Abstract
Background
The benzimidazole (BZ) anthelmintics, albendazole (ABZ) and mebendazole (MBZ) are the most common drugs used for treatment of soil-transmitted helminths (STHs). Their intensive use increases the possibility that BZ resistance may develop. In veterinary nematodes, BZ resistance is caused by a single nucleotide polymorphism (SNP) in the β-tubulin isotype 1 gene at codon position 200, 167 or 198, and these SNPs have also been correlated with poor response of human Trichuris trichiura to BZ treatment. It is important to be able to investigate the presence of resistance-associated SNPs in STHs before resistance becomes clinically established.
Methods
The objective of this study was to develop new genotyping assays to screen for the presence of β-tubulin SNPs in T. trichiura and Ascaris lumbricoides. Rapid, simple and accurate genotyping assays were developed based on the SmartAmp2 method. Primer sets were optimized and selected to distinguish the SNP-variant genotypes. After initial optimization on control plasmids, the feasibility of the assay was assessed in field samples from Haiti and Panama. Finally, spiked fecal samples were assessed to determine the tolerance of Aac polymerase to fecal inhibitors.
Findings
Rapid SNP genotyping assays were developed to target β-tubulin polymorphisms in T. trichiura and A. lumbricoides. The assays showed high sensitivity and specificity in field samples and also demonstrated high tolerance to PCR inhibitors in fecal samples.
Conclusion
These assays proved to be robust and efficient with the potential to be used as field tools for monitoring SNPs that could be associated with BZ resistance. However, further work is needed to validate the assays on large numbers of field samples before and after treatment.
Author Summary
The soil-transmitted helminths (STHs) Ascaris lumbricoides, Trichuris trichiura, and the hookworms Necator americanus and Ancylostoma duodenale, are the most prevalent intestinal helminths of humans, causing morbidity in developing countries. Large-scale preventive chemotherapy with albendazole or mebendazole is the major control strategy against STHs in mass drug administration (MDA) programs. In veterinary parasites, intensive reliance on the same anthelmintics has led to the emergence of benzimidazole (BZ) resistance, which is caused by a single nucleotide polymorphism at either codon 200, 167 or 198 in the β-tubulin isotype 1 gene. A great concern is that the intensive use of anthelmintics with suboptimal efficacy may lead to the development of resistance in human STHs. We developed novel genotyping assays to screen for β-tubulin polymorphisms in T. trichiura and A. lumbricoides based on the SmartAmp2 method. SmartAmp2 is a unique genotyping technology that detects a mutation under isothermal conditions with high specificity and sensitivity. The SNP detection assays were rapid, sensitive and highly specific for the detection of SNPs associated with BZ resistance with the potential to be used for monitoring in the field.
Introduction
Soil-transmitted helminths (STHs) are a major cause of morbidity in developing countries. Ascaris lumbricoides, Trichuris trichiura and the hookworms Necator americanus and Ancylostoma duodenale are estimated to infect more than 1.5 billion people, resulting in approximately 5.2 million disability adjusted life years (DALYs) lost worldwide [1, 2]. Pre-school and school-age children are the most at risk of heavy infection with STHs and of developing severe morbidity [3, 4], leading to malnourishment, stunted growth and intellectual retardation, with cognitive and educational deficits [5]. Recent estimates indicate that approximately 900 million children are at high risk of acquiring STH infection and in need of annual treatment [6].
The current control strategy against STHs is the regular administration of a single-dose of ABZ (400 mg) or MBZ (500 mg) as preventive chemotherapy in large-scale mass drug administration (MDA) programs [7] with the ultimate goal of elimination of STHs as a public health problem by 2020 [3]. These programs have been greatly expanded in recent years by massive donations of these drugs. A single-dose of ABZ or MBZ shows high efficacy against A. lumbricoides, but both drugs show significantly suboptimal efficacy against T. trichiura and hookworm [8–11]. Intensive and prolonged reliance on two drugs of the same anthelmintic class with the same mode of action and suboptimal efficacy greatly increases the probability that BZ resistance may develop [12–14]. This would raise serious complications for control of STHs [4].
In veterinary nematodes, resistance developed in response to heavy reliance for many years on BZ anthelmintics [13]. It was found that the BZ resistance is caused by a single nucleotide polymorphism (SNP) in the β-tubulin isotype 1 gene at codon 167, codon 200 (TTC>TAC) or at codon 198 (GAG>GCG) [15–18]. Such SNPs have already been observed in T. trichiura and N. americanus [19,20]. Additionally, the frequency of SNPs at codon 200 and 198 increased with treatment and was significantly higher in individuals who showed a poor response to ABZ than in individuals who responded well to ABZ in T. trichiura [21]. To maintain the benefits of MDA programs, it is important to have tools that can be used for large-scale screening for BZ resistance in human STHs. The lack of detection of phenotypic resistance may, in part, be due to the lack of a reliable and sensitive method to monitor for resistance genotypes before and after BZ treatment [22], a low frequency of resistance alleles, and the probability that BZ resistance is recessive, as it is in veterinary parasites [23].
PCR-based methods such as real-time PCR (RT-PCR) and pyrosequencing have been developed and applied for the detection of putative BZ resistance SNPs in human STH [19, 20, 24]. Diagnostic RT-PCR is a rapid detection method in which primers bind only to specific sequence variants, with the 3’-end overlapping the SNP of interest. Allele-specific RT-PCR was developed for monitoring for β-tubulin polymorphisms in the human hookworms A. duodenale and N. americanus [24]; however, this method lacks the capability to completely distinguish background amplification noise rising from a non-target sequence [25]. Pyrosequencing has been developed for detection of resistance-associated SNPs in many veterinary parasites [26, 27] and also in human parasites [19, 20, 28]. Compared with RT-PCR, pyrosequencing is quicker and easier to perform as it allows testing multiple SNPs. However, the equipment required is expensive and not widely available [27]. Additionally, careful DNA purification is needed as the Taq DNA polymerases can be inhibited by impurities in clinical samples [29]. Intolerance to fecal inhibitors could result in a high percentage of PCR failure, which in turn, limits the ability to identify polymorphisms and to draw a conclusion, particularly in samples that have low DNA concentration. Current PCR-based methods have been shown to be more accurate, sensitive, and convenient than conventional sequencing; however, they are still time consuming procedures, require careful DNA extraction and expensive equipment, and include multiple steps. Therefore, they are not suitable for large scale screening and are difficult to implement.
