescence intensities. Staining with an appropriate isotype control monoclonal antibody (MAb) did not result in cells within the indicated gate (data not shown).
Kinetics of epitope-specific CD8+-T-cell expansion following pCI/S immunization.
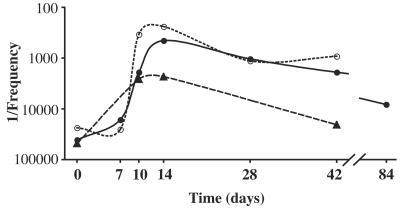
The kinetics for the generation of IFN-γ-responding KbS-specific CD8+ T cells in the spleen, liver, and draining LN were examined after a single immunization with pCI/S. On day 7 postimmunization, no significant increase in the frequency of IFN-γ-responding epitope-specific CD8+ T cells was detectable in either spleens or livers from pCI/S-immunized mice (Fig. 2). However, on day 10 postimmunization, these cells were present in the spleen and inguinal (draining) LN at similar mean frequencies (1:1,900 and 1:2,600, respectively), with the highest mean frequency being detected in the liver (1:340) (Fig. 2). No KbS-responsive cells were found in the other examined LN on day 10 (data not shown). By day 14 postimmunization, the frequencies of epitope-specific cells were further increased for the liver and spleen (n = 21), and frequencies at this time point were often high for these organs (>1:500), although intermouse variations were observed at all time points (Fig. 3). For spleens and livers, epitope-specific cells remained detectable at day 28 postimmunization, with mean frequencies that were somewhat reduced compared to those on day 14. The mean frequencies waned slowly after day 28 postimmunization, with only a slight change apparent at day 42 relative to day 28, and remained above the frequencies of specific cells detected in naïve animals, even at longer time points post-pCI/S immunization (Fig. 2). Days 14 and 42 postimmunization were selected as time points for further analyses of epitope-specific CD8+ T cells during the expansion and memory phases, respectively.
FIG. 2.
Kinetics of KbS-specific CD8+-T-cell response to pCI/S immunization in different organs. Mice were immunized with plasmid pCI/S, and the mean frequencies of IFN-γ-responding KbS-specific CD8+ T cells were determined, as described in Materials and Methods, for multiple time points. The graph shows this response in the spleen (•), liver (○), and draining (inguinal) LN (▴). Data were obtained from 62 pCI/S-immunized mice in seven independent experiments. n = 3 to 6 for each time point for each organ.
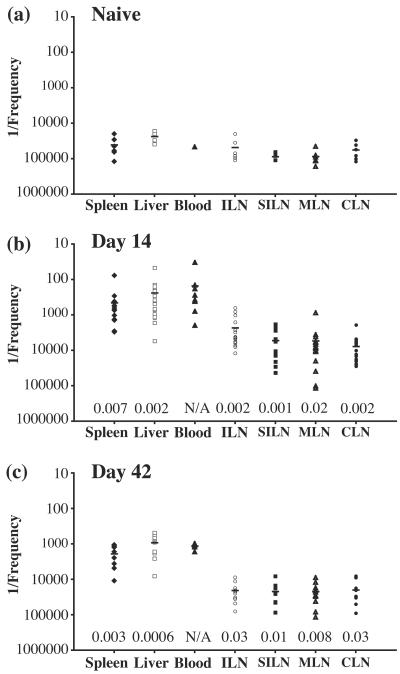
FIG. 3.
Frequencies of epitope-specific IFN-γ-responsive cells in different organs after immunization with pCI/S. Plots show 1/the frequency of cytokine-positive CD8+ cells within the gated TCRβ+ CD8+ population for each organ from naïve mice (a) or pCI/S-immunized mice at day 14 (b) or day 42 (c) postimmunization. Each symbol represents the value for an individual mouse. Horizontal lines indicate the mean values for each organ. Frequencies were quantitated by intracellular cytokine staining and flow cytometry after a 5-h restimulation with HBsAg (208-215) peptide as described in Materials and Methods. Organs were taken from 6 to 21 individuals in two to five separate experiments. For all organs, at least six individuals were examined, except for the blood of a naïve mouse (a), for which a single individual was analyzed. ILN, inguinal lymph node; SILN, superficial inguinal lymph node; MLN, mesenteric lymph node; CLN, cervical lymph node. P values obtained by the two-tailed Student's t test for unequal variances for each organ for infected mice compared to naïve mice are shown.
IFN-γ-responding KbS-specific CD8+ T cells preferentially localize to the spleen, liver, and blood during the expansion phase after DNA immunization.
The presence of KbS-specific CD8+ T cells that were capable of rapidly responding to peptide stimulation at physically distinct sites was examined with individual naïve mice or mice who were immunized 14 days previously with pCI/S DNA. On day 14 after DNA immunization, during the expansion of the epitope-specific population, the highest frequencies of KbS-specific CD8+ T cells were present in the spleen, liver, and blood (Fig. 3b), with the spleen containing the largest total number of responding cells. Within these sites, cell frequencies for the spleen and liver were consistently of comparable magnitudes for individual animals, with a slightly higher frequency associated with peripheral blood. The LN examined had an increased mean frequency of KbS-specific cells compared to those from naïve animals, with the highest frequency occurring in the draining (inguinal) LN (Fig. 3a and b). Surprisingly, the mean frequency of IFN-γ-responding KbS-specific CD8+ T cells within the draining LN was more than fivefold less than that for the spleen, liver, or blood during the expansion phase of the response. All other LN examined appeared to be equally uninvolved in the early specific response to DNA immunization (Fig. 3b).
IFN-γ-responding KbS-specific CD8+ T cells are retained within the spleen, liver, and blood, but not the LN, in DNA-immunized mice.
The frequency of KbS-specific CD8+ T cells that were capable of rapid IFN-γ production was further examined within each site on day 42 after pCI/S DNA immunization (Fig. 3c). At time points of >28 days postimmunization, the observed frequencies are likely to reflect those which will be maintained long-term within the host (Fig. 2). The mean frequencies of KbS-specific CD8+ T cells within the spleen, liver, and peripheral blood were reduced fourfold or more at this time point compared with those from day 14 (Fig. 3b and c). As observed on day 14, comparable frequencies were often present in the spleen, liver, and blood within individuals.
Within peripheral LN sites on day 42 after pCI/S immunization, there was no longer a clear distinction between the IFN-γ-responding KbS-specific CD8+-T-cell frequencies in the draining LN and other examined LN (Fig. 3c). Indeed, each of these sites had an extremely low mean frequency of these cells (<1:20,000), with a frequency of >1:10,000 observed for ∼10% of the examined LN. Together, these data indicate that rapidly responsive epitope-specific CD8+ T cells are preferentially located within a central spleen-liver-blood compartment after DNA immunization and are found within this compartment for extended times. Furthermore, a minor fraction of these cells are located within the draining LN only during the expansion phase. Other peripheral LN do not appear to have significant numbers of rapidly responsive CD8+ T cells at any time point.
Detection of specific polyclonal anti-Salmonella T-cell responses.
Procedures such as DNA immunization followed by peptide-specific restimulation facilitate the examination of one, or perhaps several, epitope-specific populations responding to an antigen. As a result, responses to an antigen may be overlooked if they are below the level of detection of a particular epitope. Moreover, most studies thus far have quantitated epitope-specific T-cell responses and have focused on either CD4+- or CD8+-T-cell populations. Indeed, few data are available on the locations and functions of the total memory cell pools among both CD4+ and CD8+ populations in the same individuals, particularly for infection models. Therefore, we applied restimulation techniques to visualize polyclonally responsive memory cell populations generated after a single bacterial challenge (Fig. 4 and 5). Although a memory cell pool is likely established 3 to 4 weeks after infection (10, 32, 37, 39), the aim of our Salmonella infection system was to examine the memory T-cell pool without any residual effects of the primary antibacterial response. We thus waited long after the primary infection to negate any possibility of remaining T-cell activity as a result of infection and to look at long-term, fully established memory cells. We were unable to detect bacteria in MLN or the liver 48 days after oral infection and thus set 9 weeks after infection as the earliest point to look at the memory cell pools of Salmonella-infected mice. Thus, both CD4+ and CD8+ T cells producing IFN-γ or TNF-α in response to Salmonella antigens were quantified for the same individuals 9 to 28 weeks after infection. No bacteria were recovered from any animal at these time points.
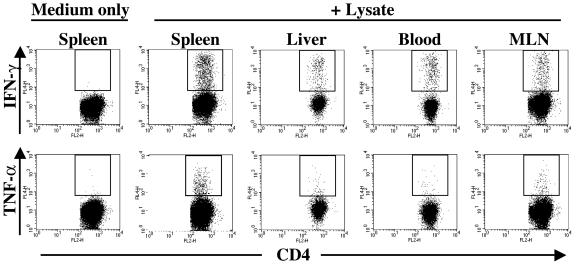
FIG. 4.
Polyclonal CD4+ memory T-cell responses to Salmonella. Single-cell suspensions of organs from Salmonella-immunized animals were restimulated either with medium alone or with Salmonella lysate and were assessed for IFN-γ and TNF-α expression as described in Materials and Methods. Plots represent the CD4+-T-cell responses of individual mice who were immunized 28 weeks previously with Salmonella. Data are representative of at least six individuals for each organ. Similar results were obtained in two to four independent experiments. Plots show gated viable TCRβ+ CD4+ cells. All axes represent log fluorescence intensities. Staining with appropriate isotype control MAbs did not result in cells within the indicated gate (data not shown).
FIG. 5.
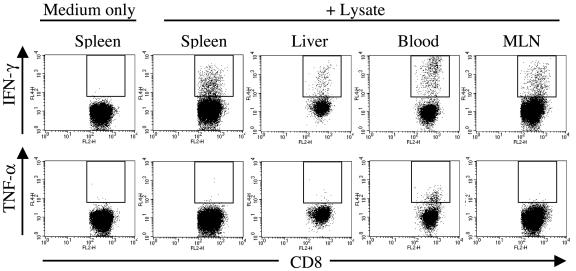
Polyclonal CD8+ memory T-cell responses to Salmonella. Single-cell suspensions of organs from Salmonella-immunized animals were restimulated either with medium alone or with Salmonella lysate and were assessed for IFN-γ and TNF-α expression. Plots represent the CD8+-T-cell responses of individual mice who were immunized 28 weeks previously with Salmonella. Data are representative of at least six individuals for each organ. Similar results were obtained in two to four independent experiments. Plots show gated viable TCRβ+ CD8+ cells. All axes represent log fluorescence intensities. Staining with appropriate isotype control MAbs did not result in cells within the indicated gate (data not shown).
For naïve animals, small polyclonal responses to the Salmonella lysate were detectable for both CD4+- and CD8+-T-cell populations. The compartments in which these responses were consistently detected at frequencies of >1:1,000 were CD4+ and CD8+ IFN-γ-producing populations in the liver and peripheral blood and CD4+ TNF-α-producing cells of the liver (Fig. 6 and 7). The tissue with the next highest frequency of IFN-γ-producing cells in naïve animals was MLN, with a frequency of ∼1:2,000 (Fig. 6 and 7).
FIG. 6.
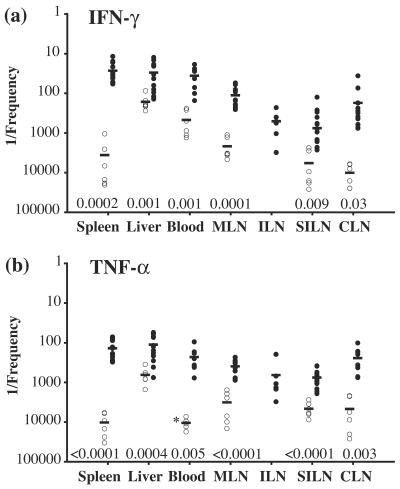
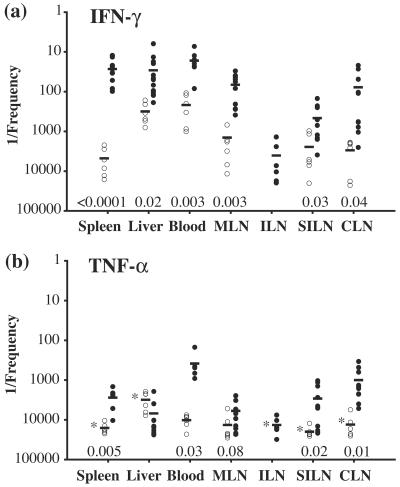
Frequencies of bacterium-specific IFN-γ- and TNF-α-producing CD4+ cells in different organs after immunization with S. enterica serovar Typhimurium. Plots show 1/the frequency of IFN-γ (a)- and TNF-α (b)-positive CD4+ cells within the gated TCRβ+ CD4+ population for each organ of naïve (○) or Salmonella-immunized mice (•) after restimulation with a Salmonella lysate. Each symbol represents the value for an individual mouse. Specific frequencies were quantitated as described in Materials and Methods. Horizontal lines indicate the mean values for each organ. The asterisk in panel b indicates that no cytokine-positive cells were detected in these samples (blood) for any naïve individual and therefore represents the mean number of cells analyzed. Organs were taken from 6 to 18 individuals in four separate experiments, with the exception of inguinal LN responses in immunized mice, which were examined in two independent experiments. Naïve inguinal LN responses were not determined. MLN, mesenteric lymph node; ILN, inguinal lymph node; SILN, superficial inguinal lymph node; CLN, cervical lymph node. P values obtained by a two-tailed Student's t test for unequal variances for each organ for infected mice compared to naïve mice are shown.