Here we report the development of a novel genotyping assay to monitor the presence or absence of these SNPs in the β-tubulin isotype 1 gene of T. trichiura and A. lumbricoides, using the SmartAmp2 method (Smart Amplification Process). This technique has been previously used to develop assays for the hookworm, N. americanus [30]. The SmartAmp2 is a DNA amplification method for detection of DNA mutations, deletions or SNPs under isothermal conditions and in a single step. This method detects a single nucleotide polymorphism with high specificity and sensitivity within 30–45 min [25, 31]. SmartAmp2 uses Aac DNA polymerase (a strand displacing polymerase) combined with asymmetric primer design and Taq MutS (Thermaus aquaticus MutS) enzyme, which promote high specificity [32, 33].
The aim of this study was therefore to develop rapid and accurate genotyping assays based on the SmartAmp2 method for the detection of the β-tubulin SNPs that are likely associated with BZ resistance in T. trichiura and A. lumbricoides, and to validate their specificity and reliability in field samples.
Materials and Methods
Study approval and ethical considerations
Ethical approval for samples from Panama was obtained from the McGill University Institutional Review Board approval number A09-M87-11A [34, 35]. A parent or legal guardian gave informed consent for every child providing a stool sample.
Ethical approval (study 2535) was obtained by Dr. Patrick Lammie, CDC, Atlanta, GA, and included the collection, examination of fecal samples from Haiti, for helminth eggs, and DNA analysis of helminth eggs. Oral informed consent was obtained from all human adult participants and from parents or legal guardians of minors, as described previously (20, 21). Based on experience, it was considered unlikely that most persons in the sample communities would be literate. A request was therefore made to the Institutional Review Board for a waiver of written documentation of informed consent on the basis that the research presented no more than minimal risk of harm to the subjects and involves no procedures for which written consent is normally required outside of the research context in this setting. The reader of the consent form and a witness were asked to sign the form to indicate the subject’s agreement. The procedure was approved by the Institutional Review Board. This approach had been successfully used as part of recent studies and the forms were based on Institutional Review Board approved protocols (CDC Protocol #1524). Subjects were offered a written copy of the consent form.
Parasite materials and DNA extraction
A. lumbricoides eggs and adult worms had been collected as previously described [19]. T. trichiura DNA from adult worms was donated by Dr. P. Nesjum, University of Copenhagen, Denmark [36], and 40 fecal samples were collected in Haiti from children that were naturally infected. From Panama, 34 fecal samples were collected. All samples were preserved in 70% ethanol after collection. Eggs were isolated under a dissecting microscope using a 10 μl pipette and genomic DNA was extracted from eggs as described [37]. Lysis buffer was prepared as follow (KCl [50 mM], Tris[10 mM] pH 8.3, MgCl2 [2.5 mM], 0.45% Nonidet P-40, 0.45% Tween 20 and 0.01% gelatine). Ten μl of proteinase-K [10 μg/ml] (Invitrogen, Life Technologies; Burlington, ON) and β-mercaptoethanol (Sigma-Aldrich, ON, Canada) were added to 1 ml of this buffer, just before use. Twenty-five μl of the resulting lysis buffer mix was added to previously isolated eggs and then tubes were incubated at 60°C for 2 h. Genomic DNA was extracted from adult A. lumbricoides using DNeasy tissue extraction kit (Qiagen, Mississauga, ON, Canada) according to the manufacture’s protocol.
Wild-type and mutant-type plasmid constructs
To assist with SmartAmp2 development, wild-type (WT) and mutant-type (MT) plasmids were engineered and used as DNA templates for assay optimization. Extracted genomic DNA from individual adult worms was used to amplify a 472 bp fragment of the T. trichiura β-tubulin isotype 1 gene, including codon positions 167, 198 and 200. Specific forward primer 5’-GGCTAAAGGGCACTATACG-3’ and specific reverse primer 5’-GGAAAGCGTAGGCATGTCG-3’ (Invitrogen) were designed in the exonic regions of T. trichiura genomic DNA sequence (GenBank accession. no. AF034219.1). Genomic DNA extracted from A. lumbricoides adult worms was used to amplify a 564 pb fragment of the Ascaris β-tubulin isotype 1 gene, including codon positions 167, 198 and 200. Specific forward primer 5’- CCAGCTGACGCACTCGCTTGG -3’ and Specific reverse primer 5’-ATGGTTGAGGTCTCCGTATGTG-3’ (Invitrogen) were designed on the mRNA sequence of A. lumbricoides (GenBank accession. no. EU814697.1).
The PCR master mix contained 2 μl 10×PCR buffer, 1 μl MgSO4 [50 mM], 1 μl dNTP [10mM], 1 μl forward and reverse primer [10μM], 1 U Platinum Taq DNA polymerase High Fidelity (Invitrogen), 2 μl genomic DNA and distilled H2O to reach a final volume of 20 μl. No-template controls were also included for quality control. The PCR reaction conditions were the same for the two species (94°C for 3 min, followed by 35 cycles at 94°C for 45 s, 59°C for 45 s and 68°C for 1 min and a final extension at 68°C for 10 min. The resulting PCR fragments were Sanger sequenced to confirm the presence of WT alleles at codon positions 167, 198, and 200. MT plasmids carrying mutations at position 167, 198, or 200 were engineered by site-directed mutagenesis. A. lumbricoides and T. trichiura primers for MT plasmid construction, including inner primers carrying the mutant alleles and outer primers, are shown in Tables 1 and 2. WT and MT amplified fragments were cloned into TOPO-TA-Cloning vector (Invitrogen). Plasmid DNAs were extracted and purified using QIAprep Spin Miniprep kit (Qiagen) and subsequently sequenced by Sanger sequencing at the McGill University/Genome Quebec Innovation Centre, Montreal, Quebec. The purity and quantity of DNA in clones was measured using a Nano Drop photometer (Implen, Munich, Germany). WT and MT plasmids were used for assay optimization and development.