FIG. 7.
Frequencies of bacterium-specific IFN-γ- and TNF-α-producing CD8+ cells in different organs after immunization with S. enterica serovar Typhimurium. Plots show 1/the frequency of IFN-γ (a)- and TNF-α (b)-positive CD8+ cells within the gated TCRβ+ CD8+ population for each organ of naïve (○) or Salmonella-immunized mice (•) after restimulation with a Salmonella lysate. Each symbol represents the value for an individual mouse. Specific frequencies were quantitated as described in Materials and Methods. Horizontal lines indicate the mean values for each organ. The asterisks in panel b indicate that no cytokine-positive cells were detected in these organs for any naïve individual and therefore represent the mean numbers of cells analyzed. Organs were taken from 6 to 18 individuals for four separate experiments, except for inguinal LN responses in immunized mice, which were examined in two independent experiments. Naïve inguinal LN responses were not determined. MLN, mesenteric lymph node; ILN, inguinal lymph node; SILN, superficial inguinal lymph node; CLN, cervical lymph node. P values obtained by a two-tailed Student's t test for unequal variances for each organ for infected mice compared to naïve mice are shown.
Widespread distribution of rapidly responsive polyclonal CD4+ T cell populations after oral Salmonella infection.
An examination of CD4+ T cells from the organs of animals who were immunized 9 to 28 weeks previously with Salmonella demonstrated minimal IFN-γ or TNF-α expression in the absence of restimulation (Fig. 4). However, these cells produced both IFN-γ and TNF-α in response to ex vivo restimulation with the Salmonella lysate (Fig. 4).
CD4+ T cells that responded to restimulation with IFN-γ expression were present at the highest mean frequencies in the spleen, liver, and peripheral blood (Fig. 6a), with 133-, 5-, and 13-fold increases, respectively, in the mean frequencies of responding cells compared with those for naïve controls. However, significantly elevated mean frequencies of IFN-γ-producing CD4+ T cells were also present in each of the examined peripheral LN sites. Within each LN, the Salmonella-responsive CD4+ cell population was increased at least 10-fold and up to >50-fold over naïve levels. The spleen contained the largest number of CD4+ T cells that produced IFN-γ in response to the Salmonella lysate, followed by MLN, the liver, and blood (Table 1). These data indicate that CD4+ memory T cells are distributed throughout the lymphoid tissues after Salmonella infection.
TABLE 1.
Absolute numbers of cytokine-producing T cells from Salmonella-immunized mice after restimulation with Salmonella lysatea
| Organ | No. of CD4+ T cells producing cytokine
|
No. of CD8+ T cells producing cytokine
|
||
|---|---|---|---|---|
| IFN-γ | TNF-α | IFN-γ | TNF-α | |
| Spleenb | (3.1 ± 1.7) × 105 | (1.1 ± 5.4) × 105 | (5.8 ± 3.2) × 105 | (6.7 ± 4.0) × 103 |
| Liverc | 2.4 × 103 | 6.5 × 102 | 5.0 × 102 | 37 |
| Bloodd | 2.6 × 102 | 2.9 × 102 | 70 | 7.3 |
| MLNe | (5.6 ± 3.2) × 103 | (1.9 ± 1.4) × 103 | (3.1 ± 2.8) × 104 | (3.8 ± 5.0) × 102 |
Mice were orally immunized ≥15 weeks earlier with Salmonella, and cytokine production was assessed after restimulation with Salmonella lysate. The absolute numbers of cytokine-producing T cells in the indicated organs are shown and are expressed as mean values ± standard deviations.
n = 7.
Cells were pooled from five mice. Values represent an estimate of the total number of cytokine-producing T cells per mouse.
Cells were pooled from four mice. Values represent an estimate of the total number of cytokine-producing T cells per mouse.
Cells were pooled from six mice into three groups. Values represent an estimate of the total number of cytokine-producing T cells per mouse.
Salmonella-responsive CD4+ T cells producing TNF-α were present in the spleen, liver, and peripheral blood at frequencies up to sixfold lower than those of cells producing IFN-γ (Fig. 6). However, compared with the frequencies in naïve animals, these frequencies represented increases of 74-, 6-, and 46-fold for the spleen, liver and peripheral blood, respectively (Fig. 6b). As observed for IFN-γ expression, TNF-α-responsive CD4+ T cells in peripheral LN also had increased frequencies (up to 19-fold). Interestingly, a larger degree of frequency equivalence was observed between IFN-γ- and TNF-α-producing CD4+ Salmonella-specific populations in peripheral LN than between those in the spleen, liver, and blood (Fig. 6). Like the case for IFN-γ-producing cells, the largest number of CD4+ T cells producing TNF-α in Salmonella-immunized mice in response to a bacterial lysate was found in the spleen (Table 1). Together, these data indicate that the numbers of both IFN-γ- and TNF-α-producing Salmonella-specific CD4+ T cells are increased in all examined organs during the memory phase, with IFN-γ-producing cells dominating and with the spleen containing the largest number of responding cells. Furthermore, the levels of retention of Salmonella-specific CD4+ T cells occurred in an organ-specific manner.
Differential distribution of Salmonella-specific IFN-γ- and TNF-α-producing CD8+ T cells after oral infection.
The distribution of CD8+ T cells producing IFN-γ after Salmonella lysate restimulation was examined for animals that were immune to Salmonella and was found to be similar to that observed for CD4+ T cells producing IFN-γ. Specifically, the highest mean frequencies of these CD8+ T cells occurred in the spleen, liver, and peripheral blood and were associated with increases of 176-, 11-, and 13-fold, respectively, for these organs compared to those from naïve mice (Fig. 7a). In addition, the frequencies of IFN-γ-expressing CD8+ T cells were also increased in the peripheral LN of these immune animals and were 5- to 38-fold higher than those in naïve mice (Fig. 7a). The total number of Salmonella-responsive cytokine-producing CD8+ T cells was largest in the spleen (Table 1).
In contrast, CD8+ T cells producing TNF-α after restimulation were limited both in frequency and in distribution (Fig. 5 and 7b). First, each examined site, with the notable exception of the liver, exhibited a Salmonella-specific CD8+ TNF-α response (Fig. 7b). The extents of these frequency increases ranged from 2-fold for MLN to 27-fold for peripheral blood compared to those for naïve mice. Second, only in the peripheral blood did the frequency of TNF-α-producing cells exceed 1:500. Finally, CD8+ TNF-α-producing cells were greatly reduced in frequency compared with their IFN-γ-producing counterparts, with TNF-α+/IFN-γ+ frequency ratios of >1:100 for the spleen and >1:20 for the peripheral blood (Fig. 7). No increase above naïve levels was observed for the Salmonella-specific CD8+ TNF-α response in the livers of immune individuals.
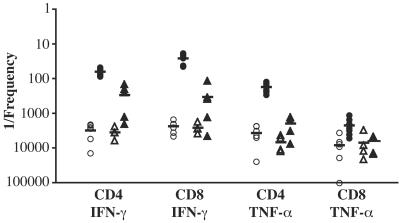
NKT cells as well as activated or memory T cells, particularly CD8+ cells, can be induced to produce IFN-γ in a non-antigen-specific fashion in response to cytokines such as IL-12, IL-18, and IFN-α/β (1, 6, 7, 20, 27). Moreover, LPS can indirectly stimulate IFN-γ production by CD8+ T cells, CD4+ T cells, and NKT cells by inducing IL-12 secretion by antigen-presenting cells (20). Since the restimulation protocol using the Salmonella lysate raised the possibility that some of the cytokine-producing cells detected responded in a non-antigen-specific fashion to LPS, we quantitated the frequencies of cytokine-producing T cells in splenocyte cultures that were restimulated with a concentration of purified Salmonella LPS equal to that in the Salmonella lysate. The restimulation of splenocytes from Salmonella-infected mice with LPS resulted in an increased frequency of cytokine-producing T cells relative to the frequency in naïve mice (Fig. 8). This was most apparent for IFN-γ-producing cells, for which there were 12- and 8-fold increases in the frequencies of TCRβ+ CD4+ and TCRβ+ CD8+ cells, respectively, producing this cytokine in response to LPS compared to those in naïve mice. In contrast, no increase was apparent for the mean frequency of TCRβ+ CD8+ cells producing TNF-α after restimulation with LPS in infected mice relative to naïve mice, and only a threefold increase in TCRβ+ CD4+ T cells was observed (Fig. 8). However, despite the presence of splenocytes from Salmonella-infected mice that produced cytokines in response to LPS, 5 and 13 times more TCRβ+ CD4+ and TCRβ+ CD8+ cells, respectively, produced IFN-γ in response to lysate than in response to LPS. In other words, only 21 or 7.8% of the total number of TCRβ+ CD4+ and TCRβ+ CD8+ cells, respectively, that produced IFN-γ in response to the Salmonella lysate appeared to be due to LPS responsiveness. Likewise, 11 times more TCRβ+ CD4+ cells produced TNF-α in response to lysate than in response to LPS, revealing that 9% of the TNF-α-producing TCRβ+ CD4+ cells from infected mice can be attributed to LPS responsiveness. The MLN had the highest fraction of cells that responded to LPS with IFN-γ production, with 44 and 17% of TCRβ+ CD4+ and TCRβ+ CD8+ cells, respectively, with lysate-responsive cytokine production appearing to be due to LPS. In the liver, 15 and 13% of TCRβ+ CD4+ and TCRβ+ CD8+ cells, respectively, with lysate-responsive IFN-γ production appeared to be due to LPS, while for the blood these values were 2.5 and 21%, respectively. Together, these data demonstrate that a fraction of the cells that produce IFN-γ in response to bacterial lysate, particularly TCRβ+ CD4+ cells, responded to LPS.
FIG. 8.
Frequencies of IFN-γ- and TNF-α-producing T cells in the spleens of Salmonella-immunized mice restimulated with LPS. The plot shows 1/the frequency of IFN-γ- and TNF-α-positive TCRβ+ CD4+ or TCRβ+ CD8+ cells in the spleens of naïve mice (open symbols) or mice who were orally immunized >15 weeks earlier with Salmonella (filled symbols) after an ex vivo restimulation with Salmonella lysate (○ or •) or with purified Salmonella LPS at the same concentration as that in the lysate (▵ or ▴). Organs were from four to seven individuals, and each symbol represents the value for an individual mouse. Specific frequencies were quantitated as described in Materials and Methods. Horizontal lines indicate the mean values for each organ.
To further investigate the contribution of non-antigen-specific cytokine production induced by the Salmonella lysate, we performed lysate restimulation assays under conditions in which IL-12 was neutralized (Table 2). Neutralizing IL-12 had little effect on the frequency of CD4+ or CD8+ TNF-α-producing T cells in Salmonella-immunized mice in response to the bacterial lysate, reducing the frequency in only one of five mice. In contrast, neutralizing IL-12 reduced the frequency of IFN-γ-producing CD4+ T cells in three of five mice, by 18 to 40%. Likewise, neutralizing IL-12 reduced the frequency of CD8+ T cells producing IFN-γ in response to the lysate in all mice tested, ranging from a 7 to 52% reduction (Table 2).
TABLE 2.
Effect of neutralizing IL-12 on T-cell cytokine production by splenocytes from Salmonella-immunized mice after restimulation with Salmonella lysatea
| Mouse no. | % CD4 IFN-γ-producing cells
|
% CD8 IFN-γ-producing cells
|
% CD4 TNF-α-producing cells
|
% CD8 TNF-α-producing cells
|
||||
|---|---|---|---|---|---|---|---|---|
| Isotype Ab | Anti-IL-12 | Isotype Ab | Anti-IL-12 | Isotype Ab | Anti-IL-12 | Isotype Ab | Anti-IL-12 | |
| 1 | 2.0 | 1.2 | 8.4 | 6.1 | 0.66 | 0.62 | 0.14 | 0.20 |
| 2 | 1.4 | 0.89 | 7.3 | 5.8 | 0.31 | 0.39 | 0.005 | 0.017 |
| 3 | 1.7 | 1.4 | 2.7 | 2.4 | 0.6 | 0.83 | 0.11 | 0.14 |
| 4 | 1.0 | 1.1 | 4.4 | 4.1 | 0.36 | 0.38 | 0.001 | 0.012 |
| 5 | 8.3 | 9.2 | 16 | 7.7 | 2.7 | 4.1 | 0.154 | 0.062 |
Splenocytes from mice orally infected with Salmonella 26 weeks earlier were stimulated with Salmonella lysate in the presence of either a neutralizing antibody to IL-12 (clone C17.8) or a rat IgG2a isotype control, as indicated. The numbers show the percentages of cytokine-producing cells after the percentages of cells staining with the isotype-matched control antibody were subtracted. The data are from five infected mice.
These data indicate that 2 to 6 months after pathogen exposure, polyclonal CD4+- and CD8+-T-cell populations that are capable of rapid IFN-γ production as well as CD4+ TNF-α responses are primarily located in a central spleen-liver-blood compartment, with the largest number of cytokine-producing cells being found in the spleen. Significant amounts of these cells also remain within peripheral lymphoid sites. However, not only are both CD4+ and CD8+ Salmonella-specific TNF-α responses numerically much more restricted than those for IFN-γ, those Salmonella-specific CD8+ T cells that are capable of rapid TNF-α production upon antigen exposure appear to be relatively restricted to the peripheral blood. Finally, a fraction of the T cells from Salmonella-immunized mice that produce cytokines, particularly IFN-γ, after restimulation with bacterial lysate can be attributed to the effects of LPS and are partially dependent on IL-12.