Table 1. Trichuris trichiura specific primers for the mutant plasmid constructs.
| Codon | Primer sequences (5′–3′) |
|---|---|
| 198 | Forward: GGCTAAAGGGCACTATACG |
| Reverse: GGAAAGCGTAGGCATGTCG | |
| SNP Fwd: GTAGAGAACACGGACGCAAC | |
| SNP Rev: GTTGCGTCCGTGTTCTCTAC | |
| 200 | Forward: GGCTAAAGGGCACTATACG |
| Reverse: GGAAAGCGTAGGCATGTCG | |
| SNP Fwd: GAACACGGACGAAACATACTG | |
| SNP Rev: CAGTATGTTTCGTCCGTGTTC |
Table 2. Ascaris lumbricoides specific primers for the mutant plasmid constructs.
| Codon | Primer sequences (5′–3′) |
|---|---|
| 167 | Forward: CCAGCTGACGCACTCGCTTGG |
| Reverse: GGTTGAGGTCTCCGTATGTG | |
| SNP Fwd: GCTCGTACTCAGTTGTTCCATC | |
| SNP Rev: GATGGAACAACTGAGTACGAGC | |
| 198 | Forward: CCAGCTGACGCACTCGCTTGG |
| Reverse: GGTTGAGGTCTCCGTATGTG | |
| SNP Fwd: GAACACCGATGCAACCTTC | |
| SNP Rev: GAAGGTTGCATCGGTGTTC | |
| 200 | Forward: CCAGCTGACGCACTCGCTTGG |
| Reverse: GGTTGAGGTCTCCGTATGTG | |
| SNP Fwd: GAACACCGATGAAACCTACTG | |
| SNP Rev: CAGTAGGTTTCATCGGTGTTC |
* SNP-Fwd: forward primer mutated for a single nucleotide; SNP-Rev: reverse primer mutated for a single nucleotide.
SmartAmp2 primer design
Primer sets were designed specifically to amplify and detect the F167Y (TTC>TAC), F200Y (TTC>TAC) and E198A (GAA>GCA) SNPs of the β-tubulin isotype 1 gene of A. lumbricoides and F200Y (TTC>TAC) and E198A (GAA>GCA) SNPs in the T. trichiura β-tubulin isotype 1 gene. The online software version 1.1 (www.SMAPDNA.com) was used initially to design several primer sets on the forward and reverse sequences with different discrimination primers which bind only to specific sequence variants, with the 3’-end overlapping the SNP of interest. Further refinements in the primer design were made by running evaluation tests and the best candidate primer sets were selected for each assay. A primer set consisting of five specific primers [the folding primer (FP), turn-back primer (TP), boost primer (BP), and two outer primers (OP1 and OP2)] were designed to recognize six different sequences on the target DNA. All primers were designed to be species-specific based on β-tubulin isotype 1 alignments (http://multalin.toulouse.inra.fr/multalin/) for A. lumbricoides, T. trichiura and N. americanus (Fig 1). The location and the sequences of primers for each SNP target are illustrated in Figs 2 and 3.
Fig 1. Partial sequence alignment of β-tubulin isotype 1 gene for Necator americanus, Ascaris lumbricoides and Trichuris trichiura.
SNP 167T>A, SNP 198A>C, and SNP 200T>A SmartAmp2 primer sets were designed with the 3’-end or 5’-end of one or two primers are overlapping inter-species nucleotide variations (species-specific).
Fig 2. SmartAmp2 primer design for β-tubulin SNP detection in Ascaris lumbricoides.
Partial sequence of the β-tubulin isotype 1 gene carrying (A) SNP 167A, (B) SNP 198C, and (C) SNP 200A as well as the sequences of primers used for the SmartAmp2 assay for the three SNP positions. The locations of SNPs indicated in bold. Both TP (turn-back primer) and FP (folding primer) were used as discrimination primers to target F167Y (TTC>TAC) SNP. BP (boost primer) was used as discrimination primer to target E198A (GAA>GCA) and F200Y (TTC>TAC) SNPs. The folding primer (FP) has a specific sequence (CCTATATATATATAGG) at the 5’ end to allow self-annealing hairpin formation.
Fig 3. SmartAmp2 primer design for β-tubulin SNP detection in Trichuris trichiura.
Partial sequence of the β-tubulin isotype 1 gene carrying (A) SNP 198C (B) SNP 200A as well as the sequences of primers used for the SmartAmp2 assay for the two SNP positions. The locations of SNPs indicated in bold. BP (boost primer) was used as discrimination primer to target E198A (GAA>GCA) and F200Y (TTC>TAC) SNPs. The folding primer (FP) has a specific sequence (CCTATATATATATAGG) at the 5’ end to allow self-annealing hairpin formation.
SmartAmp2 assay development and optimization
Control plasmids encoding WT or MT alleles were used to develop each assay and to evaluate the accuracy of genotyping between different primer sets. Accordingly, a candidate primer set was developed and optimized to distinguish MT and WT genotypes. Further optimization of assay conditions and components (concentrations of primers, MgSO4, betaine, SYBR Green, polymerase and Taq MutS) was performed. SmartAmp2 reactions were carried out in a total volume of 25 μl containing 2 μM TP/FP, 1 μM BP, 0.25 μM OP1/OP2 (Invitrogen), 1.4 mM dNTPs (Invitrogen), 1 M betaine (Sigma- Aldrich), 1x isothermal buffer [20 mM Tris-HCl (pH 8.6), 10 mM KCl, 10 mM (NH4)2SO4, (4–8 mM) MgSO4, 0.1% Tween 20], 1/100,000 dilution SYBR Green I (Invitrogen), 1 μg Taq MutS (Wako Chemicals, USA /Nippon Gene CO, Japan), and 12 U Aac DNA polymerase (KK. DNAFORM, Japan). One microliter of each WT or MT plasmid (5 ng) was heated at 95°C for 5 min before being added to the assay. Reactions were incubated at 60°C for 60 min. The Rotor-Gene Q system (Qiagen) was used to maintain isothermal conditions and to monitor the change in fluorescence intensity of SYBR Green I during the reaction. Assays were evaluated in terms of full match amplification and mismatch (non-amplification) within 60 min.
SmartAmp2 assay sensitivity and specificity
Following optimization of the assay with WT and MT plasmids, the specificity of each SNP-detection assay was verified with MT plasmids (F167Y, F200Y or E198A) of different STHs. The aim was to assess whether SNPs of another STH species would affect the specific amplification of each SNP-detection assay. Further optimization was performed to estimate the sensitivity and reproducibility of the assay. Extracted gDNA from individual adult worms, individual eggs and pools (10 eggs/pool) was tested. After DNA extraction from eggs using lysis buffer and proteinase K, 3μl of this crude lysate was added to the reactions and then tubes were incubated at 60°C for 90 min. Positive (WT and MT plasmids) and negative controls (no template) were included and the experiments were repeated twice, each time in duplicate.
Genotyping of T. trichiura and A. lumbricoides field samples
Validation for field samples was performed with pools of A. lumbricoides and T. trichiura eggs obtained from Haiti and Panama. Pools of 10 eggs previously collected under microscopy for each stool sample were analyzed. Eggs were digested using 25 μl of previous lysis buffer mix. From this crude lysate, 3 μl were added to each reaction after a DNA heating step at 95°C for 3–5 min. Assays were carried out as previously described. Positive and negative controls were always included as references in each experiment. Tubes were incubated in a RT-PCR system at 60°C for 90 min.