DISCUSSION
The present study examined the frequencies, functions, and locations of memory T cells in monoclonal and polyclonal systems in DNA- and bacterium-immunized mice, respectively. After immunization with a plasmid encoding HBsAg, a splenic Kb-restricted CD8+-T-cell response was detectable by day 10 and reached a peak frequency in the spleen on day 14 postimmunization, as reported for other plasmid immunization systems (15). Far lower frequencies of HBsAg-specific CD8+ T cells were observed for the draining LN on day 14. In a model of herpes simplex virus 1 infection, specific cytotoxic T lymphocytes (CTL) are activated within draining LN but undergo the bulk of their subsequent expansion within the spleen (11). Our results are in agreement with such a model and indicate that, in addition to being present in the spleen, KbS-specific cells are also present, at equivalent frequencies, in both the liver and the peripheral blood on day 14. Cells in the liver and blood may have been released from the draining LN or from the spleen and may accumulate within the liver, possibly prior to apoptosis and clearance (34).
On day 42 post-pCI/S immunization, rapidly responding KbS-specific CD8+ cells were present within both lymphoid (spleen) and nonlymphoid (liver and blood) organs. The spleen, liver, and blood had similar frequencies of KbS-specific cells among the total TCRβ+ CD8+ cells. Moreover, when the total number of T cells in these different immune compartments was taken into account, the largest number of epitope-specific T cells was present in the spleen. In contrast to the case for the spleen, liver, and blood, very few IFN-γ-producing, KbS-specific CD8+ T cells were detectable within the draining LN on day 42. The rapid IFN-γ response and non-LN distribution of these cells suggest that they are similar to TEM CD8+ lymphocytes in the TEM-TCM model (32, 41, 53). However, further characterization of CCR7 and CD62L expression, for example, on the memory cells is required to define both TEM and TCM populations in each location.
It has been suggested that organ-specific memory cell populations may develop, particularly within nonlymphoid tissues, after pathogen exposure (21, 31, 32, 39). Distinct functional capabilities associated with nonlymphoid CD8+-T-cell memory, such as immediate cytolytic activity (32), may aid the host in rapidly responding to antigens at sites of pathogen exposure, such as the lungs and intestinal mucosa. The retention of KbS-specific cells that are capable of rapid IFN-γ production within the spleen, liver, and blood is consistent with this model in that any antigen entering the bloodstream will traffic rapidly through the spleen and liver. For this system, it may not necessarily be the lymphoid or nonlymphoid nature of a tissue per se that determines TEM cell retention, but perhaps another feature of the tissue that influences its capacity to encounter or screen for antigens.
The frequency of specific IFN-γ-producing cells detected by short-term ex vivo peptide restimulation mirrored that detected by MHC tetramer staining (10, 32, 39), which detects antigen-specific T cells independently of any antigen-presenting cells. This supports the hypothesis that the frequencies of peptide-specific T cells detected in different organs after peptide restimulation are not influenced by different antigen-presenting-cell populations that are present in different proportions in the organs. In addition, although the off-cycling of cytokine production can influence the detection of cytokine-positive cells, the peptide restimulation protocol used here (5 h, with BFA added during the last 4 h) should allow optimal IFN-γ detection (2-4, 45).
TCRαβ T cells and cytokines that can be produced by these cells, particularly IFN-γ and TNF-α, are critical for the host defense against Salmonella (13, 36). Indeed, IFN-γ-producing TCRαβ+ CD4+ and CD8+ T cells are elicited during primary Salmonella infections (49, 55), and class Ia and Ib-restricted cytolytic CD8+ T cells are evoked in immune mice challenged with virulent Salmonella (28). However, little is known about the memory T-cell compartment in Salmonella-immunized mice, and a recent study showed that the expansion of endogenous Salmonella-specific CD4+ T cells could even prevent the survival of adoptively transferred, Salmonella-specific transgenic T cells and hinder their development into memory cells (49). We thus extended the analysis of antigen-specific memory responses to a polyclonal bacterial system and determined the frequencies and locations of CD4+ and CD8+ TCRαβ T cells that were capable of rapid, ex vivo IFN-γ or TNF-α production in mice who were orally immunized 9 to 28 weeks earlier with Salmonella.
We found large increases in the frequencies of IFN-γ- or TNF-α-producing memory CD4+ T cells responding to Salmonella stimulation associated with both lymphoid organs and nonlymphoid compartments. Significant numbers of responding CD4+ memory cells were present in the spleen, liver, blood, and LN, with liver populations exhibiting the smallest increases relative to naïve mice. Moreover, the largest absolute number of Salmonella-specific CD4+ memory cells was found in the spleen.
A feature of Salmonella-specific CD4+ memory was that the frequency of IFN-γ+ cells in immune mice was consistently higher than the TNF-α response in populations located in the spleen, liver, MLN, and peripheral blood. Recent work also showed a higher percentage of IFN-γ-producing cells relative to TNF-α-producing CD4+ T cells in the spleen during the expansion phase after Salmonella vaccination (49). Although TNF-α and IFN-γ are both important for controlling Salmonella infection (13, 36), IFN-γ production by T cells may be more important than TNF-α production by these cells considering, for example, that phagocytes are numerically dominant sources of TNF-α during infection (23). Alternatively, a robust IFN-γ response may be sufficient to control an attenuated strain such as χ4550.
In contrast to the spleen, liver, blood, and MLN, the other LN examined here had similar frequencies of TNF-α- and IFN-γ-responding CD4+ cells on day 42 postinfection. Although cytokine off-cycling can influence the detection of TNF-α-producing T cells in particular (2), the similar frequencies of cells producing TNF-α and IFN-γ in the LN suggest that functionally distinct subsets of Salmonella-specific CD4+ memory cells may be differentially represented in, for example, the spleen compared with peripheral LN.
Examining the polyclonal T-cell response by restimulation with a bacterial lysate resulted in the activation of some cells in naïve animals, which was most noticeable in the liver and peripheral blood. However, the frequencies of cytokine-positive cells from the liver and peripheral blood of naïve animals were lower than those from immune animals, indicating the presence of specific immunity in these organs. Some of the background response may be attributed to endogenous Salmonella-specific responses (49). Furthermore, some of the observed cytokine production may come from non-antigen-specific components. Indeed, a fraction of the TCRβ+ cells detected produced IFN-γ in response to LPS, and this was more apparent for CD4+ than for CD8+ T cells. Some of these cells, particularly CD4+ cells (38), could be NKT cells that produce IFN-γ in response to LPS-induced IL-12 by antigen-presenting cells (8, 20). The partial IL-12 dependence of IFN-γ production in response to the lysate may indicate that NKT cells comprise a fraction of the cells detected. However, given the very small number of NKT cells relative to the number of conventional TCRαβ T cells, IFN-γ-producing NKT cells would be only a small fraction of IFN-γ-producing cells among TCRβ+ cells (8, 23).
Adoptive transfer studies have shown that the generation of CD4+ memory to a protein antigen administered with an adjuvant results in antigen-specific memory cells in both lymphoid and nonlymphoid tissues, including the liver (40). In those experiments, memory CD4+ T cells in nonlymphoid tissues were more likely to produce IFN-γ than those from lymphoid sites (40). Since our assessment of the polyclonal response to Salmonella revealed significant frequencies of IFN-γ-producing CD4+ memory cells in lymphoid organs as well as in the liver and blood, additional studies are needed to determine whether a distinct cytokine production capacity, such as that for Salmonella-specific IL-2, correlates with cells from a particular location (40).
Our data also revealed that Salmonella-specific IFN-γ-producing CD8+ T cells were present in the organs examined, with the spleen again exhibiting the largest, and the liver the smallest, increase in the frequency of these cells relative to that for naïve mice. As for CD4+ T cells, a fraction of the CD8+ T cells detected produced IFN-γ in response to LPS, and IFN-γ production in response to the lysate showed a partial dependence on IL-12. Since the majority of NKT cells do not express CD8 (38), only very few, if any, of the CD8+ cells producing IFN-γ in response to LPS could be NKT cells. However, conventional CD8+ TCRαβ T cells can produce IFN-γ by a cytokine-dependent bystander mechanism (1, 6, 7, 20, 27), and such cells likely contribute to the non-antigen-specific response that we detected. In addition, some of the Salmonella-reactive IFN-γ-producing CD8+ T cells could be restricted to nonclassical MHC molecules (28).
Similar frequencies of IFN-γ-producing CD4+ and CD8+ T cells among memory cells were found for any given organ, with the exception of the inguinal LN. In contrast, previous work has shown that IFN-γ+ T cells in the spleen 14 days after primary Salmonella infection were present in larger numbers among CD4+ T cells than among their CD8+ equivalents (55). In addition, another study showed a massive expansion of IFN-γ-producing CD4+ T cells after Salmonella administration (49). The fact that IFN-γ-producing memory CD4+ T cells may not necessarily outnumber IFN-γ-producing memory CD8+ T cells may reflect the availability of CD4-specific epitopes during primary infection rather than, for example, differences in the clonality of the T-cell populations in the primary and memory responses (52). Alternatively, the different restimulation protocols used for these studies or the fraction of cells in the T-cell subsets that produce IFN-γ in response to LPS may influence the observations. Additional experiments to quantitate the frequencies of T cells in Salmonella-infected mice during the expansion and memory phases by using the same restimulation protocol and accounting for differences in LPS responsiveness among the T-cell subsets would directly address this issue.
Salmonella-specific CD8+ memory differed functionally from CD4+ memory in that very few rapidly responding TNF-α-expressing CD8+ T cells were detectable in most organs examined from immune animals. Indeed, Salmonella-specific TNF-α+ CD8+ memory cells were approximately 100-fold less frequent than IFN-γ-responding cells in the spleen and MLN, with a >20-fold difference in the frequencies found in the blood. The highest frequency of CD8+ TNF-α memory cells was found in the peripheral blood, with the largest absolute number of these cells being found in the spleen. We also found that little, if any, production of TNF-α by CD8+ memory T cells in Salmonella-immunized mice was due to LPS.
Other studies have also shown that the rapid antigen-specific CD8+ TNF-α response is consistently lower than the equivalent IFN-γ response for both primary and secondary antiviral and antibacterial systems (2, 4, 46). However, >90% of LCMV-specific splenic CD8+ memory cells produce both cytokines after ex vivo peptide restimulation (46). The apparent lack of a significant rapid TNF-α response among CD8+ memory cells in Salmonella compared with LCMV immunity may be a consequence of examining a polyclonal versus monoclonal (immunodominant peptide) response. Alternatively, pathogen-specific functional attributes, in addition to location- or organ-specific differences, may be a feature of memory populations generated during infections. This may reflect, for example, pathogen tropism for certain organs.
Both lymphoid and nonlymphoid CD8+ memory pools have been demonstrated in viral and bacterial infections by the use of epitope-specific responses as a readout (21, 29-32, 39). In these systems, very few memory CD8+ cells were detectable in the peripheral LN, as demonstrated here for KbS-specific CD8+ memory. The polyclonal analysis of Salmonella-specific memory cells, however, revealed that CD8+ memory populations are represented in peripheral LN at significant frequencies, albeit with less representation than in the spleen. This trend is more apparent for IFN-γ+ than for TNF-α+ cells. An analysis of memory CD8+ T cells after Listeria infection revealed a higher percentage of epitope-specific memory cells in the liver than in the spleen (39). This does not appear to be the case for Salmonella-specific memory, for which similar frequencies are retained in each organ into the memory phase. The frequency of antigen-specific memory cells among the total CD8+ T cells in a given organ may thus differ depending on the pathogen. For both infection models, however, the absolute number of memory CD8+ T cells was largest in the spleen.
Different antigen-presenting-cell populations, such as dendritic cell subsets, are present in different proportions in the tissues examined in this study (18, 44). This raises the possibility that differences in antigen-presenting cells in each tissue may influence the frequency of antigen-specific T cells detected after ex vivo restimulation with bacterial lysate. However, this is likely not the case for several reasons. First, dendritic cells, which are likely the most relevant antigen-presenting cell for stimulating T cells, from MLN and the spleen have a similar capacity to process and present a Salmonella-encoded antigen to T cells, and dendritic cells from the liver are also fully competent in this respect (18, 56). Moreover, no difference in the capacities of the CD8α− or CD8α+ dendritic cell subsets from either MLN or the spleen to process and present a Salmonella-encoded antigen to T cells was found (56). The lack of an overt difference in the ability of dendritic cells in the spleen, MLN, or liver to process and present Salmonella antigens and the similar capacities of CD8α− and CD8α+ cells in MLN and the spleen to stimulate bacterium-specific T cells suggest that the T-cell frequencies detected with the restimulation assay reflect the presence of Salmonella-specific T cells in the different organs.
In summary, the data presented here for a Salmonella infection model suggest that the polyclonal memory cell populations of CD4+ and CD8+ TCRαβ T cells directed against the same pathogen in the same individuals occupy distinct physical distributions according to their functional capabilities.
Acknowledgments
This work was supported by the Swedish Cancer Foundation (4884-B03-01XAB) and the European Union (PL970002).