Assessment of SmartAmp2 polymerase tolerance to fecal inhibitors
To evaluate the tolerance of Aac polymerase to inhibitors in fecal samples, fecal samples that were confirmed to be negative for STH eggs were spiked with a known number of T. trichiura or A. lumbricoides eggs and then DNA was extracted according to the following modified protocol. Approximately 1 g of feces, preserved in 70% ethanol, was centrifuged and the fecal pellet was washed 3 times in PBS (phosphate buffered saline) and centrifuged. PBS solution was added to the fecal pellet to a final volume of 1 ml. Ten aliquots of 100 μl of fecal homogenate were transferred to new tubes. Tubes were centrifuged and excess PBS was removed. Each fecal sample was spiked with either A. lumbricoides or T. trichiura eggs (~10 eggs/tube). Other fecal samples were not spiked and were used as negative fecal controls.
DNA was extracted as follow: Fecal samples were frozen at -80°C for 30 min, and 10 μl of buffer A [NaOH (200 mM) + 2% Tween-20] was added to each tube. After a 15 min incubation period at 25°C, tubes were heated at 99°C for 10 min. Tubes were allowed to cool down, and then 10 μl of Buffer B [Tris-HCl (100 mM) and 2mM EDTA] were added and a second heat shock at 98°C for 2 min was performed. Finally, samples were centrifuged and the supernatants were transferred to a new PCR tube. Sample preparations were diluted 4-fold with distilled H2O, heated at 95°C for 3 min, cooled on ice and then 1 μl was added to the SmartAmp2 reaction mixture (total volume of 10 μl) containing 2 μM TP/FP, 1μM BP, 0.25 μM OP1/OP2 1.4 mM dNTPs, 1 M betaine, 1x isothermal buffer [20 mM Tris-HCl (pH 8.6), 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.1% Tween 20], 2 μg bovine serum albumin (BSA) (Sigma- Aldrich,), 1/100,000 dilution SYBR Green I, 0.5 μg Taq MutS, and 5 U Aac DNA polymerase. BSA was added to the SmartAmp2 reaction mix to stabilise the DNA polymerase and to neutralise fecal inhibitors. Reactions were incubated at 60°C for 90 min in a RT-PCR system to maintain isothermal conditions and to monitor the change in fluorescence intensity of SYBR Green I during the reaction. Assays were evaluated in terms of full match amplification and mismatch amplification (non-amplification).
Results
SmartAmp2 primer design
Various sets of primers were designed to genotype F167Y (TTC>TAC), F200Y (TTC>TAC) and E198A (GAA>GCA) SNPs of the β-tubulin isotype 1 gene. Screening of these primer combinations under a variety of assay conditions identified an ideal primer set for each assay which achieved the best yield, speed and efficiency to discriminate between MT and WT genotypes. The location and sequences of primers are shown in Figs 2 and 3. For A. lumbricoides, a primer set was developed and optimized specifically to target the SNP F167Y (TTC>TAC) found in some A. lumbricoides samples [21]. Both the 5’-end of TPs and the 3’-end of FPs were employed to overlap and discriminate the F167Y SNP. Primer sets were optimized specifically to target the F200Y (TTC>TAC) or E198A (GAA>GCA) SNPs in A. lumbricoides and T. trichiura. The 3’-end of BPs were employed to discriminate the MT and the WT genotypes.
SmartAmp2 assay optimization
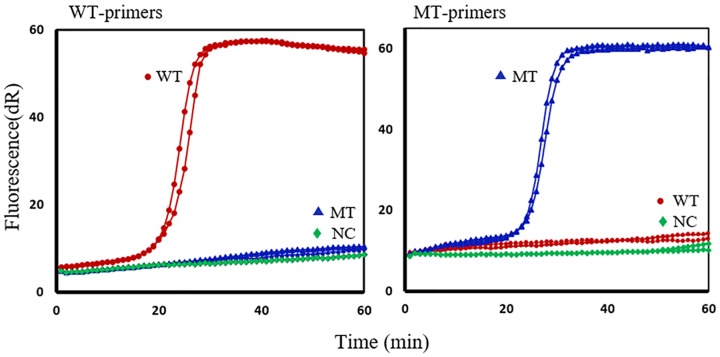
Sequencing of the WT and MT plasmids revealed that the desired mutations at codon positions 167, 198 and 200 of the β-tubulin gene were generated. WT and MT plasmids were used initially as DNA templates for assay optimization and development. Primer sets that exhibited delayed full match amplification or displayed a short delay between the full-match and mismatch amplification were omitted. Optimal amplification results were obtained when the reaction mixture contained 2 μM TP/FP, 1 μM BP and 0.25 μM OP1/OP2 with 1 M of betaine and 8 mM MgSO4. Mutant primer sets designed specifically to target the F167Y (TTC>TAC), F200Y (TTC>TAC) or E198A (GAA>GCA) SNPs rapidly amplified the MT plasmids within 20–30 min, whereas the same primer sets failed to amplify the WT plasmids (complete suppression of the mismatch amplification) by 60 min. WT primer sets designed to target the WT genotypes failed to amplify the MT plasmids. Each assay was run in duplicate and all negative control reactions included in the experiments showed no amplification within 60 min. These results confirmed that the SmartAmp2 assays were optimized, as they were able to accurately discriminate the full match amplification from the mismatch with complete suppression of mismatch amplification. The assay optimization with plasmids for T. trichiura is shown in (Fig 4). A similar amplification profile was obtained with plasmids for A. lumbricoides.
Fig 4. SmartAmp2 assay optimization for Trichuris trichiura β- tubulin polymorphisms.
Left graph, wild-type (WT)-primer amplification (full match) of WT plasmid (red circle) with no amplification (mismatch) with mutant-type (MT) plasmid (blue triangle) or NC (no template) (green diamond). Right graph, MT-primer amplification of MT plasmid (blue triangle) with no amplification with WT plasmid (red circle) or NC (green diamond). All experiments were run in duplicates. dR, difference of relative fluorescence unit.
SmartAmp2 assay sensitivity and specificity
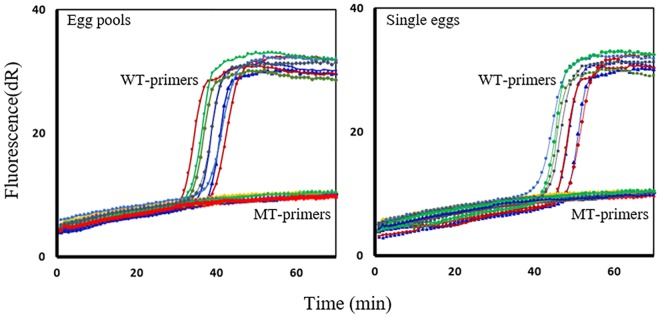
A. lumbricoides and T. trichiura-specific SmartAmp2 primers were designed based on β-tubulin sequence alignment of A. lumbricoides, T. trichiura, and N. americanus. The specificity of each SNP-detection assay was tested on MT plasmids (F167Y, F200Y or E198A) and no amplification was detected from the WT or the MT genotype of non-target sequences. This indicates that the SmartAmp2 assay has a high level of specificity for detecting only SNPs of a target sequence and inhibiting amplification from non-target sequences. To test reproducibility and the sensitivity, each assay was applied to analyze egg pools (10–50 eggs/pool) and single eggs for A. lumbricoides and T. trichiura. In the SmartAmp2 assays, the WT-primer set amplified the DNA target from egg pools within 30–40 min, and from single eggs within 40–50 min, but no amplification was observed with the MT-primer set or the negative controls. Positive controls (adult gDNA) were always included and showed full match amplification with only the WT-primer sets (Fig 5).
Fig 5. SmartAmp2 assay evaluation with pools and single eggs of Ascaris lumbricoides and Trichuris trichiura.
Wild-type (WT)-specific primer amplification of gDNA from egg pools (30–40 min) with complete suppression of amplification using the mutant-type (MT)-primer sets (left). WT-specific primer amplification of gDNA from single eggs (40–50 min) (right) with complete suppression of amplification with the MT-primer sets. Negative controls (no template control) were included in each run. Both graphs show replicated runs for either A. lumbricoides and T. trichiura egg pools or single eggs, respectively. Multiple assays (different colors) are shown in each plot showing reproducibility of the assay to distinguish between WT and MT DNA. dR, difference of relative fluorescence unit.
Genotyping of T. trichiura field samples
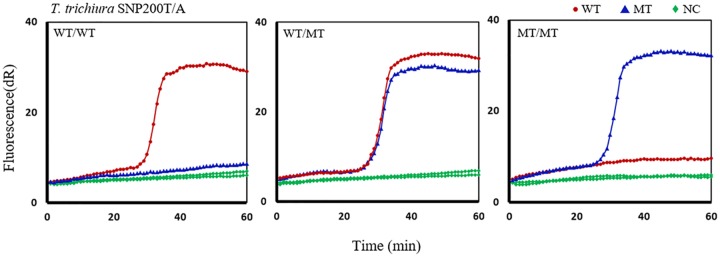
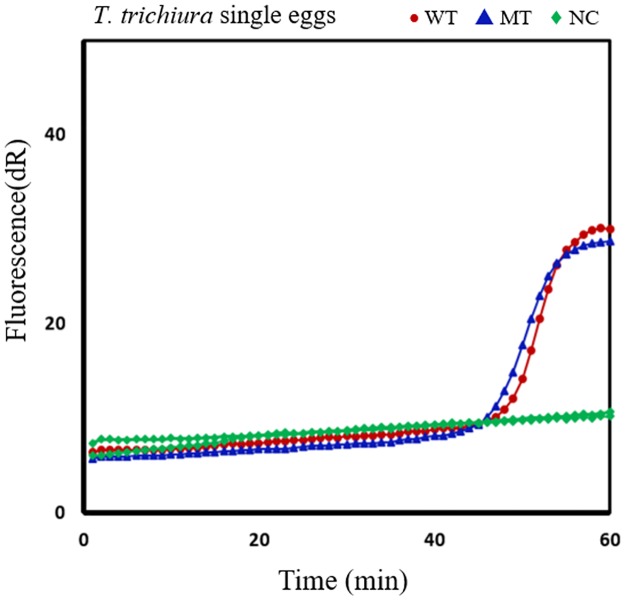
Twenty-five of 40 field samples examined under the microscope, collected from children in Haiti, had T. trichiura eggs. From each sample, pools of 10 eggs were placed in a PCR tube, and DNA was extracted as described in the Methods. SmartAmp2 assays were fully optimized to target F200Y (TTC>TAC) and E198A (GAA>GCA) SNPs in T. trichiura and were able to detect the presence or absence of WT and MT genotypes within 30 min after incubation at 60°C. No amplification was observed from negative controls. Positive controls (adult gDNA) were always included and showed full match amplification with only the WT-primer sets, as expected. For codon position 198, the WT-primer set allowed amplification of the DNA target within 30 min, but no amplification was observed with the MT-primer set. This indicates that no polymorphism was found at codon position 198 in the T. trichiura eggs analyzed. However, a polymorphism F200Y (TTC>TAC) was identified and the MT-primer set allowed amplification of the DNA target within 30 min (full match amplification). The 200SNP detection primer set recognized the F200Y (TTC>TAC) and discriminated homozygous 200T/T (WT), mixed 200 T/A (WT/MT), and homozygous 200A/A (MT) in Trichuris egg pools (Fig 6). To verify the genotyping results for codon 200 obtained with Trichuris egg pools, all experiments were repeated twice and performed in duplicate from the same samples and assays were always run with positive and negative controls. Additionally, single eggs from the same samples that had MT genotypes were analysed and the results confirmed the presence of the same MT genotypes obtained with egg pools. By analyzing single eggs, samples that had mixed WT and MT alleles were verified to have heterozygous genotypes at F200Y (TTC/TAC) SNP (Fig 7).
Fig 6. SmartAmp2 genotyping results of Trichuris trichiura samples at codon 200.
SmartAmp2 amplification of F200Y (TTC>TAC) using wild type (WT) and mutant type (MT)-primer sets. Left, center, and right panels show assay results for homozygous WT (WT/WT), mixed (WT/MT), and homozygous MT (MT/MT) pooled samples, respectively. dR, difference of relative fluorescence unit.
Fig 7. SmartAmp2 genotyping results of Trichuris trichiura samples at codon 200.
SmartAmp2 amplification of F200Y (TTC/TAC) with wildtype (WT) and mutant type (MT)-primer sets. The results showed heterozygous (WT/MT) genotypes in Trichuris single egg DNA. dR, difference of relative fluorescence unit.