We gratefully acknowledge Jörg Reimann, University of Ulm, Ulm, Germany, for pCI/S and pCI/C as well as for helpful discussions. The technical assistance of Kristina Lindgren is gratefully acknowledged.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Andersson, Å., W. J. Dai, J. P. Di Santo, and F. Brombacher. 1998. Early IFN-γ production and innate immunity during Listeria monocytogenes infection in the absence of NK cells. J. Immunol. 161:5600-5606. [PubMed] [Google Scholar]
- 2.Badovinac, V. P., G. A. Corbin, and J. T. Harty. 2000. Cutting edge: OFF cycling of TNF production by antigen-specific CD8+ T cells is antigen independent. J. Immunol. 165:5387-5391. [DOI] [PubMed] [Google Scholar]
- 3.Badovinac, V. P., and J. T. Harty. 2000. Adaptive immunity and enhanced CD8+ T cell response to Listeria monocytogenes in the absence of perforin and IFN-γ. J. Immunol. 164:6444-6452. [DOI] [PubMed] [Google Scholar]
- 4.Badovinac, V. P., and J. T. Harty. 2000. Intracellular staining for TNF and IFN-gamma detects different frequencies of antigen-specific CD8(+) T cells. J. Immunol. Methods 238:107-117. [DOI] [PubMed] [Google Scholar]
- 5.Badovinac, V. P., B. B. Porter, and J. T. Harty. 2002. Programmed contraction of CD8 T cells after infection. Nat. Immunol. 3:619-626. [DOI] [PubMed] [Google Scholar]
- 6.Berg, R. E., C. J. Cordes, and J. Forman. 2002. Contribution of CD8+ T cells to innate immunity: IFN-γ secretion induced by IL-12 and IL-18. Eur. J. Immunol. 32:2807-2816. [DOI] [PubMed] [Google Scholar]
- 7.Berg, R. E., E. Crossley, S. Murray, and J. Forman. 2003. Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J. Exp. Med. 198:1583-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brigl, M., L. Bry, S. C. Kent, J. E. Gumperz, and M. B. Brenner. 2003. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat. Immunol. 4:1230-1237. [DOI] [PubMed] [Google Scholar]
- 9.Busch, D. H., I. M. Pilip, S. Vijh, and E. G. Pamer. 1998. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity 8:353-362. [DOI] [PubMed] [Google Scholar]
- 10.Cauley, L. S., T. Cookenham, T. B. Miller, P. S. Adams, K. M. Vignali, D. A. A. Vignali, and D. L. Woodland. 2002. Virus-specific CD4+ memory T cells in nonlymphoid tissues express a highly activated phenotype. J. Immunol. 169:6655-6658. [DOI] [PubMed] [Google Scholar]
- 11.Coles, R. M., S. N. Mueller, W. R. Heath, F. R. Carbone, and A. G. Brooks. 2002. Progression of armed CTL from draining lymph node to spleen shortly after localized infection with herpes simplex virus 1. J. Immunol. 168:834-838. [DOI] [PubMed] [Google Scholar]
- 12.Doherty, P. C., and J. P. Christensen. 2000. Accessing complexity: the dynamics of virus-specific T cell responses. Annu. Rev. Immunol. 18:561-592. [DOI] [PubMed] [Google Scholar]
- 13.Eckmann, L., and M. F. Kagnoff. 2001. Cytokines in host defense against Salmonella. Microb. Infect. 3:1191-1200. [DOI] [PubMed] [Google Scholar]
- 14.Foulds, K. E., L. A. Zenewicz, D. J. Shedlock, J. Jian, A. E. Troy, and H. Shen. 2002. Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J. Immunol. 168:1528-1532. [DOI] [PubMed] [Google Scholar]
- 15.Hassett, D. E., M. K. Slifka, J. Zhang, and J. L. Whitton. 2000. Direct ex vivo kinetic and phenotypic analyses of CD8(+) T-cell responses induced by DNA immunization. J. Virol. 74:8286-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan, R. J., L. S. Cauley, K. H. Ely, T. Cookenhan, A. D. Roberts, J. W. Brennan, S. Monard, and D. L. Woodland. 2002. Long-term maintenance of virus-specific effector memory CD8+ T cells in the lung airways depends on proliferation. J. Immunol. 169:4976-4981. [DOI] [PubMed] [Google Scholar]
- 17.Jankovic, D., M. C. Kullberg, S. Hieny, P. Caspar, C. M. Collazo, and A. Sher. 2002. In the absence of IL-12, CD4+ T cell responses to intracellular pathogens fail to default to a Th2 pattern and are host protective in an IL-10−/− setting. Immunity 16:429-439. [DOI] [PubMed] [Google Scholar]
- 18.Johannson, C., and M. J. Wick. 2004. Liver dendritic cells present bacterial antigens and produce cytokines upon Salmonella encounter. J. Immunol. 172:2496-2503. [DOI] [PubMed] [Google Scholar]
- 19.Kaech, S. M., and R. Ahmed. 2001. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat. Immunol. 2:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kambayashi, T., D. Assarsson, A. E. Lukacher, H.-G. Ljunggren, and P. E. Jensen. 2003. Memory CD8+ T cells provide an early source of IFN-γ. J. Immunol. 170:2399-2408. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S.-K., K. S. Schluns, and L. Lefrançois. 1999. Induction and visualization of mucosal memory CD8 T cells following systemic virus infection. J. Immunol. 163:4125-4132. [PubMed] [Google Scholar]
- 22.Kirby, A. C., U. Yrlid, M. Svensson, and M. J. Wick. 2001. Differential involvement of dendritic cell subsets during acute Salmonella infection. J. Immunol. 166:6802-6811. [DOI] [PubMed] [Google Scholar]
- 23.Kirby, A. C., U. Yrlid, and M. J. Wick. 2002. The innate immune response differs in primary and secondary Salmonella infection. J. Immunol. 169:4450-4459. [DOI] [PubMed] [Google Scholar]
- 24.Kuhober, A., H. P. Pudollek, K. Reifenberg, F. V. Chisari, H. J. Schlicht, J. Reimann, and R. Schirmbeck. 1996. DNA immunization induces antibody and cytotoxic T cell responses to hepatitis B core antigen in H-2b mice. J. Immunol. 156:3687-3695. [PubMed] [Google Scholar]
- 25.Kursar, M., K. Gonhagen, A. Köhler, T. Kamradt, S. H. E. Kaufmann, and H.-W. Mittrücker. 2002. Organ-specific CD4+ T cell response during Listeria monocytogenes infection. J. Immunol. 168:6382-6387. [DOI] [PubMed] [Google Scholar]
- 26.Lefrançois, L., and D. Masopust. 2002. T cell immunity in lymphoid and non-lymphoid tissues. Curr. Opin. Immunol. 14:503-508. [DOI] [PubMed] [Google Scholar]
- 27.Lertmemongkolchai, G., G. Cai, C. A. Hunter, and G. J. Bancroft. 2001. Bystander activation of CD8+ T cells contributes to the rapid production of IFN-γ in response to bacterial pathogens. J. Immunol. 166:1097-1105. [DOI] [PubMed] [Google Scholar]