Genotyping of A. lumbricoides field samples
Thirty-four fecal samples collected from children in Panama were examined under the microscope and 20 samples had A. lumbricoides eggs. For each individual sample, pools of 10 eggs were placed in a PCR tube and DNA was extracted as described in the Methods. SmartAmp2 assays were fully optimized to target the F167Y (TTC>TAC), F200Y (TTC>TAC) and E198A (GAA>GCA) SNPs in A. lumbricoides and were able to detect the presence or absence of WT and MT genotypes within 30 min. In the Panama samples, the WT-primer sets allowed full match amplification targeting the three codon positions, but no amplification was observed with the MT primer sets. This indicated that there was no polymorphism in these samples at codons 167, 198, or 200. However, in A. lumbricoides samples obtained from Haiti [21] (4 pools of 10 eggs each), the MT primer set allowed full match amplification within 30 min targeting the F167Y (TTC>TAC) SNP, but no amplification was observed with the WT-primer set. To verify the results, 40 individual eggs were tested in SmartAmp2 assays and were found to have the homozygous MT alleles at codon 167 (Fig 8). Genotyping results were confirmed by both conventional Sanger sequencing and pyrosequencing.
Fig 8. SmartAmp2 genotyping results of Ascaris lumbricoides samples at codon 167.
SmartAmp2 amplification of β-tubulin isotype 1 gene targeting the F167Y (TTC>TAC) SNP using the wild-type (WT) (red circle) and mutant-type (MT) (blue triangle)-primer sets. The MT-primer set allowed the full match amplification from egg pools (left) and single eggs (right). No amplification was observed from the WT-primer set or the negative control (green diamond). dR, difference of relative fluorescence unit.
SmartAmp2 polymerase tolerance to fecal inhibitors
To assess the tolerance of Aac polymerase to fecal inhibitors, fecal samples (spiked with eggs) and negative fecal samples were assessed in triplicate in the SmartAmp2 assay. High amplification efficiency was achieved when DNA samples were diluted 4-fold. Full-match amplification was obtained only from positive fecal samples using the WT-primer set within 40–45 min while the negative fecal samples and the negative controls remained at the base line for at least 90 min (Fig 9). This indicates that the Aac polymerase is highly tolerant (resistant) to fecal inhibitors even when crude sample preparations were used.
Fig 9. Evaluation of polymerase tolerance in fecal samples spiked with Trichuris trichiura or Ascaris lumbricoides eggs.
SmartAmp2 amplification using the wildtype (WT)-primer set on fecal samples spiked with A. lumbricoides eggs (red circle), T. trichiura eggs (blue circle) and negative fecal samples (green triangle) without eggs. Negative controls (no template) were included in each run (green diamond). dR, difference of relative fluorescence unit.
Discussion
BZ-resistance is a serious problem in veterinary parasites, and the intensive use of these drugs in MDA against human parasites raises concern that resistance may be selected in human STH. The development of rapid and sensitive methods for the detection of resistance-associated SNPs is needed for monitoring the presence and extent of BZ-resistant nematodes. Isothermal diagnostic methods have proven rapidity, sensitivity and specificity for the diagnosis of many parasitic, viral, bacterial and fungal infections [38, 39]. Among these methods, SmartAmp2 is a unique DNA amplification method for rapid detection of genetic polymorphisms under isothermal conditions in a single step, which eliminates the need for PCR amplification, a thermocycler or electrophoresis [31]. This technique uses asymmetrical primer design and the Aac DNA polymerase. This polymerase is highly resistant to cellular contaminants in clinical samples and works on crude sample preparations after a simple heating step to degrade RNA and denature proteins [29, 33]. Moreover, inclusion of the Thermus aquaticus MutS (Taq MutS) enzyme in the isothermal assay completely suppresses the exponential back-ground amplification (mismatch amplification), resulting in high specificity in distinguishing a specific target sequence based on one nucleotide difference [25]. Thus SmartAmp2 has unique advantages over PCR-based methods such as diagnostic real time PCR and pyrosequencing.
In this study, we developed a new SNP genotyping assay based on the SmartAmp2 method for monitoring β-tubulin polymorphisms. Optimal primer sets were selected and optimized specifically to target the F200Y (TTC>TAC) and E198A (GAA>GCA) SNPs in T. trichiura and A. lumbricoides, in addition to the F167Y (TTC>TAC) SNP in A. lumbricoides, SNP-detection primer sets efficiently and rapidly discriminated MT and WT genotypes using plasmids as DNA templates. The SmartAmp2 assay has high specificity for detecting only SNPs of a specific target sequence. A unique advantage of SmartAmp2 is the ability to genotype a SNP in highly homologous regions without cross-amplification of closely related genes. However, this requires intensive primer modification and optimization, particularly when targeting a highly conserved gene such as β-tubulin.
The assays showed high reproducibility and sensitivity for detecting genomic DNA from single egg DNA within 40–50 min in a single amplification and detection step. To detect amplified fragment from samples with low DNA concentrations, using a PCR based method, a nested PCR consisting of two consecutive rounds of amplification using the same primers, followed by gel electrophoresis is required [21]. This multistep technique is time consuming and also increases the risk of contamination as a result of manipulating the PCR product.
SmartAmp2 assays were developed to identify the F200Y (TTC>TAC) SNP in T. trichiura. The MT-detection primer set detected MT homozygous and mixed genotypes in T. trichiura samples from Haiti. This finding was consistent with previous studies in which the SNP 200 was identified in T. trichiura samples from Kenya [19] and other Haitian samples [21]. In A. lumbricoides, SmartAmp2 assays were developed to target the F167Y (TTC>TAC), F200Y (TTC>TAC) and E198A (GAA>GCA) SNPs and the MT-detection primer set detected the MT genotype F167Y (TTC>TAC) in A. lumbricoides samples from Haiti. This genotyping result was consistent with previous analyses on Ascaris samples from Haiti, Panama and Kenya using pyrosequencing and conventional sequencing [21]. No polymorphism was identified at codon 167 in the A. lumbricoides samples from Panama. Previous analysis of other A. lumbricoides samples from Panama found polymorphism at codon 167 [21].
Furthermore, fecal samples spiked with STH eggs were processed in the SmartAmp2 assay and the results showed the high tolerance of the Aac polymerase to fecal inhibitors in crude sample preparations. The Bst 2.0 DNA polymerase was initially used and was also found to exhibit high tolerance to fecal inhibitors. However, more suppression of mismatch amplification was observed with the Aac polymerase.
We were not successful in the optimization of a SNP detection assay to identify a F167Y (TTT>TAT) SNP in T. trichiura. As optimal primer design mainly depends on the target sequence, we were not able to optimize a set that could specifically distinguish the MT genotype from the WT genotype.
Compared with PCR-based methods, the SmartAmp2 assay is considered relatively inexpensive; the main costs are for the Taq MutS and the polymerase. Primers used in this study were not HPLC purified. Additionally, reducing the reaction mixture to 10 μl and using in-house prepared buffers and reagents can also reduce costs. Our data were generated on a RT-PCR system to follow the formation of double stranded DNA in real-time, using SYBR green. However, end point detection system for monitoring fluorescence that would allow high-throughput analysis of samples in a 96-well microplate format could be employed. Other approaches for visualizing the formation of DNA residues could be applied using fluorescence dyes that allow colorimetric screening of the results by the naked eye.
A limitation of our study is that the assay can detect only one SNP at a time, but SmartAmp2 offers simplicity of sample and reaction preparation, rapidity in detection and the convenience of isothermal amplification, which are significant advantages over other genotyping technologies. Additionally, in this study, we simplified the detection of the 198 and 200 SNPs by designing one assay with only a specific discrimination primer for each SNP.
The present study provides evidence that the SmartAmp2 method targeting β-tubulin polymorphisms in STHs allowed direct detection of SNPs of a target DNA sequence in field samples. Additionally, these results indicate that our SNP genotyping assays are rapid, simple, very sensitive and highly specific, which provide unique tools for investigating BZ resistance in STHs. The development of rapid and sensitive tools for detecting resistance-associated alleles would therefore be a key for monitoring the appearance and spread of potential BZ-resistant STHs.
Acknowledgments
Dr. Peter Nesjum, University of Copenhagen, Denmark is thanked for donating DNA from T. trichiura, and Dr. Patrick Lammie, Centers for Disease Control and Prevention, Atlanta, GA, USA for facilitating the collection of samples in Haiti. Dr. Äissatou Diawara is thanked for collecting the samples from Haiti and Dr. Rachel Krause for collecting samples from Panama. Research at the Institute of Parasitology is enabled by the Fonds de recherche du Québec—Nature et technologies, Centre for Host-Parasite Interactions, Québec, Canada.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) (Grant No. RGPIN/2777-2012), and the Fonds de recherche du Québec – Nature et technologies (FRQNT) through the Centre for Host-Parasite Interactions, Québec, Canada. NR received a fellowship from FRQNT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Murray CJL, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. The Lancet. 2012;380(9859):2197–223. [DOI] [PubMed] [Google Scholar]
- 2.Pullan RL, Smith JL, Jasrasaria R, Brooker SJ. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasites & vectors. 2014;7:37. Epub 2014/01/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Soil-transmitted Helminthiases: STH: Eliminating Soil-transmitted Helminthiases as a Public Health Problem in Children: Progress Report 2001–2010 and Strategic Plan 2011–2020: World Health Organization; 2012.
- 4.Prichard RK, Basanez MG, Boatin BA, McCarthy JS, Garcia HH, Yang GJ, et al. A research agenda for helminth diseases of humans: intervention for control and elimination. PLoS neglected tropical diseases. 2012;6(4):e1549 Epub 2012/05/01. 10.1371/journal.pntd.0001549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D, et al. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet (London, England). 2006;367(9521):1521–32. Epub 2006/05/09. [DOI] [PubMed] [Google Scholar]
- 6.McCarty TR, Turkeltaub JA, Hotez PJ. Global progress towards eliminating gastrointestinal helminth infections. Current opinion in gastroenterology. 2014;30(1):18–24. Epub 2013/11/19. 10.1097/MOG.0000000000000025 [DOI] [PubMed] [Google Scholar]
- 7.WHO. Helminth control in school age children: a guide for managers of control programmes. 2011.
- 8.Keiser J, Utzinger J. Efficacy of current drugs against soil-transmitted helminth infections: systematic review and meta-analysis. Jama. 2008;299(16):1937–48. Epub 2008/04/24. 10.1001/jama.299.16.1937 [DOI] [PubMed] [Google Scholar]
- 9.Vercruysse J, Behnke JM, Albonico M, Ame SM, Angebault C, Bethony JM, et al. Assessment of the anthelmintic efficacy of albendazole in school children in seven countries where soil-transmitted helminths are endemic. PLoS neglected tropical diseases. 2011;5(3):e948 Epub 2011/04/07. 10.1371/journal.pntd.0000948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett A, Guyatt H. Reducing intestinal nematode infection: efficacy of albendazole and mebendazole. Parasitology today (Personal ed). 2000;16(2):71–4. Epub 2000/02/01. [DOI] [PubMed] [Google Scholar]
- 11.Bennett AB, Anderson TJ, Barker GC, Michael E, Bundy DA. Sequence variation in the Trichuris trichiura beta-tubulin locus: implications for the development of benzimidazole resistance. International journal for parasitology. 2002;32(12):1519–28. Epub 2002/10/24. [DOI] [PubMed] [Google Scholar]
- 12.Geerts S, Gryseels B. Anthelmintic resistance in human helminths: a review. Tropical medicine & international health: TM & IH. 2001;6(11):915–21. Epub 2001/11/13. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan RM. Drug resistance in nematodes of veterinary importance: a status report. Trends in parasitology. 2004;20(10):477–81. Epub 2004/09/15. 10.1016/j.pt.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 14.Vercruysse J, Albonico M, Behnke JM, Kotze AC, Prichard RK, McCarthy JS, et al. Is anthelmintic resistance a concern for the control of human soil-transmitted helminths? International journal for parasitology Drugs and drug resistance. 2011;1(1):14–27. Epub 2011/12/01. 10.1016/j.ijpddr.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwa MS, Veenstra JG, Roos MH. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Molecular and biochemical parasitology. 1994;63(2):299–303. Epub 1994/02/01. [DOI] [PubMed] [Google Scholar]