- 28.Lo, W.-F., H. Ong, E. S. Metcalf, and M. J. Soloski. 1999. T cell responses to gram-negative intracellular bacterial pathogens: a role for CD8+ T cells in immunity to Salmonella infection and the involvement of MHC class Ib molecules. J. Immunol. 162:5398-5406. [PubMed] [Google Scholar]
- 29.Marshall, D. R., S. J. Turner, G. T. Belz, S. Wingo, S. Andreansky, M. Y. Sangster, J. M. Riberdy, T. Liu, M. Tan, and P. C. Doherty. 2001. Measuring the diaspora for virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA 98:6313-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzo, A. L., V. Vezys, K. Williams, D. F. Tough, and L. Lefrançois. 2002. Tissue-level regulation of Th1 and Th2 primary and memory CD4 T cells in response to Listeria infection. J. Immunol. 168:4504-4510. [DOI] [PubMed] [Google Scholar]
- 31.Masopust, D., J. Jiang, H. Shen, and L. Lefrançois. 2001. Direct analysis of the dynamics of the intestinal mucosa CD8 T cell response to systemic virus infection. J. Immunol. 166:2348-2356. [DOI] [PubMed] [Google Scholar]
- 32.Masopust, D., V. Vezys, A. Marzo, and L. Lefrançois. 2001. Preferential localization of effector memory cells in nonlymphoid tissue. Science 291:2413-2417. [DOI] [PubMed] [Google Scholar]
- 33.McSorley, S. J., S. Asch, M. Costalonga, R. L. Reinhardt, and C. Jenkins. 2002. Tracking Salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity 16:365-377. [DOI] [PubMed] [Google Scholar]
- 34.Mehal, W. Z., A. E. Juedes, and I. N. Crispe. 1999. Selective retention of activated CD8+ T cells by the normal liver. J. Immunol. 163:3203-3210. [PubMed] [Google Scholar]
- 35.Mercado, R., S. Vijh, S. E. Allen, K. Kerksiek, I. M. Pilip, and E. G. Pamer. 2000. Early programming of T cell populations responding to bacterial infection. J. Immunol. 165:6833-6839. [DOI] [PubMed] [Google Scholar]
- 36.Mittrücker, H.-W., and S. H. E. Kaufmann. 2000. Immune response to infection with Salmonella typhimurium in mice. J. Leukoc. Biol. 67:457-462. [DOI] [PubMed] [Google Scholar]
- 37.Murali-Krishna, K., J. D. Altman, M. Suresh, D. J. D. Sourdive, A. J. Zajac, J. D. Miller, J. Slansky, and R. Ahmed. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8:177-187. [DOI] [PubMed] [Google Scholar]
- 38.Ohteki, T., and H. R. MacDonald. 1994. Major histocompatibility complex class I related molecules control the development of CD4+8− and CD4−8− subsets of natural killer 1.1+ T cell receptor-α/β+ cells in the liver of mice. J. Exp. Med. 180:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pope, C., S.-K. Kim, A. Marzo, K. Williams, J. Jiang, H. Shen, and L. Lefrançois. 2001. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J. Immunol. 166:3402-3409. [DOI] [PubMed] [Google Scholar]
- 40.Reinhardt, R. L., A. Khoruts, R. Merica, T. Zell, and M. K. Jenkins. 2001. Visualizing the generation of memory CD4 T cells in the whole body. Nature 410:101-105. [DOI] [PubMed] [Google Scholar]
- 41.Sallusto, F., D. Lenig, R. Förster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708-712. [DOI] [PubMed] [Google Scholar]
- 42.Schirmbeck, R., W. Bohm, K. Ando, F. V. Chisari, and J. Reimann. 1995. Nucleic acid vaccination primes hepatitis B virus surface antigen-specific cytotoxic T lymphocytes in nonresponder mice. J. Virol. 69:5929-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schirmbeck, R., J. Wild, and J. Reimann. 1998. Similar as well as distinct MHC class I-binding peptides are generated by exogenous and endogenous processing of hepatitis B virus surface antigen. Eur. J. Immunol. 28:4149-4161. [DOI] [PubMed] [Google Scholar]
- 44.Shortman, K., and Y.-J. Liu. 2002. Mouse and human dendritic cell subsets. Nat. Rev. Immunol. 2:151-161. [DOI] [PubMed] [Google Scholar]
- 45.Slifka, M. K., F. Rodriguez, and J. L. Whitton. 1999. Rapid on/off cycling of cytokine production by virus-specific CD8+ T cells. Nature 401:76-79. [DOI] [PubMed] [Google Scholar]
- 46.Slifka, M. K., and J. L. Whitton. 2000. Activated and memory CD8+ T cells can be distinguished by their cytokine profiles and phenotypic markers. J. Immunol. 164:208-216. [DOI] [PubMed] [Google Scholar]
- 47.Slifka, M. K., and J. L. Whitton. 2000. Antigen-specific regulation of T cell-mediated cytokine production. Immunity 12:451-457. [DOI] [PubMed] [Google Scholar]
- 48.Sprent, J., and C. D. Surh. 2002. T cell memory. Annu. Rev. Immunol. 20:551-579. [DOI] [PubMed] [Google Scholar]
- 49.Srinivasan, A., J. Foley, and S. J. McSorley. 2004. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated Salmonella causes interclonal competition. J. Immunol. 172:6884-6893. [DOI] [PubMed] [Google Scholar]
- 50.Trobonjaca, Z., F. Leithäuser, P. Moller, R. Schirmbeck, and J. Reimann. 2001. Activating immunity in the liver. I. Liver dendritic cells (but not hepatocytes) are potent activators of IFN-gamma release by liver NKT cells. J. Immunol. 167:1413-1422. [DOI] [PubMed] [Google Scholar]
- 51.Tuma, R. A., and E. G. Pamer. 2002. Homeostasis of naive, effector and memory CD8 T cells. Curr. Opin. Immunol. 14:348-353. [DOI] [PubMed] [Google Scholar]
- 52.Turner, S. J., G. Diaz, R. Cross, and P. C. Doherty. 2003. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity 18:549-559. [DOI] [PubMed] [Google Scholar]
- 53.Wherry, E. J., V. Teichgräber, T. C. Becker, D. Masopust, S. M. Kaech, R. Antia, U. H. von Andrian, and R. Ahmed. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4:225-234. [DOI] [PubMed] [Google Scholar]
- 54.Wong, P., and E. G. Pamer. 2003. Feedback regulation of pathogen-specific T cell priming. Immunity 18:499-511. [DOI] [PubMed] [Google Scholar]
- 55.Yrlid, U., M. Svensson, B. Chambers, H.-G. Ljunggren, and M. J. Wick. 2001. In vivo activation of dendritic cells and T cells during Salmonella enterica serovar Typhimurium infection. Infect. Immun. 69:5726-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yrlid, U., and M. J. Wick. 2002. Antigen presentation capacity and cytokine production by murine splenic dendritic cell subsets upon Salmonella encounter. J. Immunol. 169:108-116. [DOI] [PubMed] [Google Scholar]