- 16.Silvestre A, Cabaret J. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Molecular and biochemical parasitology. 2002;120(2):297–300. Epub 2002/03/19. [DOI] [PubMed] [Google Scholar]
- 17.Ghisi M, Kaminsky R, Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Veteterinary parasitology. 2007;144(3–4):313–20. Epub 2006/11/15. [DOI] [PubMed] [Google Scholar]
- 18.Mottier M, Prichard RK. Genetic analysis of a relationship between macrocyclic lactone and benzimidazole anthelmintic selection on Haemonchus contortus. Pharmacogenetics and genomics. 2008;18(2):129–40. Epub 2008/01/15. [DOI] [PubMed] [Google Scholar]
- 19.Diawara A, Drake LJ, Suswillo RR, Kihara J, Bundy DA, Scott ME, et al. Assays to detect beta-tubulin codon 200 polymorphism in Trichuris trichiura and Ascaris lumbricoides. PLoS neglected tropical diseases. 2009;3(3):e397 Epub 2009/03/25. 10.1371/journal.pntd.0000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diawara A, Schwenkenbecher JM, Kaplan RM, Prichard RK. Molecular and biological diagnostic tests for monitoring benzimidazole resistance in human soil-transmitted helminths. The American journal of tropical medicine and hygiene. 2013;88(6):1052–61. Epub 2013/03/06. 10.4269/ajtmh.12-0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diawara A, Halpenny CM, Churcher TS, Mwandawiro C, Kihara J, Kaplan RM, et al. Association between response to albendazole treatment and beta-tubulin genotype frequencies in soil-transmitted helminths. PLoS neglected tropical diseases. 2013;7(5):e2247 Epub 2013/06/06. 10.1371/journal.pntd.0002247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vercruysse J, Levecke B, Prichard R. Human soil-transmitted helminths: implications of mass drug administration. Current opinion in infectious diseases. 2012;25(6):703–8. Epub 2012/09/12. 10.1097/QCO.0b013e328358993a [DOI] [PubMed] [Google Scholar]
- 23.Prichard R. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends in parasitology. 2001;17(9):445–53. Epub 2001/09/01. [DOI] [PubMed] [Google Scholar]
- 24.Schwenkenbecher JM, Albonico M, Bickle Q, Kaplan RM. Characterization of beta-tubulin genes in hookworms and investigation of resistance-associated mutations using real-time PCR. Molecular and biochemical parasitology. 2007;156(2):167–74. 10.1016/j.molbiopara.2007.07.019 [DOI] [PubMed] [Google Scholar]
- 25.Mitani Y, Lezhava A, Kawai Y, Kikuchi T, Oguchi-Katayama A, Kogo Y, et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nature Methods. 2007;4(3):257–62. Epub 2007/02/27. 10.1038/nmeth1007 [DOI] [PubMed] [Google Scholar]
- 26.Barrere V, Keller K, von Samson-Himmelstjerna G, Prichard RK. Efficiency of a genetic test to detect benzimidazole resistant Haemonchus contortus nematodes in sheep farms in Quebec, Canada. Parasitology international. 2013;62(5):464–70. 10.1016/j.parint.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 27.von Samson-Himmelstjerna G, Walsh TK, Donnan AA, Carriere S, Jackson F, Skuce PJ, et al. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136(3):349–58. Epub 2009/01/22. 10.1017/S003118200800543X [DOI] [PubMed] [Google Scholar]
- 28.Schwab AE, Boakye DA, Kyelem D, Prichard RK. Detection of benzimidazole resistance-associated mutations in the filarial nematode Wuchereria bancrofti and evidence for selection by albendazole and ivermectin combination treatment. The American journal of tropical medicine and hygiene. 2005;73(2):234–8. Epub 2005/08/17. [PubMed] [Google Scholar]
- 29.Azuma K, Lezhava A, Shimizu M, Kimura Y, Ishizu Y, Ishikawa T, et al. Direct genotyping of Cytochrome P450 2A6 whole gene deletion from human blood samples by the SmartAmp method. Clinica chimica acta; international journal of clinical chemistry. 2011;412(13–14):1249–51. Epub 2011/03/23. 10.1016/j.cca.2011.03.017 [DOI] [PubMed] [Google Scholar]
- 30.Rashwan N, Bourguinat C, Keller K, Gunawardena NK, de Silva N, Prichard RK. Isothermal diagnostic assays for monitoring single nucleotide polymorphisms in Necator americanus associated with benzimidazole drug resistance. PLoS neglected tropical diseases. 2016;10(12):e0005113 Epub 2016/12/09. 10.1371/journal.pntd.0005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitani Y, Lezhava A, Sakurai A, Horikawa A, Nagakura M, Hayashizaki Y, et al. Rapid and cost-effective SNP detection method: application of SmartAmp2 to pharmacogenomics research. Pharmacogenomics. 2009;10(7):1187–97. Epub 2009/07/17. 10.2217/pgs.09.39 [DOI] [PubMed] [Google Scholar]
- 32.Biswas IHP. Identification and Characterization of a Thermostable MutS Homolog from Thermus aquaticus. Journal of biological chemistry. 1996;271(9):5040–8. [DOI] [PubMed] [Google Scholar]
- 33.Hoshi K, Takakura H, Mitani Y, Tatsumi K, Momiyama N, Ichikawa Y, et al. Rapid detection of epidermal growth factor receptor mutations in lung cancer by the SMart-Amplification Process. Clinical cancer research. 2007;13(17):4974–83. Epub 2007/09/06. 10.1158/1078-0432.CCR-07-0509 [DOI] [PubMed] [Google Scholar]
- 34.Krause RJ, Koski KG, Pons E, Sandoval N, Sinisterra O, Scott ME. Ascaris and hookworm transmission in preschool children from rural Panama: role of yard environment, soil eggs/larvae and hygiene and play behaviours. Parasitology. 2015;142(12):1543–54. Epub 2015/08/26. 10.1017/S0031182015001043 [DOI] [PubMed] [Google Scholar]
- 35.Krause RJ, Koski KG, Pons E, Sinisterra O, Scott ME. Ascaris and hookworm transmission in preschool children in rural Panama: role of subsistence agricultural activities. Parasitology. 2016;143(8):1043–54. Epub 2016/03/24. 10.1017/S0031182016000366 [DOI] [PubMed] [Google Scholar]
- 36.Hansen TV, Nejsum P, Olsen A, Thamsborg SM. Genetic variation in codons 167, 198 and 200 of the beta-tubulin gene in whipworms (Trichuris spp.) from a range of domestic animals and wildlife. Veterinary parasitology. 2013;193(1):141–9. [DOI] [PubMed] [Google Scholar]
- 37.Lake SL, Matthews JB, Kaplan RM, Hodgkinson JE. Determination of genomic DNA sequences for beta-tubulin isotype 1 from multiple species of cyathostomin and detection of resistance alleles in third-stage larvae from horses with naturally acquired infections. Parasites & vectors. 2009;2 Suppl 2:S6. Epub 2009/09/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. Journal of infection and chemotherapy. 2013;19(3):404–11. 10.1007/s10156-013-0590-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic acids research. 2000;28(12):E63 Epub 2000/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.